Large Genetic Intraspecific Diversity of Autochthonous Lactic Acid Bacteria and Yeasts Isolated from PDO Tuscan Bread Sourdough
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms
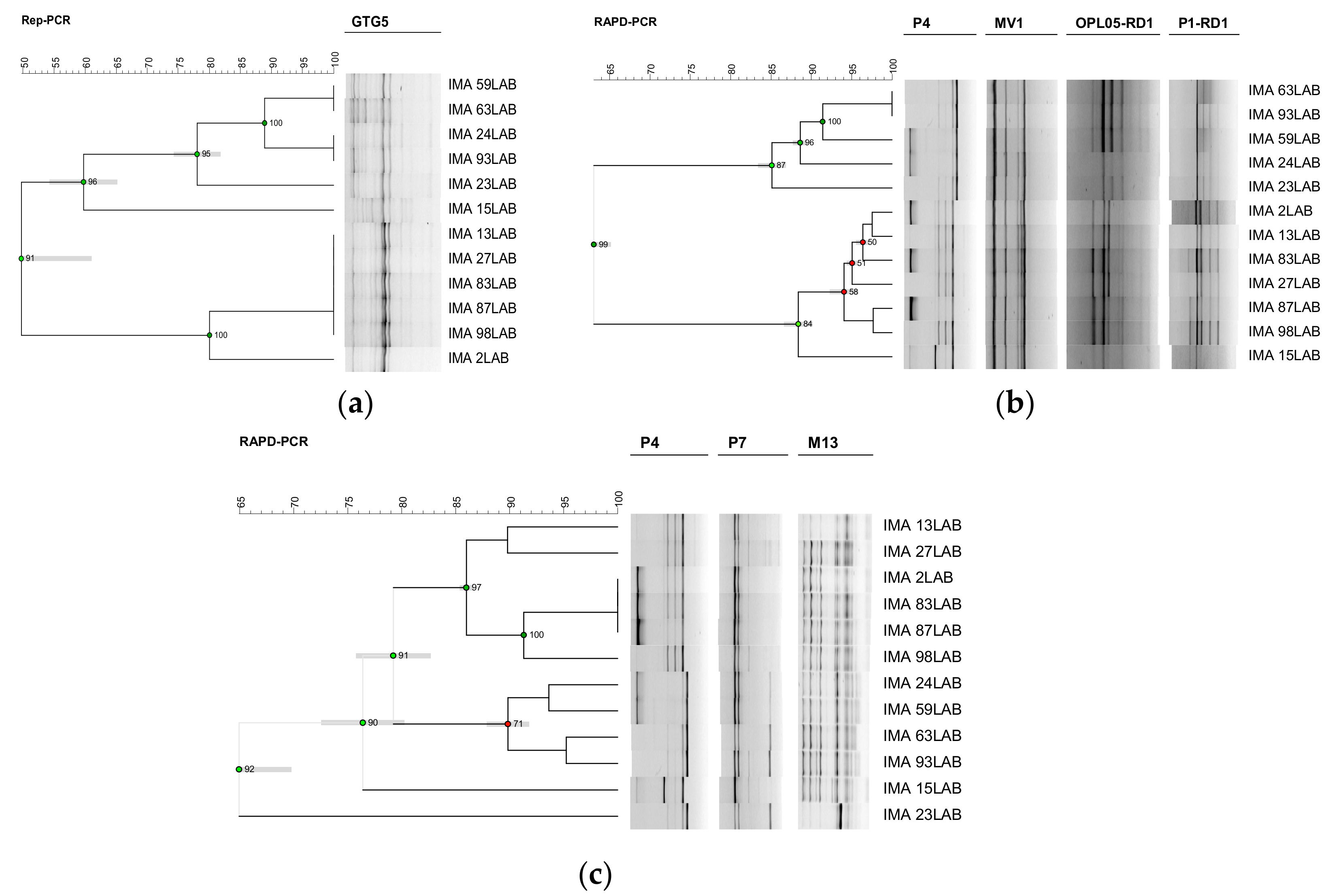
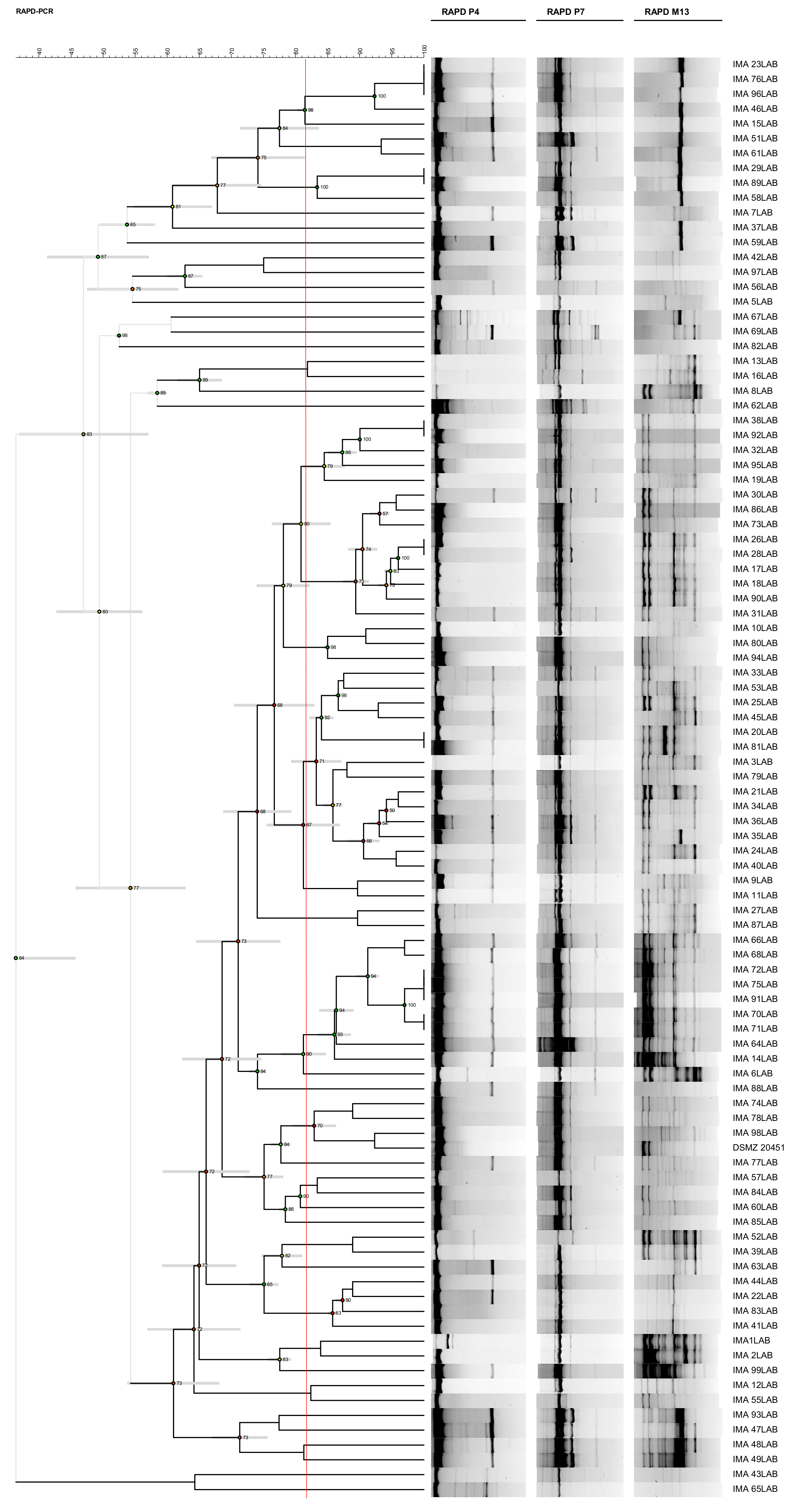
2.2. Molecular Intraspecific Diversity of LAB Isolates Characterizing the PDO Tuscan Bread Sourdough
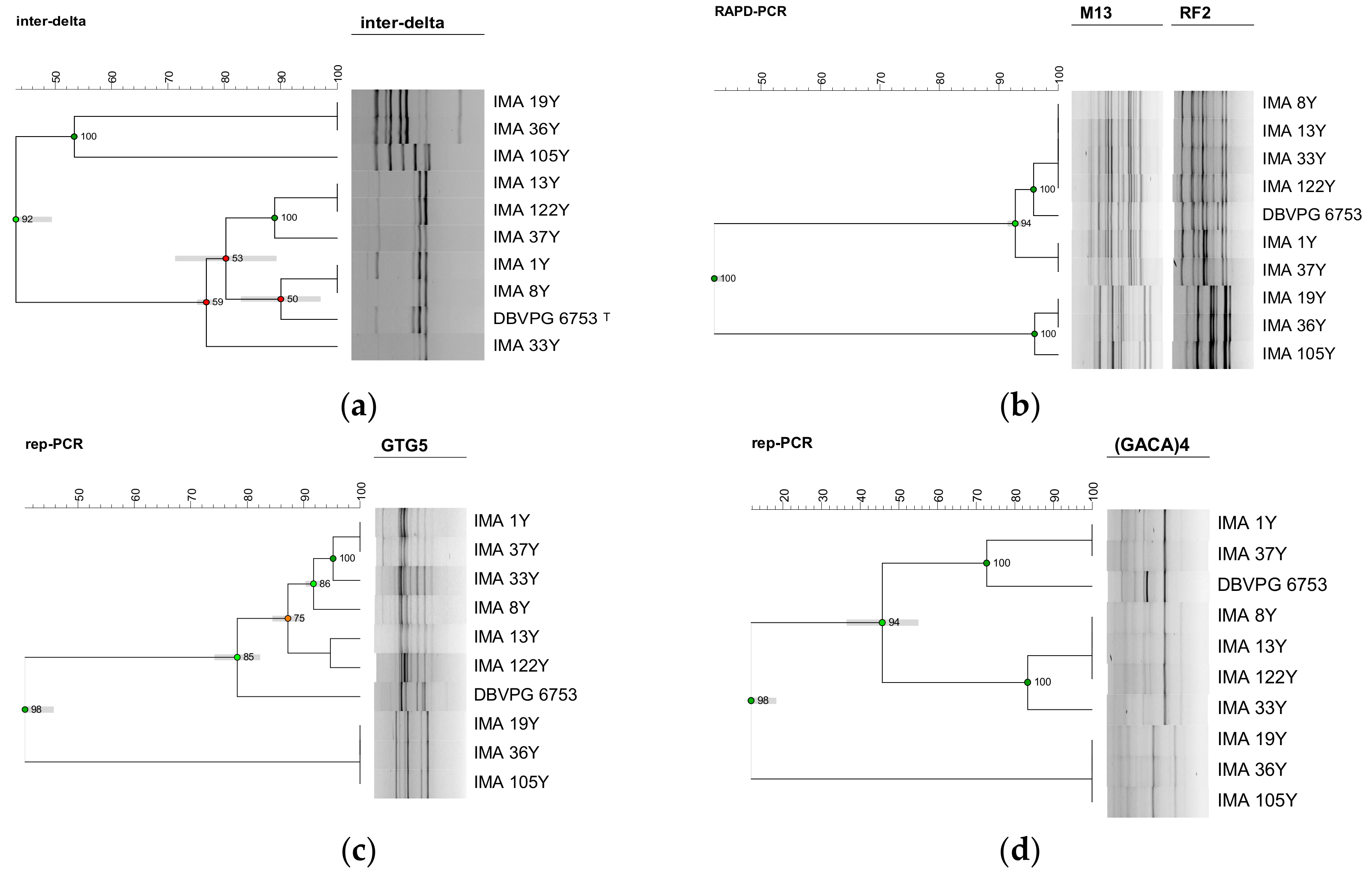
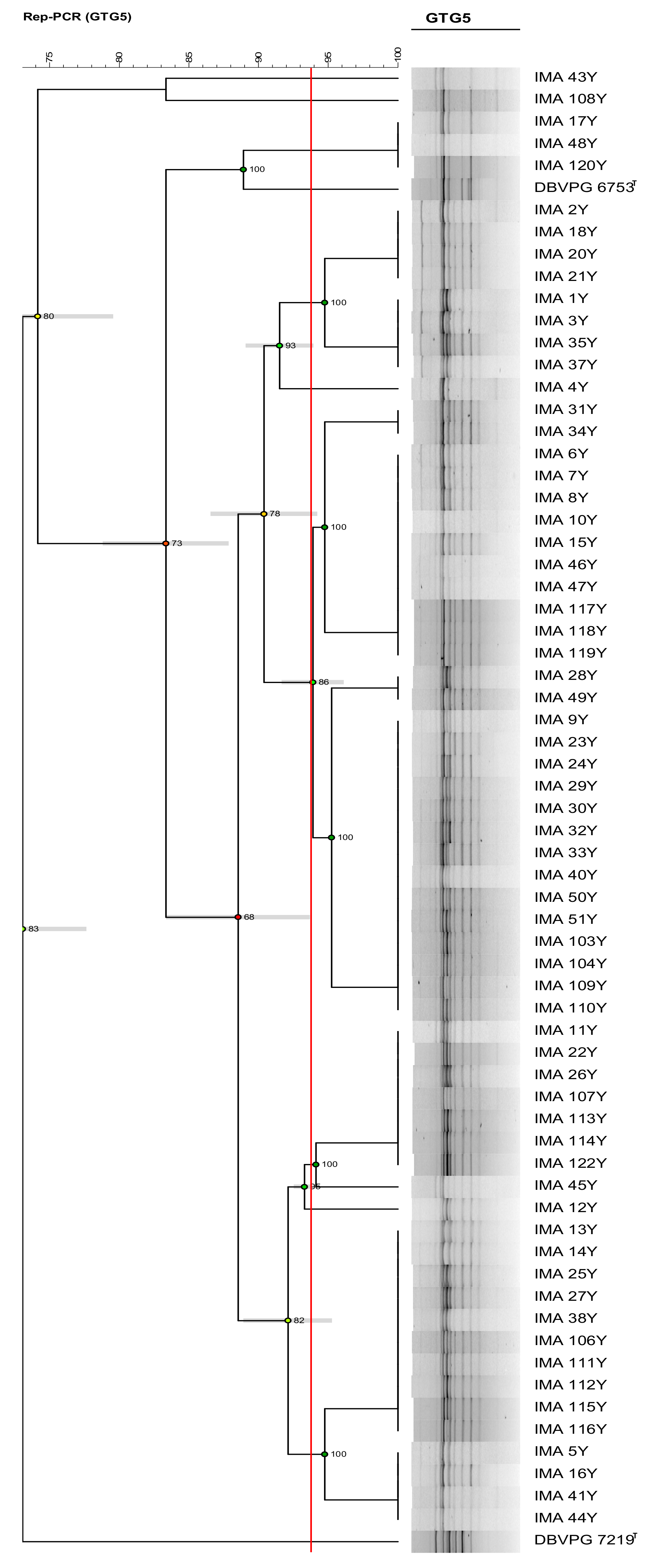
2.3. Molecular Intraspecific Diversity of Yeast Isolates Characterizing the PDO Tuscan Bread Sourdough
3. Results
3.1. Molecular Intraspecific Diversity of LAB Isolates Characterizing the PDO Tuscan Bread Sourdough
3.2. Molecular Intraspecific Diversity of Yeast Isolates Characterizing the PDO Tuscan Bread Sourdough
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Hutkins, R. Fermented vegetables. In Microbiology and Technology of Fermented Foods, 1st ed.; Hutkins, R., Ed.; IFT Press Blackwell Publishing: Oxford, UK, 2006; Volume 22, pp. 223–259. [Google Scholar]
- Dolci, P.; Alessandria, V.; Rantsiou, K.; Cocolin, L. Advanced methods for the identification, enumeration, and characterization of microorganisms in fermented foods. In Advances in fermented foods and beverages: Improving quality, technologies and health benefits; Holzapfel, W., Ed.; Woodhead Publishing Series in Food Science; Technology and Nutrition: Cambridge, UK, 2015; Volume 265, pp. 157–176. [Google Scholar]
