Abstract
Peptide nucleic acid can recognise sequences in double-stranded DNA (dsDNA) through the formation of a double-duplex invasion complex. This double-duplex invasion is a promising method for the recognition of dsDNA in cellula because peptide nucleic acid (PNA) invasion does not require the prior denaturation of dsDNA. To increase its applicability, we developed PNAs modified with a nuclear localisation signal (NLS) peptide. In this study, the characteristics of NLS-modified PNAs were investigated for the future design of novel peptide-modified PNAs.
1. Introduction
The sequence-specific recognition of double-stranded DNA (dsDNA) has been an important research subject owing to its wide range of potential applications [1,2,3,4,5,6,7]. For this purpose, various methods have been developed, including those that make use of DNA-binding proteins [1,3,4,5,8,9,10,11,12], small molecules such as minor groove binders [13,14,15,16], and artificial DNA [17,18,19,20,21,22,23,24]. In addition, the recognition of dsDNA by peptide nucleic acid (PNA), an artificial nucleic acid mimic, has been reported by Nielsen et al. [7,21,24,25,26,27,28,29,30,31]. In 1991, PNA was first designed and synthesised as an analogue of natural nucleic acids, where the negatively charged sugar-phosphate backbone of DNA was substituted with an electrostatically neutral artificial N-(2-aminoethyl)glycine backbone (Figure 1a) [25]. Consequently, electrostatic repulsion between PNA and DNA is absent, enabling PNA to form more stable duplexes with complementary DNA via Watson–Crick base pairing than those between complementary DNA strands [31]. Furthermore, when pseudo-complementary PNAs (pcPNAs: see Figure S1)—where conventional adenine (A) and thymine (T) have been replaced by 2,6-diaminopurine (D) and 2-thiouracil (U)—are used, an effective invasion into dsDNA becomes possible, with two strands of pcPNA forming a double-duplex invasion complex (or, simply, invasion complex, Figure 1b) with dsDNA [26,32]. The introduction of D and U into PNA destabilises the formation of PNA/PNA duplexes through steric repulsions between the amino group of D and the thioketone group of U. The thermal stability of the PNA/DNA duplex is enhanced by the formation of stable base pairs between D/U and the natural nucleobases. PNA’s unique DNA-recognition mode has been used in many applications, including site-directed mutagenesis [2,7], inhibition of enzymatic activity [26,33,34], and the development of artificial DNA cutters [35], with all of these depending on the sequence specificity of the invasion complex. The pseudopeptide backbone of PNA facilitates easy oligomer synthesis and modification for further improvements and imparts enhanced resistance to enzymatic degradation. Thus, double-duplex invasion is a promising method for the targeting of dsDNA in cellula.
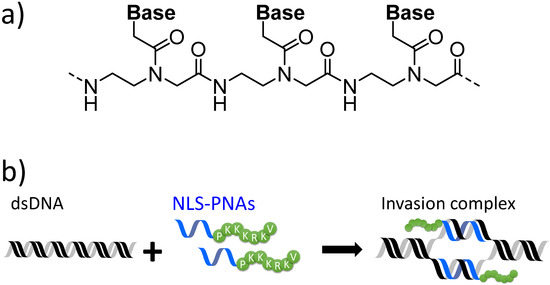
Figure 1.
(a) Chemical structure of peptide nucleic acid (PNA). (b) Stable invasion complex formation using nuclear-localisation-signal-modified PNAs (NLS-PNAs).
However, to realise such applications, an important issue must be overcome—namely, the high salt concentrations encountered in the cellular environment. High salt concentrations increase the stability of the DNA/DNA duplexes whilst slightly destabilising the PNA/DNA duplexes [36], which results in an overall decrease in invasion efficiency [37]. Several approaches have been attempted to enhance invasion efficiency in physiological salt concentrations, such as chemical modification of PNA and conjugation with functional molecules [37,38,39,40,41]. Our group developed conjugates of PNA with Ru-complexes and PNAs modified with a nuclear localisation signal (NLS) peptide to increase their applicability for DNA recognition at high salt concentrations (Figure 1b). These two modified PNAs displayed higher DNA affinity than corresponding unmodified pcPNAs and effectively formed an invasion complex even at physiological salt concentrations, whereunder unmodified PNAs stop functioning efficiently. It is important to understand the properties and limitations of these modified PNAs in order to further improve PNAs. Of these two methods, NLS modification is the simpler PNA modification method as the resulting NLS-modified PNAs (NLS-PNAs) can be procured by simply conjugating PNA with the functional peptide, and can be readily introduced during the synthesis of PNAs. Thus, in this study, we focused on NLS-PNAs and investigated their characteristics to obtain insight into how they can be improved.
