Abstract
The freshwater lakes of southwestern France are subject to the development of invasive macrophytes which are associated with mercury (Hg) contamination of the food web. The aim of this study was to determine the bioavailability of methylmercury (MeHg) produced by plant roots in aquatic ecosystems. A microcosm experiment was performed using isotopically enriched inorganic Hg at environmental concentrations (1 µg 199IHg·L−1). For all conditions, total Hg in fish as well as Hg species associated with different compartments (water, sediments, plant roots, fish) were analyzed by gas chromatography-inductively coupled plasma-mass spectrometry (GC-ICP-MS). In addition, sediment, plants, and fish gut microbiota were studied by MiSEQ sequencing. Some strains were isolated and tested for their ability to methylate Hg. The results revealed 199MeHg production in plant roots and the presence of this form in fish (tissues and gut), highlighting a MeHg trophic transfer. Moreover, methylator bacteria were identified from the gut contents of the fish when they were in the presence of plants. Some of them were related to bacteria found in the plant roots. On the basis of these results, the transfer of MeHg and bacteria from plants to fish is highlighted; in addition, Hg methylation is strongly suspected in the fish gut, potentially increasing the Hg bioaccumulation.
1. Introduction
Mercury (Hg) is considered to be an important pollutant for aquatic systems since its organic form, i.e., methylmercury (MeHg), is easily bioaccumulated and biomagnified along food webs. It has been demonstrated that the aquatic plant rhizosphere are favorable ecological niches for Hg transformations. Several studies have demonstrated high Hg methylation and demethylation potentials in the periphyton associated with floating macrophyte roots in tropical ecosystems [1,2,3,4,5], in boreal regions [6], and in temperate areas [7]. Microorganisms, especially sulfate-reducing prokaryotes (SRP), were identified as the main elements responsible for MeHg production. Indeed, SRP were detected in periphyton by 16S rDNA probing [3,8], real-time polymerase chain reaction (RT-PCR) [4], and by the cultural method [8]. However, other functional groups such as iron reducers [9,10], methanogens [11], and some fermenters [12] seem to also methylate Hg. Moreover, microorganisms able to methylate Hg seem to have the specific gene cluster hgcAB [13], which was spotted in a large diversity of prokaryotes and environments [12,14]. For the time being, the link between the level of expression of these genes and the biotic MeHg production remains to be seen [15,16], still requiring a cultural method to decide on the capacity of a strain to methylate.
Aquatic roots supply a physical and organic-rich support for the development of microbial communities. Since periphyton accumulates metals through adsorption on inorganic and organic particulate material [17], including Hg [18,19], the activity of these microorganisms in such a matrix can be a source of contamination for food webs [20,21]. This conclusion is supported by the fact that macrophytes are considered to be an important food source for herbivorous and detritivorous invertebrates and fish [22].
In southwestern France, invasive macrophytes like Ludwigia sp. colonize several freshwater lakes and rivers, causing damage to aquatic ecosystems [23,24] and interacting with Hg biogeochemistry and bioaccumulation. The evaluation of net MeHg budgets in these temperate ecosystems suggests that the aquatic rhizosphere is a hotspot for MeHg production due to SRP activity, and may represent an important source of contamination for the aquatic food chain [7]. In this context, the first objective of this study was to determine the bioaccumulation and bioavailability of Hg species, particularly MeHg formed in the rhizosphere of this invasive macrophyte in the food web, especially for fish. In this work, isotopically enriched inorganic mercury 199IHg was used to monitor the distribution and conversion into 199MeHg in different reconstituted aquatic compartments (water, sediment, plant, fish) within microcosm experimentations. The use of isotopically enriched tracers is a sensitive and reliable technique [25,26] to follow Hg reactivity during laboratory experimentations, allowing researchers to work at low concentrations of Hg, close to environmental levels.
Since ten years ago, the intestinal microbiota has been recognized as a major player in the health of its host as it takes part in major biological functions, such as nutrition [27], immunity [28], and the metabolism of xenobiotics [29]. Indeed, it could play a major role in the exposure of the host to certain chemical pollutants. Thus, the intestinal microbiota of aquatic organisms arose very recently as a potential compartment involved in the contamination of these organisms, via Hg methylation [30]. This endogenous methylation is therefore a major question in relation to the environmental problem of Hg (exposure of species and impacts) and to the understanding of the dynamics of this pollutant in ecosystems. However, there is currently a significant lack of knowledge on the subject [31]; the intestinal microbiota is therefore not taken into account today in Hg risk assessments, either in terms of the environment or human health. Thus, the second objective of the study was to investigate the possible involvement of intestinal bacteria, especially SRP, in the Hg methylation in vivo process. We analyzed the prokaryotic diversity through a molecular approach (16S rRNA gene) in the fish gut, macrophyte floating roots (feeding resource), and in sediment (habitat), with a focus on SRP diversity. The combination with a cultivation based method allowed us to reach culturable diversity and to study the ability of SRP strains to methylate Hg through a biosensor based bioassay [32].
2. Materials and Methods
2.1. Study Site and Sampling Procedure
Water, sediment, and plants were collected in June 2011 in Sanguinet Lake (south western France), a freshwater lake colonized by Ludwigia sp., an invasive macrophyte selected as the model plant in this study [7]. Field water was sampled in a water can. The first two centimeters of a sandy sediment, poor in organic matter, were sampled and sieved (size: 2.5 mm, RETSCHER) to remove any macroorganisms (invertebrates) which could interfere with the experiment [33], and transported in plastic bags. This sandy sediment represents the majority of the sediment in this lake [34]. Macrophytes (with periphyton fixed on stems and roots) were collected with special attention by hand. All the equipment for field and laboratory sampling, storage, and analysis was carefully cleaned using the ultraclean procedure.
2.2. Experimental Design
The experimental design is summarized in Figure 1 and included 16 experimental units (dimension: 30 cm high, 25 cm large, and 12.5 cm long). At the time of this study, no ethical permit was required to perform experiments on fish. Three conditions were tested: i/”sediment” (n = 3 control and n = 3 contaminated units); ii/”plant” (n = 2 control and n = 2 contaminated units); iii/”plant+sediment” (n = 3 control and n = 3 contaminated units). In the conditions “sediment” and “plant+sediment”, 2 kg of sediment were added by unit. In the conditions “plant” and “plant+sediment,” one single Ludwigia sp. plant (average size: 127 ± 21 cm; average weight: 76 ± 21 g wet weight) was added by unit. Each unit contained 6 L of nonfiltered water from the field. In order to minimize the adsorption of Hg in the walls of the experimental units, Teflon pockets (Electroplast enterprise), especially sized for these units and cleaned by the ultraclean method, were placed inside each tank. A stabilization time of 5 days was respected before the beginning of the experiment. The photoperiod was 12/12 h. Then, 1 µg 199IHg·L−1 was added to the water column in the contaminated conditions. To avoid direct contamination of the fish, microcosms were first incubated without fish during 7 days until the [THg] level in water was below 10 ng·L−1. After that, four adult fish Pseudancistrus sp., a periphytophageous species from laboratory breeding, were introduced in each unit (mean value ± SD at final time Tf: body weight = 3.0 ± 0.4 g ww; standard length = 64 ± 2 mm, n = 48). A food supplement (vegetable) was added in each condition to avoid undernourishment (especially for the sediment condition). During the experiment and in all units, the average of oxygen concentration in water was 7.9 ± 0.8 mL·L−1, pH was 6.4 ± 0.1, and the temperature was 24.9 ± 0.3 °C (daily monitoring).