- Gänzle, M.G.; Vermeulen, N.; Vogel, R.F. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 2007, 24, 128–138. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Tassios, P.T.; Dijkshoorn, L.; Haeggman, S.; Cookson, B.; Fry, N.K.; Fussing, V.; Green, J.; Feil, E.; Gerner-Smidt, P.; et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 2007, 13, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Lattanzi, A.; Minervini, F.; Di Cagno, R.; Diviccaro, A.; Antonielli, L.; Cardinali, G.; Cappelle, S.; De Angelis, M.; Gobbetti, M. The lactic acid bacteria and yeast microbiota of eighteen sourdoughs used for the manufacture of traditional Italian sweet leavened baked goods. Int. J. Food Microbiol. 2013, 163, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Venturi, M.; Guerrini, S.; Granchi, L.; Vincenzini, M. Typing of Lactobacillus sanfranciscensis isolates from traditional sourdoughs by combining conventional and multiplex RAPD-PCR profiles. Int. J. Food Microbiol. 2012, 156, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, Y.; Chen, J.; Sadiq, F.A.; Zhang, G.; Li, Y.; He, G. Prevalence and diversity of lactic acid bacteria in Chinese traditional sourdough revealed by culture dependent and pyrosequencing approaches. LWT Food Sci. Technol. 2016, 68, 91–97. [Google Scholar] [CrossRef]