2. Materials and Methods
2.1. Materials
Solvents and reagents were purchased from FUJIFILM Wako Pure Chemical Co. (Tokyo, Japan); Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan); Merck (Darmstadt, Germany); Sigma-Aldrich Co., LLC. (St. Louis, MO, USA); nacalai tesque Inc. (Kyoto, Japan); and Kanto Chemical Co., Inc. (Tokyo, Japan); and were used without further purification. PNA monomers were purchased from ASM Research Chemicals (Hannover, Germany). Oligonucleotides were purchased from Fasmac (Kanagawa, Japan).
PNAs with or without an NLS peptide were synthesised by standard Boc-chemistry-based solid-phase peptide synthesis according to a literature procedure [42], purified by reversed-phase high-performance liquid chromatography (HPLC) (Jasco Co.; Tokyo, Japan), and characterised by matrix-assisted laser desorption/ionization–time of flight mass spectrometer (MALDI-TOF MS) (ultraflex III, Bruker Daltonics; Billerica, MA, USA) (Figures S2 and S3). The concentration of PNAs and DNAs was determined based on their absorbance at 260 nm by using a UV-visible spectrophotometer (molar extinction coefficients for PNA monomers (in M−1 cm−1): ε(D) = 7600; ε(C) = 6600; ε(G) = 11,700; ε(U) = 10,200).
2.2. Invasion Experiments
Target 130-bp DNA from pBR322 was incubated with a pair of pcPNAs in a 5 mM HEPES buffer (pH 7.0) at 50 °C for 1 h. After incubation, the solutions were subjected to a microchip electrophoresis system (MultiNA, Shimadzu; Kyoto, Japan) and the invasion efficiency was evaluated.
3. Results and Discussion
3.1. A Positive Effect on Invasion Efficiency by the Introduction of an NLS into Short PNAs
In a previous report, we successfully demonstrated that conjugation of an NLS to pcPNAs results in enhanced invasion efficiency [37]. Specifically, an NLS peptide with the sequence PKKKRKV found in the SV40 virus, which has been widely studied, was conjugated at the C-terminus of a pentadecamer PNA. If the introduction of an NLS, even in short PNAs, exerts a positive effect upon invasion efficiency, then the properties of NLS-PNAs should be more easily elucidated by examining them with short PNAs, whose synthesis requires less effort than longer PNAs. Thus, we checked the effectiveness of the NLS in decamer PNAs and employed two types of PNA: (1) unmodified PNAs (U-PNAs), i.e., a pair of decamer pcPNAs without an NLS (with only a few lysine residues for solubility) and (2) C-PNAs, which are identical in sequence to U-PNAs but with an NLS at their C-termini.
U-PNAs and C-PNAs were synthesised by standard tert-butoxycarbonyl (Boc)-based solid-phase peptide synthesis. The PNA sequences are listed in Table 1. PNA1 and PNA2 represent each strand of pcPNA that is complementary to 10 bp in the pBR322 plasmid (PNA1: GUUDCUGDUG and PNA2: CDUCDGUDDC). All PNAs have a free N-terminal amino group, with the C-terminal carboxylic acid being converted to an amide. The efficiency of invasion complex formation was evaluated by an electrophoresis mobility shift assay (EMSA). The formation of the invasion complex results in a lower electrophoretic mobility, mainly due to changes in the local structure of the dsDNA at the invasion site [43]. This allows the separation of the invasion complex from the free dsDNA. An EMSA of the invasion complex formation in the presence of 75 mM NaCl is shown in Figure 2.