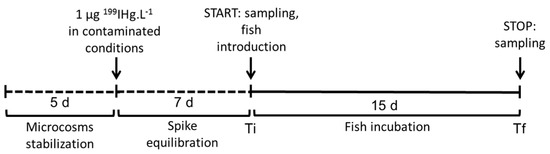
Figure 1.
Experimental design to monitor the repartition of 199IHg added and the formation of 199MeHg in different aquatic compartments (water, sediment, plant, and fish). The same design was used for control units (without the addition of 199IHg).
At initial time (Ti) and final time (Tf, after 15 days of incubation), 100 mL of water (acidified with 1% HCl v/v), 10 g ww of sediment, and 5 g ww of floating plant roots were collected for Hg speciation analysis by gas chromatography-inductively coupled plasma-mass spectrometry (GC-ICP-MS). Water samples were stored at 4 °C, and the sediment and plants at −20 °C until analysis. Root and sediment samples for molecular biology were collected in sterile cryotubes and stored at −80 °C in the laboratory until analysis. At Tf, fish were removed and killed within seconds by immersion in melting ice. Skeletal muscle, liver, gills, and gut (empty) were independently sampled for total Hg analysis. A subsample of skeletal muscle was sampled for GC-ICP-MS analysis. The intestinal contents of fish were collected in sterile cryotubes and stored at −80 °C: one part being used to study the microbial diversity by MISEQ analysis; a second part was stored with glycerol (30% v/v) in order to characterize the SRP diversity by a cultural method.
2.3. Total Mercury Analysis
Skeletal muscle, liver, gills, and gut samples were first dried at 44 °C during 48 h before being analyzed by flameless atomic absorption spectrometry (Analyseur de Mercure Altec AMA 254). The detection limit of this method is 0.01 ng. The analytical method was validated using a certified reference material TORT2 (National Research Council of Canada, lobster hepatopancreas) every ten samples. The accuracy averaged 104.7%.
2.4. GC-ICP-MS(Gas Chromatography-Inductively Coupled Plasma-Mass Spectrometry) Analysis
Sediment and Ludwigia sp. roots were digested with nitric acid 6N and fish muscle with TMAH (Tetramethylammonium hydroxide) under microwave radiation (no extraction for water) and all samples were analyzed by GC-ICP-MS (gas chromatography-inductively coupled plasma-mass spectrometry) as described elsewhere [35,36].
For nonspiked samples, the quantification of endogeneous Hg species was performed by species specific isotope dilution, by adding the appropriate amount of isotopically enriched Hg standards (199IHg and 201MeHg) [35]. Each assay was analyzed three times and blanks were also performed to check for any contamination. Results were validated using certified reference materials IAEA 405 (International Atomic Energy Agency, estuarine sediment) and BCR 464 (Institute for Reference Materials and Measurements, tuna fish).
For 199IHg-spiked samples, the amount of mercury species deriving from the enriched isotope 199 after the incubation period was calculated by isotopic pattern deconvolution methodology, as previously described in [37]. The methylation potential (M) was calculated by dividing the total amount of 199MeHg formed by the sum of the amount of 199IHg and 199MeHg.
2.5. MiSEQ Analysis
DNA extractions were performed according to the PowerSoil® DNA Isolation kit (MO BIO) following the manufacturer instructions from 500 mg of sieved soil. The V4–V5 hypervariable region of the 16S rRNA gene targeting bacteria and archaea was amplified using the primers 515F, 5′-GTG YCA GCM GCC GCG GTA-3′ and 928R, 5′-CCC CGY CAA TTC MTT TRA GT-3′. The reaction mixture included 1.40 μL of each primer (20 µM each), 28 µl AmpliTaq Gold Master mix (AmpliTaq Gold; 360 Master Mix Applied Biosystems), 2 μL of the sample DNA template at a concentration of 5 ng·µL−1, and water qsp 55 µL. The cycle conditions included initial denaturation at 94 °C for 10 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 65 °C for 30 s, and an extension at 72 °C for 40 s, and an extension step at 72 °C for 10 min after cycling was complete. Illumina MiSeq sequencing was performed with the GeT core facility, Toulouse, France (http://get.genotoul.fr). MiSEQ sequences obtained were deposited in the GenBank DNA database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) under accession numbers SAMN14117438- SAMN14117453 (BioProject PRJNA606946). Sequence analysis was done with the pipeline FROGS from the Galaxy portal of the Toulouse Midi-Pyrenees bioinformatics platform [38]. After a preprocessing step including quality filter, read trimming, and read assembly, sequences were clustered with Swarm [39] with an aggregation distance of 3 and a denoising clustering step. Operational taxonomic units (OTU) with an abundance lower than 0.005% were removed [40] in order to delete singletons. The SILVA database 128 (release date 29.09.2016) was used to perform the OTU affiliations [41].
2.6. Sulfate-Reducing Prokaryotes (SRP) Culture and Identification
Samples from fish gut content sampled in the “plant” condition were pooled before isolation using the high throughput cultivation method under anoxic conditions [42]. For 16S rRNA gene sequencing, PCR amplifications were performed with the unlabelled universal bacterial primers 63F and 1387R according to [43]. All sequencing reactions were performed by GATC Biotech. The 16S rRNA gene partial sequences were compared to sequences deposited in the GenBank DNA database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) by the BLAST (Basic Local Alignment Search Tool) algorithm. Alignments of all sequences were achieved and assembled with Bioedit 7.1.3 software, and phylogenetic trees were constructed with MEGA v5 [44] using the neighbor-joining method [45]. More details concerning these procedures are described in [8]. 16S rRNA gene sequences obtained for the strains were deposited in the NCBI under accession numbers MT071512-MT071543.
2.7. Hg Methylation Capacity by Isolated Strains
The evaluation of Hg methylation capacities was based on the formation of MeHg detected by the biosensor Escherichia coli MC1061 previsouly developed by [46] and adapted to pure culture samples by [32]. Details about the method are available elsewhere [8]. Briefly, 100 µg·L−1 of IHg was spiked in the culture supernatant of each strain. During this addition, an aliquot was sampled (T0) and stored at −20 °C until analysis. At the end of the incubation period (5 h), another aliquot was sampled and stored at −20 °C until analysis. Uninoculated media were incubated under the same conditions as controls for abiotic methylation. The strain Desulfovibrio BerOc1 was also incubated as positive control [47] for Hg methylation. Then, 100 µL of each aliquot (samples at T0 and T5 h) and 100 µL of E. coli culture were added in a microplate in triplicate. After an incubation period of 1h at 37 °C and 200 rpm, bioluminescence was measured at 482 nm using a multi-well plate reader (Series 4000 Perspective Biosystem). Results were indicated in light intensity (LI) and calculated as follows:
where Tf denotes final time of incubation; T0 denotes initial time of incubation; and OD denotes the optical density measured at T0.