- Succi, M.; Reale, A.; Andrighetto, C.; Lombardi, A.; Sorrentino, E.; Coppola, R. Presence of yeasts in Southern Italian sourdough from Triticum aestivum flour. FEMS Microbiol. Lett. 2003, 225, 143–148. [Google Scholar] [CrossRef][Green Version]
- Palla, M.; Agnolucci, M.; Calzone, A.; Giovannetti, M.; Di Cagno, R.; Gobbetti, M.; Rizzello, C.G.; Pontonio, E. Exploitation of autochthonous Tuscan sourdough yeasts as potential starters. Int. J. Food Microbiol. 2019, 302, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Minervini, F.; Di Cagno, R.; Lattanzi, A.; De Angelis, M.; Antonielli, L.; Cardinali, G.; Cappelle, S.; Gobbetti, M. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: interactions between ingredients and microbial species diversity. Appl. Environ. Microbiol. 2012, 78, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Vigentini, I.; Antoniani, D.; Roscini, L.; Comasio, A.; Galafassi, S.; Picozzi, C.; Corte, L.; Compagno, C.; Dal Bello, F.; Cardinali, G.; et al. Candida milleri species reveals intraspecific genetic and metabolic polymorphisms. Food Microbiol. 2014, 42, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Palla, M.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Identification and characterization of lactic acid bacteria and yeasts of PDO Tuscan bread sourdough by culture dependent and independent methods. Int. J. Food Microbiol. 2017, 250, 19–26. [Google Scholar] [CrossRef] [PubMed]
- De Man, J.C.; Rogosa, D.; Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Eriksson, J.; Löfströma, C.; Aspán, A.; Gunnarsson, A.; Karlsson, I.; Borch, E.; De Jonge, B.; Rådström, P. Comparison of genotyping methods by application to Salmonella livingstone strains associated with an outbreak of human salmonellosis. Int. J. Food Microbiol. 2005, 104, 93–103. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed]
- Agnolucci, M.; Vigentini, I.; Capurso, G.; Merico, A.; Tirelli, A.; Compagno, C.; Foschino, R.; Nuti, M. Genetic diversity and physiological traits of Brettanomyces bruxellensis strains isolated from Tuscan Sangiovese wines. Int. J. Food Microbiol. 2009, 130, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Lopez, V.; Fernández-Espinar, M.T.; Barrio, E.; Ramón, D.; Querol, A. A new PCR-based method for monitoring inoculated wine fermentations. Int. J. Food Microbiol. 2003, 81, 63–71. [Google Scholar] [CrossRef]