Table 1.
PNAs synthesised and used in this research.
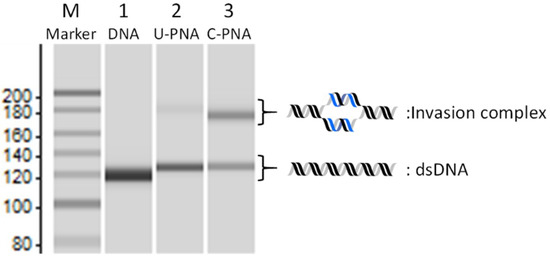
Figure 2.
Comparison of U-PNA and C-PNA invasion efficiency in the presence of 75 mM NaCl. M: 20-bp DNA Ladder (TaKaRa); lane 1: 130-bp DNA only; lane2 U-PNA; lane 3: C-PNA. Invasion conditions: [DNA] = [each PNA] = 50 nM, [HEPES (pH 7.0)] = 5 mM, and [NaCl] = 75 mM at 50 °C for 1 h.
In lane 1, a single band corresponding to free dsDNA was observed between 100 and 150 bp. In the presence of U-PNA, the invasion complex was barely formed under the high-salt conditions employed, giving only a faint band with lower mobility between 180 and 200 bp (lane 2), which was assignable to the invasion complex. However, the C-PNAs with NLS at their C-termini gave an intense band corresponding to the invasion complex (lane 3). It is clear from these results that the invasion efficiency was improved by the conjugation of an NLS to the short PNAs, leading to the formation of a stable invasion complex even at elevated salt concentrations. As a result, it was confirmed that the NLS modifications in short PNAs showed a beneficial effect on invasion. Thus, we decided to investigate the properties of NLS-PNAs by using these short decamer PNAs. The effect of NLS modification appears to be mainly attributable to electrostatic interactions. However, the conjugation of PNA with repeating cationic peptides (e.g., polylysine) also increases non-specific interactions with the DNA backbone, resulting in a negative effect on the formation of the invasion complex [44]. Presumably, in biological motifs, unfavourable sequences (e.g., those provoking non-specific binding to DNA) have been evolutionarily excluded. Thus, we can expect that NLS peptides possess an ideal amino acid sequence that shows a moderate affinity to DNA as well as a positive effect on invasion efficiency.
3.2. Changing the Location of the NLS from the C-Terminus to the N-Terminus of the PNAs
As mentioned above, in the previous report on NLS-PNAs, the NLS peptide was conjugated to the C-termini of the PNAs (C-PNAs). The synthesis of NLS-PNA was carried out from the C-terminus to the N-terminus by solid-phase peptide synthesis, and the NLS peptide was introduced to the resin first, followed by the PNA. When examining various functional peptides as alternatives to NLS for future research, N-terminally modified PNA conjugates, rather than C-terminally modified PNA conjugates, would significantly reduce the time and effort required for investigation. This is because it is possible to synthesise a large number of peptide-modified PNAs at once by elongating the PNAs first and then splitting the resin followed by peptide conjugation. Therefore, we evaluated whether changing the location of the NLS from the C-terminus to the N-terminus of the PNA (N-PNA) would have a detrimental effect upon invasion efficiency. PNAs modified with NLS peptide at the N-termini (N-PNAs) were synthesised using the same method as C-PNAs, and the invasion experiments with a series of PNAs were carried out under a broad range of salt concentrations, ranging from 0 to 125 mM NaCl.