(LI Tf − LI T0)/OD
2.8. Statistical Analysis
Factorial ANOVAs were used to study the differences in THg concentrations in different organs and conditions after checking the assumptions (normality and homoscedasticity of the error term). Log transformation was needed to fulfill the statistical assumptions. If the assumption was met, the parametric Fisher’s Least Significant Difference (LSD) test was applied. If the assumption was not met, the nonparametric Kruskall–Wallis test was used. In each test, p < 0.05 was considered significant. All statistical investigations were performed using STATISCA version 6.1 software. Values are mean ± SE.
3. Results
3.1. Organisms’ Health
During the experiment, fish appeared healthy on external inspection (no injury, no fungoid growth) and no alteration of animals’ motility was observed. Despite this, the death rate was 4% (two fish from 48: one in the condition “plant-control”, one in “plant-Hg”). No difference in standard length and weight was observed between control and contaminated fish at the end of experiment (Table S1).
An overall average growth rate of 12% was observed for plants between Ti and Tf, as well as a flowering, in all units.
3.2. Mercury Speciation in Control Units
Natural concentrations of IHg and MeHg in the control units were measured at initial time (Ti, just before the fish introduction) and at final time (Tf, after 15 days) in water, sediment, plant, and fish muscle by GC-ICP-MS. For each species, the Ti and Tf results were averaged by condition since they were similar (Table 1). The first observation was that each compartment had similar concentrations in different conditions (Kruskall–Wallis test, p < 0.05), except for the fish compartment, which had relatively higher concentrations of both species in conditions with plants (“plant” and “plant+sediment”). The second observation was that, whatever the condition and the Hg species, the same gradient of Hg concentration was measured in different compartments: water ≤ sediment < plant < skeletal muscle. IHg and MeHg concentrations were respectively 5–8-fold and 1–6-fold higher in the plant compartment than in the sediment compartment, depending on the condition.

Table 1.
Mercury speciation (ng·g−1 dry weight dw) and methylmercury proportion (%) in different aquatic compartments from control units (-C; background concentrations from field). Measurements were performed at the Ti and Tf of the experiment and were pooled in this table.
3.3. Distribution and Transformation of Isotopic Tracer 199Hg in Contaminated Units
199IHg and 199MeHg concentrations were determined in different aquatic compartments at Ti and Tf, and in addition in fish skeletal muscle at Tf by GC-ICP-MS (Table 2). At Ti (after 7 days of spike equilibration and just before fish introduction), the majority of 199Hg was under its inorganic form in all compartments. However, methylation potentials were measured because of the formation of 199MeHg (maximum in the “plant-Hg” condition: 18% in water and 13% in plant). The highest [199IHg] and [199MeHg] were observed in plants (maximum in “plant-Hg”: 2573 ng 199IHg·g−1 and 311 ng 199MeHg·g−1), followed by water and sediment. At Tf, after fish introduction, [199IHg] and [199MeHg] decreased in all compartments (water, sediment, plant) in all conditions compared to results at Ti. However, methylation potentials increased in the plant and sediment compartments only in conditions with sediment (“plant+sediment” and “sediment”), with the maximum value observed in plant compartment from the “plant+sediment” condition (27%), whereas no change was observed in the “plant” condition. In fish skeletal muscle, similar [199MeHg] levels were measured in three experimental conditions (154 ± 40 to 181 ± 70 ng 199MeHg·g−1) and higher [199IHg] levels were observed in the “plant” condition (64 ± 25 ng 199IHg·g−1) than in the two others. Methylation potentials in fish were the highest of all aquatic compartments (77%–90%).

Table 2.
Concentrations of 199IHg and 199MeHg (ng·g−1 dw), Hg methylation potentials (%) in the water, sediment, plants, and Pseudancistrus sp. fish muscle for different conditions before fish introduction in units (Ti) and after 15 days of incubation (Tf).
For each unit, a mass budget of 199Hg species was calculated (each Hg species was compared to the total quantity of Hg species in a unit) and the results are expressed in percentage (Figure 2). For all conditions, % of 199IHg and 199MeHg in water were very low (0% in the “plant+sediment” condition to 4% in the “plant” condition). 199IHg was mainly distributed in sediment (99% in the “sediment” condition; 83% in the “plant+sediment” condition) and plant (87% in the “plant” condition and 16% in the “plant+sediment” condition), and significantly lower in fish muscle for all conditions (minimum value: 0.7 and 0.8% in the “sediment” condition and the “plant+sediment” condition, respectively). 199MeHg was significantly higher in fish muscle for the “plant” condition (75%) than in fish for the other conditions (Kruskall–Wallis test, p < 0.05), but also for the other compartments. The rest of the 199MeHg was distributed between sediment and plants (maximum value: sediment = 61% in the “sediment” condition). In the “plant” condition, 199MeHg in plants presented a high heterogeneity between replicates (21.4 ± 20.1%).
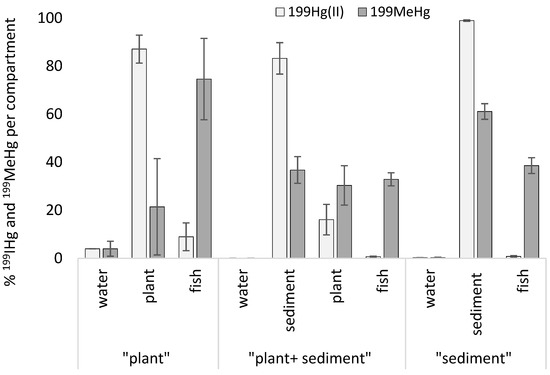
Figure 2.
Proportion of 199IHg and 199MeHg in water, sediment, plants, and Pseudancistrus sp. fish muscle from three contaminated conditions (“plant” (n = 2), “plant+sediment” (n = 3), “sediment” (n = 3)). Error bars represent standard errors.
3.4. Organostropism of THg in Control and Contaminated Fish
At Tf, total Hg (THg) concentrations were determined by AMA 254 in the skeletal muscle, liver, gills, and gut of Pseudancistrus sp. fish for control (Figure 3a) and contaminated (Figure 3b) conditions. In the control units, for each organ, Hg bioaccumulation was relatively similar between conditions (background concentrations), except for the muscle and gut where [THg] levels were slightly higher in the “plant” condition (0.97 ± 0.21 µg·g−1 and 0.72 ± 0.21 µg·g−1, respectively), and also just for muscle in the “plant+sediment” condition (0.98 ± 0.07 µg·g−1), compared to the others units (test LSD Fisher, p < 0.05).