- Andrade, M.J.; Rodriguez, M.; Sánchez, B.; Aranda, E.; Córdoba, J.J. DNA typing methods for differentiation of yeasts related to dry-cured meat products. Int. J. Food Microbiol. 2006, 107, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, M.; Sakata, S.; Benno, Y. Biodiversity of Lactobacillus sanfranciscensis strains isolated from five sourdoughs. Lett. Appl. Microbiol. 2005, 40, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Osimani, A.; Zannini, E.; Aquilanti, L.; Mannazzu, I.M.; Comitini, F.; Clementi, F. Lactic acid bacteria and yeasts from wheat sourdoughs of the Marche region. Ital. J. Food Sci. 2009, 21, 269–286. [Google Scholar]
- Pulvirenti, A.; Solieri, L.; Gullo, M.; De Vero, L.; Giudici, P. Occurrence and dominance of yeast species in sourdough. Lett. Appl. Microbiol. 2004, 38, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Venturi, M.; Guerrini, S.; Vincenzini, M. Stable and non-competitive association of Saccharomyces cerevisiae, Candida milleri and Lactobacillus sanfranciscensis during manufacture of two traditional sourdough baked goods. Food Microbiol. 2012, 31, 107–115. [Google Scholar] [CrossRef] [PubMed]
| Strains a | Source of Isolation |
|---|---|
| Lactobacillus sanfranciscensis IMA 1LAB-3LAB; 5LAB-49LAB; 51LAB-53LAB; 55LAB-99LAB | PDO Tuscan bread sourdough |
| Lactobacillus sanfranciscensis DSMZ 20451T | San Francisco sourdough |
| Kazachstania humilis IMA 1Y-18Y; 20Y-35Y; 37Y-38Y; 40Y-41Y; 43Y-51Y; 103Y-104Y; 106Y-120Y; 122Y | PDO Tuscan bread sourdough |