As a control, the invasion efficiency of U-PNAs was also examined at increasing salt concentrations. Figure 3 shows that the band intensity of the invasion complex with U-PNA decreased with increasing salt concentration and almost disappeared when the NaCl concentration exceeded 50 mM. This result reaffirms the aforementioned phenomenon, wherein elevated salt concentrations stabilise dsDNA whilst weakening PNA/DNA interaction, resulting in a lower invasion efficiency. When the same experiment was conducted using C-PNAs, however, a high invasion efficiency beyond 50 mM NaCl was observed. Unlike in our previous report with pentadecamer NLS-PNAs, in the present study, a decrease in invasion efficiency was observed above 75 mM NaCl. This is due to the lower concentration of PNA, which we selected herein to amplify any effect. The introduction of NLS, even in shorter PNAs, was once more confirmed to be beneficial for invasion at high salt concentrations. Next, the effect of NLS position on invasion efficiency was investigated using N-PNAs. At first glance, both C-PNAs and N-PNAs exhibited similar behaviour in terms of invasion efficiency. The invasion efficiencies of both NLS-PNAs remained high, up to about 75 mM NaCl, although this value gradually decreased with increasing salt concentration. This comparison confirms that, regardless of the location of the NLS, PNA-NLS conjugates display similar performance. However, N-PNAs were slightly better in terms of resistance to higher salt concentrations (over 75 m>M) than C-PNAs, with a more pronounced effect at very high salt concentrations of 125 mM.
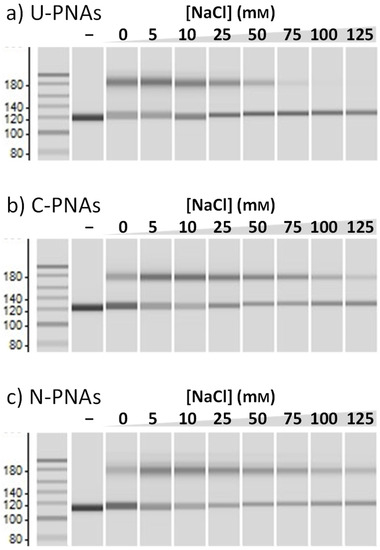
Figure 3.
Comparison of (a) U-PNAs’, (b) C-PNAs’, and (c) N-PNAs’ invasion efficiency. Invasion conditions: [DNA] = [each PNA] = 50 nM, [HEPES (pH 7.0)] = 5 mM, and [NaCl] = 0–125 mM at 50 °C for 1 h.
3.3. Insertion of Proline between the PNAs and Valine of the NLS
Besides the location of the NLS itself, there is a further difference between C-PNAs and N-PNAs. In order to maintain the directionality of the NLS, in C-PNAs, proline (P) of the NLS connects to the PNA, whereas in N-PNAs, the NLS is connected via valine (V). Unlike valine, proline possesses a unique cyclic structure bestowing it with a much higher backbone rigidity than any of the other canonical amino acids. In order to verify its significance, we synthesised N-Pro-PNAs, which are identical to N-PNAs except that a proline residue has been inserted between the PNA and valine. Interestingly, the insertion of proline proved unfavourable for invasion efficiency, and N-Pro-PNAs displayed invasion properties between those of N-PNAs and C-PNAs (Figure 4). These results strongly indicate that the rigid amino acid connecting the NLS to PNA may unfavourably affect invasion.
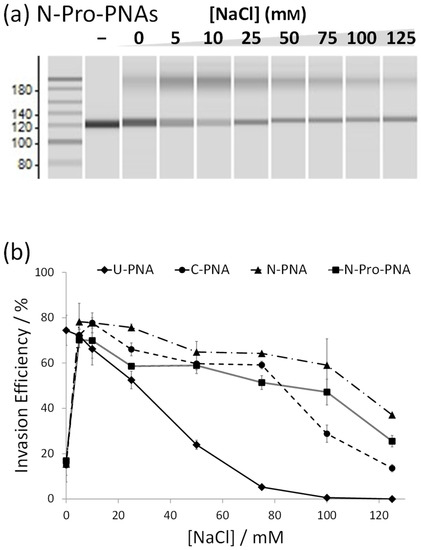
Figure 4.
(a) Invasion experiments for N-Pro-PNAs to evaluate the effect of an amino acid between NLS and PNA. Invasion conditions: [DNA] = [each PNA] = 50 nM, [HEPES (pH 7.0)] = 5 mM, and [NaCl] = 0–125 mM at 50 °C for 1 h. (b) Effect of salt concentration on invasion efficiency with various PNAs (U-, C-, N-, and N-Pro-PNAs). The value for invasion efficiency was obtained by comparing the band intensities in a microchip electrophotogram.