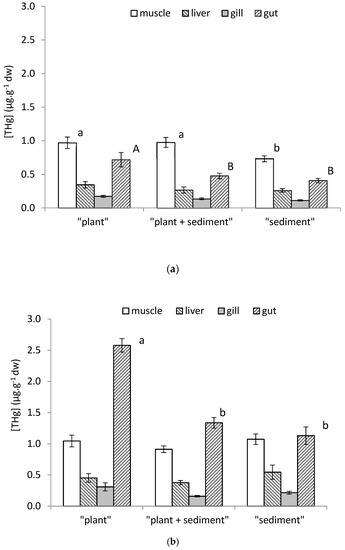
Figure 3.
The average of total mercury concentrations (µg THg·g−1 dw) measured by AMA in the muscle, liver, gill, and (empty) gut of Pseudancistrus sp. fish from control conditions (a) and contaminated conditions (b) at Tf. Error bars represent standard errors, n = 9 for each condition and organ, except for the “plant” condition (n = 6 for muscle, liver, and gill; and n = 4 for the gut). Letters indicate statistical differences (p < 0.05, lower case and capital letter must be considered separately).
Comparison of the control and contaminated muscle, liver, and gills showed no statistical differences between them. Only the [THg] levels in the fish gut were significantly higher in the contaminated conditions as compared to the control conditions. Moreover, Hg bioaccumulation in the fish gut from the contaminated “plant” condition (2.6 ± 0.2 µg·g−1) was significantly higher than those from the “sediment” and “sediment+plant” conditions (1.1 ± 0.4 µg·g−1 and 1.3 ± 0.3 µg·g−1, respectively).
3.5. Microorganisms Global Diversity
16S rRNA diversity from the fish gut of the “plant” and “plant+sediment” conditions, and from plant and sediment was assessed using the MiSeq approach (Figure 4a). The maximum specific richness was observed in the sediment, followed by plant and fish gut (both conditions). However, the bacterial composition was relatively similar in all conditions, with a prevalence of proteobacteria (maximum: fish gut from the “plant” condition = 52%). In both fish gut conditions, the second phylum which dominated was planctomycetes (maximum: fish gut from the “plant+sediment” condition = 28%), followed by cyanobacteria (13% in fish gut from the “plant+sediment” condition) and firmicutes (11% fish gut from the “plant” condition). In the plant and sediment, it was cyanobacteria (24% in the sediment), followed by bacteroidetes (14% in the plant), and chloroflexi (12% in the plant).
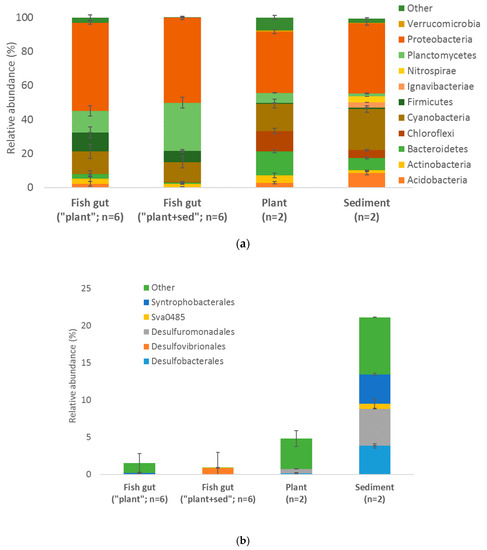
Figure 4.
Comparison of the relative abundance of bacterial diversity (16S rRNA genes sequencing) (a) and Deltaproteobacteria known to be potential methylators (b), from the fish gut of the “plant” units and “plant+sediment” units, and from plant and sediment. Error bars represent standard errors.
An assessment of the relative abundance of sulfate reducers (SRP), known as potential Hg methylator in these environments [7], was then performed (Figure 4b). The highest diversity and relative abundance of SRP was observed in the sediment, followed by the plant and fish gut. In the “others”, Deltaproteobacteria were dominant in the fish gut from the “plant” condition (1.3%) but some Syntrophobacterales were detected (0.2%), whereas the fish gut from the “plant+sediment” condition was mainly composed of Desulfovibrionales (0.9%).
3.6. Sulfate Reducers Isolation and Hg Methylation Capacity
Since SRP are considered to be the principal microorganisms responsible for Hg methylation, SRP strains were isolated from Pseudancistrus sp. gut contents of the contaminated “plant” condition. The study of gene encoding 16S rRNA allowed us to affiliate 32 isolated pure strains to the Desulfovibrionales order (Figure 5), and more precisely to the Desulfovibrionaceae (30 strains) and Desulfobulbaceae (two strains) families. Concerning Desulfovibrionaceae, six strains (B11 to H3) were close to Desulfovibrio aerotolerans (strain DvO5), seven strains (F7 to F2) were close to Desulfovibrio carbinolicus (strain DSM 3852), 17 strains (G6 to G3) were close to Desulfovibrio sp. LG-2009. The maximum bootstrap percentage was 82% (for D. carbinolicus), indicating that the isolated strains were not yet described in the literature. Two strains that belong to Desulfobulbaceae (E1, E4) were close to Desulfobulbaceae bacterium (strain PR8 A05), but considering the bootstrap percentages, they were probably also new species.
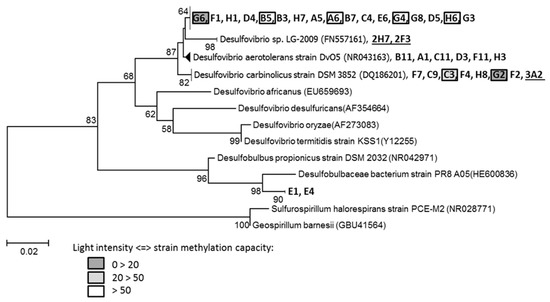
Figure 5.
Phylogenetic relationships based on the alignment of 16S rRNA gene sequences of strains isolated from Pseudancistrus sp. gut and Ludwigia sp. rhizosphere, and belong to Desulfovibrionales. Strains originated from the Ludwigia sp. rhizosphere are underlined. Methylating strains are framed. Capacity of isolated strains to methylate Hg was estimated by the light intensity produced by the biosensor E. coli MC 1061. Bootstrap percentages above 50% (1000 bootstrap replicates) are indicated at the branching points. The scale bar represents nucleotide substitutions. Gene bank accession numbers are shown in parentheses. Tree was rooted with E. coli (M25588).
The capacity of these 32 isolated strains to methylate Hg was tested, using the biosensor E. coli MC 1061. About 18.8% of isolated strains were able to methylate Hg (7 of 32 strains) at variable rates. Maximum light intensity was measured for strain G2 (103.7 ± 25.7), followed by strain G6 (53.9 ± 6.7), strain C3 (33.5 ± 6.1), and strain G4 (11.5 ± 5.0). Strains B5, A6, and H6 showed positive results and were considered as methylating strains although with a high standard deviation (6.3 ± 2.7; 5.1 ± 4.4; 4.4 ± 3.8, respectively).