| Saccharomyces cerevisiae IMA 19Y, 36Y; 105Y | PDO Tuscan bread sourdough |
| Kazachstania humilis DBVPG 6753T | San Francisco sourdough |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palla, M.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Large Genetic Intraspecific Diversity of Autochthonous Lactic Acid Bacteria and Yeasts Isolated from PDO Tuscan Bread Sourdough. Appl. Sci. 2020, 10, 1043. https://doi.org/10.3390/app10031043
Palla M, Cristani C, Giovannetti M, Agnolucci M. Large Genetic Intraspecific Diversity of Autochthonous Lactic Acid Bacteria and Yeasts Isolated from PDO Tuscan Bread Sourdough. Applied Sciences. 2020; 10(3):1043. https://doi.org/10.3390/app10031043
Chicago/Turabian StylePalla, Michela, Caterina Cristani, Manuela Giovannetti, and Monica Agnolucci. 2020. "Large Genetic Intraspecific Diversity of Autochthonous Lactic Acid Bacteria and Yeasts Isolated from PDO Tuscan Bread Sourdough" Applied Sciences 10, no. 3: 1043. https://doi.org/10.3390/app10031043
APA StylePalla, M., Cristani, C., Giovannetti, M., & Agnolucci, M. (2020). Large Genetic Intraspecific Diversity of Autochthonous Lactic Acid Bacteria and Yeasts Isolated from PDO Tuscan Bread Sourdough. Applied Sciences, 10(3), 1043. https://doi.org/10.3390/app10031043