4. Conclusions
In summary, we investigated the characteristics of NLS-PNA by employing a series of NLS-modified PNAs. These NLS-PNA conjugates were found to be superior to unmodified PNAs at elevated salt concentrations. The difference between C-terminally modified and N-terminally modified NLS-PNAs was not very significant, signifying that N-terminally modified PNA-NLS conjugates can be readily used for research purposes using the same method as previously reported C-terminally modified conjugates. This facilitates the development of new functional peptide-modified PNA conjugates, since solid-phase peptide synthesis requires synthesis from the C-terminus to the N-terminus. As an example, for C-terminally modified PNAs, when multiple functional peptides bound to the same PNA need to be screened, each functional peptide must be synthesised separately. In the case of N-terminal modification, the synthesis of peptide-modified PNAs can be simplified by synthesising the individual PNAs only once, separating the resin beads into as many samples as needed and continuing the synthesis of each NLS. In addition to the finding that the position of the NLS has some influence on invasion, we demonstrated that the amino acids between the NLS and PNA also play an important role. These findings also provide new perspectives for the future design of novel peptide-modified PNAs.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/23/8663/s1, Figure S1: chemical structures of 2,6-diaminopurine (D) and 2-thouracil (U), Figure S2: HPLC charts of purified PNAs, Figure S3: MALDI-TOF MS spectra of purified PNAs.
Author Contributions
Investigation, G.U. and M.S.; writing—original draft preparation, Y.A. and G.U.; writing—review and editing, Y.A., G.U., M.S. and O.S.; supervision, Y.A. and O.S.; project administration, Y.A.; funding acquisition, Y.A. and O.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by JSPS KAKENHI grant no. 19K05730 to Y.A. from the Ministry of Education, Culture, Sports, Science, and Technology (Japan). This work was also supported in part by the Izumi Science and Technology Foundation to Y.A., Kato Memorial Bioscience Foundation to Y.A., and Integrated Research Consortium on Chemical Sciences to Y.A.
Acknowledgments
We thank Joshua Kyle Stanfield for checking the English of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Nielsen, P.E.; Glazer, P.M. Site-directed gene mutation at mixed sequence targets by psoralen-conjugated pseudo-complementary peptide nucleic acids. Nucleic Acids Res. 2007, 35, 7604–7613. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Komiyama, M.; Yoshimoto, K.; Sisido, M.; Ariga, K. Chemistry Can Make Strict and Fuzzy Controls for Bio-Systems: DNA Nanoarchitectonics and Cell-Macromolecular Nanoarchitectonics. Bull. Chem. Soc. Jpn. 2017, 90, 967–1004. [Google Scholar] [CrossRef]
- Economos, N.G.; Oyaghire, S.; Quijano, E.; Ricciardi, A.S.; Saltzman, W.M.; Glazer, P.M. Peptide Nucleic Acids and Gene Editing: Perspectives on Structure and Repair. Molecules 2020, 25, 735. [Google Scholar] [CrossRef]
- Wolfe, S.A.; Nekludova, L.; Pabo, C.O. DNA recognition by Cys(2)His(2) zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 183–212. [Google Scholar] [CrossRef]
- Jantz, D.; Amann, B.T.; Gatto, G.J.; Berg, J.M. The design of functional DNA-binding proteins based on zinc finger domains. Chem. Rev. 2004, 104, 789–799. [Google Scholar] [CrossRef]
- Dhanasekaran, M.; Negi, S.; Sugiura, Y. Designer zinc finger proteins: Tools for creating artificial DNA-binding functional proteins. Acc. Chem. Res. 2006, 39, 45–52. [Google Scholar] [CrossRef]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.S.; Oi, L.S. A CRISPR-dCas Toolbox for Genetic Engineering and Synthetic Biology. J. Mol. Biol. 2019, 431, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Dervan, P.B.; Burli, R.W. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999, 3, 688–693. [Google Scholar] [CrossRef]