SRP strains were also isolated from Ludwigia sp. roots sampled in Sanguinet Lake [8] and three of them (2H7, 2F3, 3A2) are added in Figure 5. Strains 2H7 and 2F3 were close to Desulfovibrio sp. (LG-2009), which is relatively close to 17 strains isolated from fish gut contents. Strains 3A2 were close to Desulfovibrio carbinolicus, like seven other strains from gut microbiota.
4. Discussion
4.1. Periphyton Associated with Root’s Macrophytes: Main Source of MeHg for Organisms
4.1.1. High Methylation Potential in Aquatic Rhizosphere of Invasive Macrophyte
In our study, IHg is mainly trapped in plant roots and MeHg is mainly synthetized in this compartment, both in the contaminated (Table 2, Ti) and control units (Table 1). The rhizosphere of aquatic plants is effectively known to participate significantly in storage and Hg transformations [1]. Sediment is also known to participate to these processes [48,49,50], especially if it is an organic-rich sediment. Here, the sandy sediment used to constitute the microcosms probably explains the low Hg concentrations measured and the low reactivity of this compartment with Hg. However, our study supports a previous work where significantly higher Hg methylation and demethylation potentials were determined by incubations in the same plant (Ludwigia sp.) compared to the same sediment and in the same ecosystem [7]. In this previous study, the main quantity of MeHg was found in the periphyton separated from Ludwigia sp. roots compared to Ludwigia sp. roots with its periphyton. Analysis of periphytic particles from “plant” units at Tf indicates a large amount of Hg species with a high MeHg proportion (1278.3 ng 199IHg·g−1 dw and 88.2 ng 199MeHg·g−1 dw) compared to plant roots (Table 2, Tf), confirming that the periphyton is the main compartment responsible for the Hg transformation processes. Periphyton is an organic-rich matrix, considered to be a hotspot for microbial activity within the water column, with many heterotrophic aerobes, facultative anaerobes, or even strict anaerobes [8,51,52]. During sampling, a possible disruption of the system could be created causing a detachment of these periphytic particles. Such a disruption could then potentially increase the amount of 199MeHg measured in the sediment, thus leading to an overestimation of [199MeHg] in the sediment of the “sediment+plant” condition. Other studies also observed higher Hg methylation potentials in the periphyton of floating macrophyte roots in tropical areas [2,3] and temperate lakes [11,53] than in sediment associated or not with roots [54]. A similar proportion of 199MeHg was observed between the plant and sediment in the “plant+sediment” condition (Figure 2), while the proportion of 199IHg is significantly higher in sediment than in the plants. The lowest proportion of 199IHg observed in plants indicates that IHg trapped in plants is effectively more bioavailable for methylation than inorganic Hg trapped in sediment.
4.1.2. MeHg Produced in Aquatic Rhizosphere is Bioavailable for Organisms
The trophic pathway is considered to be the major contamination pathway in aquatic food webs for Hg bioaccumulation and biomagnification [55,56,57]. The “plant” condition allows for the evaluation of the role played by aquatic plants in MeHg bioavailability to aquatic organisms without sediment influence. Aquatic plants are regarded as an important food source for herbivorous and scavengers of some aquatic ecosystems [22,58,59]. In this study, [THg] is significantly higher in the gut of periphytophageous fish for the control and contaminated “plant” conditions compared to the “plant+sediment” and “sediment” conditions (Figure 3), and between the control and contaminated units. This indicates that (1) the periphyton associated with aquatic plant roots could be a food source for fish—this result is generally observed with periphytophageous fish due to their diet [60], and (2) there is a trophic contamination pathway for Hg. However, a low direct contamination could be possible at Ti (just before the fish introduction) in the three contaminated conditions since [199IHg] levels in water were not negligible (Table 2) and [199MeHg] levels were also very low in terms of proportion (Figure 2). The maximum levels of [199IHg] and [199MeHg] were observed in the plant compartment compared to the sediment compartment. However, 199MeHg proportion and concentrations in fish muscle were similar in both the “sediment+plant” and “sediment” conditions. Sediment could act as a limiting factor of Hg bioavailability for aquatic organisms. In [61], the ability of mangrove sediment to trap Hg, limiting its bioavailability is highlighted.
In all conditions (control and contaminated), the MeHg % in fish is always higher than the IHg %, showing the higher bioavailability of MeHg compared to IHg (Table 1 and Table 2). The significantly higher 199MeHg proportion observed in fish from the “plant” condition supports the fact that plants (and sediment to a lesser extent) play a role in fish Hg contamination, despite a heterogeneity observed between replicates of fish muscle. The difference in plants in each unit could explain this heterogeneity. Indeed, only a single plant of Ludwigia sp. was planted by aquarium (average weight ww = 40 g). Periphyton could not have the same microbial composition from one root to another, even if they are spatially close. Studies of SRP diversity (dsrAB and 16S rRNA genes) in Ludwigia sp. roots from Sanguinet Lake revealed heterogeneous microbial communities between replicates of samples [7,8]. This could be attributed to the different stages of biofilm development. In this case, Hg methylation could vary significantly from one plant to another, and hence to one microcosm to another, leading to differences in MeHg production and transfer to fish. Another explanation could be the variable amount of food eaten by the fish, some feeding more than others and therefore bioaccumulating more Hg. The duration of the experiment (15 days) may also have played a role and may have been too short to obtain clear information about it.
4.2. Involvement of Fish Gut Microbiota in Hg Methylation
The relative abundance of Deltaproteobacteria assessed using the molecular approach showed a low abundance and diversity in the fish gut (1%) with a high variability depending on the condition, whereas it is 5-fold higher in macrophyte roots and 20-fold in sediment (Figure 4B). Strains isolated from the fish gut (the contaminated “plant” condition) belonged to Desulfovibrionaceae and Desulfobulbaceae families. Some of the isolates obtained probably represent new taxa among the family Desulfovibrionaceae and should be further characterized and described. The strain isolation overlooked the dominant Deltaproteobacteria since the bacterial diversity seen through the cultural method is different and lower than that seen from using the molecular method. Indeed, members of Desulfovibrionales were exclusively detected through the cultivation method in the fish gut from the “plant” condition. Some of them were detected in the fish gut from the “plant+sediment” condition by MISEQ, showing the potential influence of the external environment (plant roots and sediment) on the diversity of such a matrix. Their capacity to methylate Hg was assessed using a biosensor (the light intensity produced by the biosensor directly reflected the methylation potential of a strain) and seven were able to methylate Hg at variable rates (Figure 5). Few studies have demonstrated the ability of SRP and Enterobacteria to methylate Hg in the digestive tract of terrestrial organisms [62,63] or fish [31] and the results are quite controversial [30,64]. IHg assimilation by intestinal epithelium is limited [65,66,67]. However, IHg can be methylated before passing the intestinal barrier into the blood. A mechanism of Hg assimilation was suggested in [68], involving both amiloride sensitive, and energy-dependent pathways to explain IHg absorption across the vertebrate gut. However, the mechanisms of Hg absorption through the intestinal cells have not yet been elucidated in fish or in mammals. In our work, a part of IHg could be methylated in the rhizosphere of aquatic plants before being ingested as MeHg by fish. Another part could be ingested as IHg, then methylated in the digestive tract, increasing the contamination risk. In addition, methylating strains also affiliated with Desulfovibrio carbinolicus were isolated from Ludwigia sp. roots from the Sanguinet Lake [8]. This indicates that SRP isolated from intestinal contents of fish are derived from the aquatic rhizosphere, which is a food resource for fish. These strains could be responsible for Hg methylation in the gut. Variability in bacterial diversity of gut microbiota could explain the variability of 199MeHg production measured in fish muscle at the end of the experiment. This hypothesis remains to be checked and quantified since the methylation capacity of strains could be stimulated or inhibited within the intestine (due to the unfavorable physicochemical growth medium, competition with gut microbiota). It would be interesting to further investigate the gut microbiota of fish and confirm if Hg methylation is possible in vivo in the gut. Moreover, it is also possible that these strains are able to demethylate MeHg [47,69,70]. Another incubation experiment with Hg stable isotopes (199IHg and 201MeHg) would be important to simultaneously and precisely quantify both processes.
5. Conclusions
This study suggests the important role of invasive macrophytes, resulting from sulfate reducing prokaryote activity, in the Hg biogeochemistry cycle and in the Hg contamination of the aquatic food chain in a freshwater temperate lake. The results highlight the transfer of MeHg (and bacteria) from plants to fish. MeHg formed in the periphyton could significantly participate in food web contamination, since aquatic plants are often considered to be an important food source. The occurrence of new methylator strains belonging to Desulfovibrionales in fish gut contents, probably from Ludwigia sp. roots, is important both in terms of microbial ecology and ecotoxicity in aquatic organisms. The involvement of intestinal microbiota in Hg transformations would need to be investigated in future research, especially to know if microbiota should be considered as a new reactive biological compartment for Hg.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/4/1500/s1, Table S1: Biometry of fish in different experimental conditions at Tf.
Author Contributions
R.G., S.G. designed research; J.-M.A., R.G. acquired funding; S.G., A.L., R.M.-B., R.G., J.-M.A. collected data; C.G., M.M., S.G. analyzed data; S.G. wrote the paper, M.M., R.G., A.L., R.M.-B. reviewed and edited the paper. All authors have read and agreed with the published version of the manuscript.
Funding
This research was funded by the Conseil Général des Landes and the INSU (EC2C0 DIRECT project).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guimaraes, J.R.D.; Meili, M.; Hylander, L.D.; Castro e Silva, E.; Roulet, M.; Mauro, J.B.N.; Lemos, R.A. Net mercury methylation in five tropical flood plain regions of Brazil, high in the root zone of floating macrophyte mats but low in surface sediments and flooded soils. Sci. Total Environ. 2000, 261, 99–107. [Google Scholar] [CrossRef]
- Mauro, J.B.; Guimaraes, J.R.; Hintelmann, H.; Watras, C.J.; Haack, E.A.; Coelho-Souza, S.A. Mercury methylation in macrophytes, periphyton, and water, comparative studies with stable and radio-mercury additions. Anal. Bioanal. Chem. 2002, 374, 983–989. [Google Scholar]
- Achá, D.; Iñiguez, V.; Roulet, M.; Guimarães, J.R.D.; Luna, R.; Alanoca, L.; Sanchez, S. Sulfate-reducing bacteria in floating macrophyte rhizospheres from an Amazonian floodplain lake in Bolivia and their association with Hg methylation. Appl. Environ. Microbiol. 2005, 71, 7531–7535. [Google Scholar] [CrossRef]
- Achá, D.; Hintelmann, H.; Yee, J. Importance of sulfate reducing bacteria in mercury methylation and demethylation in periphyton from Bolivian Amazon Region. Chemosphere 2011, 82, 911–916. [Google Scholar] [CrossRef]
- Correia, R.R.S.; Miranda, M.R.; Guimaraes, J.R.D. Mercury methylation and the microbial consortium in periphyton of tropical macrophytes, Effect of different inhibitors. Environ. Res. 2012, 112, 86–91. [Google Scholar] [CrossRef]
- Desrosiers, M.; Planas, D.; Mucci, A. Mercury methylation in the epilithon of boreal shield aquatic ecosystems. Environ. Sci. Technol. 2006, 40, 1540–1546. [Google Scholar] [CrossRef]
- Gentès, S.; Monperrus, M.; Legeay, A.; Maury-Brachet, R.; Davail, S.; André, J.M.; Guyoneaud, R. Incidence of invasive macrophytes on methylmercury budget in temperate lakes, Central role of bacterial periphytic communities. Environ. Pollut. 2013, 172, 116–123. [Google Scholar] [CrossRef]
- Gentès, S.; Taupiac, J.; Colin, Y.; André, J.M.; Guyoneaud, R. Bacterial periphytic communities related to mercury methylation within aquatic plant roots from a temperate freshwater lake (South-Western France). Environ. Sci. Pollut. Res. 2017, 24, 19223–19233. [Google Scholar] [CrossRef]
- Flemming, E.J.; Mack, E.E.; Green, P.G.; Nelson, D.C. Mercury methylation from unexpected sources, Molybdate-inhibited freshwater sediments and an iron-reducing bacterium. Appl. Environ. Microbiol. 2006, 72, 457–464. [Google Scholar] [CrossRef]
- Kerin, E.J.; Gilmour, C.C.; Roden, E.; Suzuki, M.T.; Coates, J.D.; Mason, R.P. Mercury methylation by dissimilatory iron-reducing bacteria. Appl. Environ. Microbiol. 2006, 72, 7919–7921. [Google Scholar] [CrossRef]
- Hamelin, S.; Amyot, M.; Barkay, T.; Wang, Y.; Planas, D. Methanogens, Principal methylators of mercury in lake periphyton. Environ. Sci. Technol. 2011, 45, 7693–7700. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, C.C.; Podar, M.; Bullock, A.L.; Graham, A.M.; Brown, S.D.; Somenahally, A.C.; Elias, D. Mercury methylation by novel microorganisms from new environments. Environ. Sci. Technol. 2013, 47, 11810–11820. [Google Scholar] [CrossRef] [PubMed]
- Parks, J.M.; Johs, A.; Podar, M.; Bridou, R.; Hurt, R.A., Jr.; Smith, S.D.; Tomanicek, S.J.; Qian, Y.; Brown, S.D.; Brandt, C.C.; et al. The genetic basis for bacterial mercury methylation. Science 2013, 339, 1332–1335. [Google Scholar] [CrossRef] [PubMed]
- Podar, M.; Gilmour, C.C.; Brandt, C.C.; Soren, A.; Brown, S.D.; Crable, B.R.; Palumbo, A.V.; Somenahally, A.C.; Elias, D.A. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci. Adv. 2015, 1, 1–13. [Google Scholar] [CrossRef]
- Goñi-Urriza, M.; Corsellis, Y.; Lanceleur, L.; Tessier, E.; Gury, J.; Monperrus, M.; Guyoneaud, R. Relationships between bacterial energetic metabolism, mercury methylation potential, and hgcA/hgcB gene expression in Desulfovibrio dechloroacetivorans BerOc1. Environ. Sci. Pollut. Res. 2015, 22, 13764–13771. [Google Scholar] [CrossRef]
- Bravo, A.G.; Loizeau, J.L.; Dranguet, P.; Makri, S.; Björn, E.; Ungureanu, V.G.; Cosio, C. Persistent Hg contamination and occurrence of Hg-methylating transcript (hgcA) downstream of a chlor-alkali plant in the Olt River (Romania). Environ. Sci. Pollut. Res. 2016, 23, 10529–10541. [Google Scholar] [CrossRef]
- Guasch, H.; Admiraal, W.; Sabater, S. Contrasting effects of organic and inorganic toxicants on freshwater periphyton. Aquat. Toxicol. 2003, 64, 165–175. [Google Scholar] [CrossRef]
- King, J.K.; Harmon, S.M.; Fu, T.T.; Gladden, J.B. Mercury removal, methylmercury formation, and sulfate-reducing bacteria profiles in wetland mesocosms. Chemosphere 2002, 46, 859–870. [Google Scholar] [CrossRef]
- Göthberg, A.; Greger, M. Formation of methyl mercury in an aquatic macrophyte. Chemosphere 2006, 65, 2096–2105. [Google Scholar]
- Roulet, M.; Lucotte, M.; Guimarães, J.R.; Rheault, I. Methylmercury in water, seston, and epiphyton of an Amazonian river and its floodplain, Tapajós River, Brazil. Sci. Total Environ. 2000, 261, 43–59. [Google Scholar] [CrossRef]
- Molina, C.I.; Gibon, F.M.; Duprey, J.L.; Dominguez, E.; Guimaraes, J.R.; Roulet, M. Transfer of mercury and methylmercury along macroinvertebrate food chains in a floodplain lake of the Beni River, Bolivian Amazonia. Sci. Total Environ. 2010, 408, 3382–3391. [Google Scholar] [CrossRef] [PubMed]
- Leite, R.G.; Araújo-Lima, C.A.R.M.; Victoria, R.L.; Martinelli, L.A. Stable isotope analysis of energy sources for larvae of eight fish species from the Amazon floodplain. Ecol. Freshw. Fish. 2002, 11, 56–63. [Google Scholar] [CrossRef]
- Dutartre, A. De la régulation des plantes aquatiques envahissantes à la gestion des hydrosystèmes. Ingénierie Ecol. 2004, 87–100. [Google Scholar]
- Lambert, E.; Dutartre, A.; Coudreuse, J.; Haury, J. Relationships between the biomass production of invasive Ludwigia species and physical properties of habitats in France. Hydrobiologia 2010, 656, 173–186. [Google Scholar] [CrossRef]
- Hintelmann, H.; Keppel-Jones, K.; Evans, R.D. Constants of mercury methylation and demethylation rates in sediments and comparison of tracer and ambient mercury availability. Environ. Toxicol. Chem. 2000, 19, 2204–2211. [Google Scholar] [CrossRef]
- Monperrus, M.; Tessier, E.; Amouroux, D.; Leynaert, A.; Huonnic, P.; Donard, O.F.X. Mercury methylation, demethylation and reduction rates in coastal and marine surface waters of the Mediterranean Sea. Mar. Chem. 2007, 107, 49–63. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Biddinger, S.B. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Dietert, R.R.; Silbergeld, E.K. Biomarkers for the 21st century, listening to the microbiome. Toxicol. Sci. 2015, 144, 208–216. [Google Scholar] [CrossRef]
- Wang, R.; Feng, X.B.; Wang, W.X. In vivo mercury methylation and demethylation in freshwater tilapia quantified by mercury stable isotopes. Environ. Sci. Technol. 2013, 47, 7949–7957. [Google Scholar] [CrossRef]
- Rudd, J.W.; Furutani, A.; Turner, M.A. Mercury methylation by fish intestinal contents. Appl. Environ. Microbiol. 1980, 40, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Colin, Y.; Gury, J.; Monperrus, M.; Gentès, S.; Ayala, P.; Goñi-Urriza, M.; Guyoneaud, R. Biosensor for screening bacterial mercury methylation, example within the Desulfobulbaceae. Res. Microbiol. 2018, 169, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Ciutat, A.; Boudou, A. Bioturbation effects on cadmium and zinc transfers from contaminated sediment and on metal bioavailability of benthic bivalves. Environ. Toxicol. Chem. 2003, 22, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Canredon, A.; Anschutz, P.; Buquet, D.; Charbonnier, C.; Gentès, S.; Legeay, A.; Feurtet-Mazel, A.; Poirier, D.; Bujan, S.; Devaux, L.; et al. Sulphate and organic matter control on mercury biogeochemistry and production of methylmercury in Aquitaine coastal lakes. Appl. Geochem. 2019, 104, 135–145. [Google Scholar] [CrossRef]
- Rodriguez Martin-Doimeadios, R.C.; Krupp, E.; Amouroux, D.; Donard, O.F.X. Application of isotopically labeled methylmercury for isotope dilution analysis of biological samples using gas chromatography/ICPMS. Anal. Chem. 2002, 74, 2505–2512. [Google Scholar] [CrossRef]
- Clémens, S.; Monperrus, M.; Donard, O.F.; Amouroux, D.; Guérin, T. Mercury speciation analysis in seafood by species-specific isotope dilution, method validation and occurrence data. Anal. Bioanal. Chem. 2011, 401, 2699. [Google Scholar] [CrossRef]
- Rodriguez-Gonzalez, P.; Monperrus, M.; Alonso, J.I.G.; Amouroux, D.; Donard, O.F.X. Comparison of different numerical approaches for multiple spiking species-specific isotope dilution analysis exemplified by the determination of butyltin species in sediments. J. Anal. At. Spectrom. 2007, 22, 1373–1382. [Google Scholar] [CrossRef]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS, Find; Rapidly; OTUs with Galaxy Solution. Bioinformatics 2017, 34, 1287–1294. [Google Scholar] [CrossRef]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm, robust and fast clustering method for amplicon-based studies. PeerJ 2014, 2, e593. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2012, 10, 57–59. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project, improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Colin, Y.; Goñi-Urriza, M.; Caumette, P.; Guyoneaud, R. Combination of high throughput cultivation and dsrA sequencing for assessment of sulfate-reducing bacteria diversity in sediments. FEMS Microbiol. Ecol. 2012, 83, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4, Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbour-joining method, a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Ivask, A.; Hakkila, K.; Virta, M. Detection of organomercurials with sensor bacteria. Anal. Chem. 2001, 73, 5168–5171. [Google Scholar] [CrossRef]
- Ranchou-Peyruse, M.; Monperrus, M.; Bridou, R.; Duran, R.; Amouroux, D.; Salvado, J.C.; Guyoneaud, R. Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers, Implications for environmental studies. Geomicrobiol. J. 2009, 26, 1–8. [Google Scholar] [CrossRef]
- Hammerschmidt, C.R.; Fitzgerald, W.F. Photodecomposition of methylmercury in an arctic Alaskan lake. Environ. Sci. Technol. 2006, 40, 1212–1216. [Google Scholar] [CrossRef]
- Engstrom, D.R.; Balogh, S.J.; Swain, E.B. History of mercury inputs to Minnesota lakes, influences of watershed disturbance and localized atmospheric deposition. Limnol. Oceanogr. 2007, 52, 2467–2683. [Google Scholar] [CrossRef]
- Muir, D.; Wang, X.; Yang, F.; Nguyen, N.; Jackson, T.A.; Evans, M.S.; Douglas, M.; Köck, G.; Lamoureux, S.F.; Pienitz, R.; et al. Spatial trends and historical deposition of mercury in eastern and northern Canada inferred from lake sediment cores. Environ. Sci. Technol. 2009, 43, 4802–4809. [Google Scholar] [CrossRef]
- Christensen, P.B.; Revsbech, N.P.; Sand-Jensen, K. Microsensor analysis of oxygen in the rhizosphere of the aquatic macrophyte littorella uniflora. Plant. Physiol. 1994, 105, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Van Gemerden, H. Microbial mats, a joint-venture. Mar. Geol 1993, 113, 3–25. [Google Scholar] [CrossRef]
- Hamelin, S.; Planas, D.; Amyot, M. Mercury methylation and demethylation by periphyton biofilms and their host in a fluvial wetland of the St. Lawrence River (QC, Canada). Sci. Total Environ. 2015, 512–513, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.Q.; Adatto, I.; Montesdeoca, M.R.; Driscoll, C.T.; Hines, M.E.; Barkay, T. Mercury methylation in Sphagnum moss mats and its association with sulfate-reducing bacteria in an acidic Adirondack forest lake wetland. FEMS Microbiol. Ecol. 2010, 74, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.D.; Bodaly, R.A.; Fudge, R.J.P.; Rudd, J.W.M.; Rosenberg, D.M. Food as the dominant pathway of methylmercury uptake by fish. Water Air Soil Pollut. 1997, 100, 13–24. [Google Scholar]
- Harris, R.; Bodaly, R.A. Temperature, growth and dietary effects on fish mercury dynamics in two Ontario lakes. Biogeochemistry 1998, 40, 175–187. [Google Scholar] [CrossRef]
- Pickhardt, P.C.; Stepanova, M.; Fisher, N.S. Contrasting uptake routes and tissue distributions of inorganic and methylmercury in mosquitofish (Gambusia affinis) and redear sunfish (Lepomis microlophus). Environ. Toxicol. Chem. 2006, 25, 2132–2142. [Google Scholar] [CrossRef]
- Junk, W.J.; Piedade, M.T.F. Plant life in the floodplain with special reference to herbaceous plants. In The Central Amazon Floodplain; Springer: Berlin, Germany, 1997; pp. 147–185. [Google Scholar]
- Gentès, S.; Maury-Brachet, R.; Guyoneaud, R.; Monperrus, M.; André, J.M.; Davail, S.; Legeay, A. Mercury bioaccumulation along food webs in temperate aquatic ecosystems colonized by aquatic macrophytes in south western France. Ecotoxicol. Environ. Saf. 2013, 91, 180–187. [Google Scholar] [CrossRef]
- Maury-Brachet, R.; Durrieu, G.; Dominique, Y.; Boudou, A. Mercury distribution in fish organs and food regimes, significant relationships from twelve species collected in French Guiana (Amazonian basin). Sci. Total Environ. 2006, 368, 262–270. [Google Scholar]
- Silva, L.F.F.; Machado, W.; Filho, S.D.L.; Lacerda, L.D. Mercury accumulation in sediments of a mangrove ecosystem in SE Brazil. Water Air Soil Pollut. 2003, 145, 67–77. [Google Scholar] [CrossRef]
- Kaschak, E.; Knopf, B.; Petersen, J.H.; Bings, N.H.; König, H. Biotic methylation of mercury by intestinal and sulfate-reducing bacteria and their potential role in mercury accumulation in the tissue of the soil-living Eisenia foetida. Soil Biol. Biochem. 2014, 69, 202–211. [Google Scholar] [CrossRef]
- Martín-Doimeadios, R.R.; Mateo, R.; Jiménez-Moreno, M. Is gastrointestinal microbiota relevant for endogenous mercury methylation in terrestrial animals? Environ. Res. 2017, 152, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, F.; Wang, W. In vivo mercury demethylation in a marine fish (Acanthopagrus schlegeli). Environ. Sci. Technol. 2017, 51, 6441–6451. [Google Scholar] [CrossRef] [PubMed]
- Boudou, A.; Ribeyre, F. Experimental study of trophic contamination of Salmogairdneri by two mercury compounds—HgCl2 and CH3HgCl—Analysis at the organism and organ levels. Water Air Soil Pollut. 1985, 26, 137–148. [Google Scholar] [CrossRef]
- Ribeiro, O.C.A.; Belger, L.; Pelletier, E.; Rouleau, C. Histopathological evidence of inorganic mercury and methyl mercury toxicity in the arctic charr (Salvelinus alpinus). Environ. Res. 2002, 90, 217–225. [Google Scholar] [CrossRef]
- Berntssen, M.H.G.; Hylland, K.; Julshamn, K.; Lundebye, A.K.; Waagbo, R. Maximum limits of organic and inorganic mercury in fish feed. Aquacult. Nutr. 2004, 10, 83–97. [Google Scholar] [CrossRef]
- Hoyle, I.; Handy, R.D. Dose-dependent inorganic mercury absorption by isolated perfused intestine of rainbow trout, Oncorhynchus mykiss, involves both amiloride-sensitive and energy-dependent pathways. Aquat. Toxicol. 2005, 72, 147–159. [Google Scholar] [CrossRef]
- Pak, K.; Bartha, R. Products of mercury demethylation by sulfidogens and methanogens. Bull. Environ. Contam. Toxicol. 1998, 61, 690–694. [Google Scholar] [CrossRef]
- Bridou, R.; Monperrus, M.; Rodriguez Gonzalez, P.; Guyoneaud, R.; Amouroux, D. Simultaneous determination of mercury methylation and demethylation capacities of various sulfate reducing bacteria using species-specific isotopic tracers. Environ. Toxicol. Chem. 2011, 30, 337–344. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).