- Dervan, P.B. Molecular recognition of DNA by small molecules. Biorg. Med. Chem. 2001, 9, 2215–2235. [Google Scholar] [CrossRef]
- Dervan, P.B.; Edelson, B.S. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003, 13, 284–299. [Google Scholar] [CrossRef]
- Bando, T.; Sugiyama, H. Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole-imidazole polyamides. Acc. Chem. Res. 2006, 39, 935–944. [Google Scholar] [CrossRef]
- Thuong, N.T.; Hélène, C. Sequence-Specific Recognition and Modification of Double-Helical DNA by Oligonucleotides. Angew. Chem. Int. Ed. Engl. 1993, 32, 666–690. [Google Scholar] [CrossRef]
- Praseuth, D.; Guieysse, A.L.; Hélène, C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta Gene Struct. Expr. 1999, 1489, 181–206. [Google Scholar] [CrossRef]
- Fox, K.R. Targeting DNA with triplexes. Curr. Med. Chem. 2000, 7, 17–37. [Google Scholar] [CrossRef]
- Rapireddy, S.; He, G.; Roy, S.; Armitage, B.A.; Ly, D.H. Strand invasion of mixed-sequence B-DNA by acridine-linked, gamma-peptide nucleic acid (gamma-PNA). J. Am. Chem. Soc. 2007, 129, 15596–15600. [Google Scholar] [CrossRef]
- Vilaivan, T. Pyrrolidinyl PNA with alpha/beta-Dipeptide Backbone: From Development to Applications. Acc. Chem Res. 2015, 48, 1645–1656. [Google Scholar] [CrossRef] [PubMed]
- Hrdlicka, P.J.; Karmakar, S. 25 years and still going strong: 2′-O-(pyren-1-yl) methylribonucleotides-versatile building blocks for applications in molecular biology, diagnostics and materials science. Org. Biomol. Chem. 2017, 15, 9760–9774. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Kawabata, H.; Fujimoto, K. Double duplex invasion of DNA induced by ultrafast photo-cross-linking using 3-cyanovinylcarbazole for antigene methods. Chem. Commun. 2017, 53, 7616–7619. [Google Scholar] [CrossRef] [PubMed]
- Muangkaew, P.; Vilaivan, T. Modulation of DNA and RNA by PNA. Bioorg. Med. Chem. Lett. 2020, 30, 127064. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Lohse, J.; Dahl, O.; Nielsen, P.E. Double duplex invasion by peptide nucleic acid: A general principle for sequence-specific targeting of double-stranded DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 11804–11808. [Google Scholar] [CrossRef] [PubMed]
- Demidov, V.V.; Frank-Kamenetskii, M.D. Two sides of the coin: Affinity and specificity of nucleic acid interactions. Trends Biochem. Sci. 2004, 29, 62–71. [Google Scholar] [CrossRef]
- Nielsen, P.E. Peptide Nucleic Acids (PNA) in Chemical Biology and Drug Discovery. Chem. Biodivers. 2010, 7, 786–804. [Google Scholar] [CrossRef]
- Sharma, C.; Awasthi, S.K. Versatility of peptide nucleic acids (PNAs): Role in chemical biology, drug discovery, and origins of life. Chem. Biol. Drug Des. 2017, 89, 16–37. [Google Scholar] [CrossRef]
- Manicardi, A.; Rozzi, A.; Korom, S.; Corradini, R. Building on the peptide nucleic acid (PNA) scaffold: A biomolecular engineering approach. Supramol. Chem. 2017, 29, 784–795. [Google Scholar] [CrossRef]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Haaima, G.; Lohse, A.; Buchardt, O.; Nielsen, P.E. Peptide Nucleic Acids (PNAs) Containing Thymine Monomers Derived from Chiral Amino Acids: Hybridization and Solubility Properties of D-Lysine PNA. Angew. Chem. Int. Ed. 1996, 35, 1939–1942. [Google Scholar]
- Izvolsky, K.I.; Demidov, V.V.; Nielsen, P.E.; Frank-Kamenetskii, M.D. Sequence-specific protection of duplex DNA against restriction and methylation enzymes by pseudocomplementary PNAs. Biochemistry 2000, 39, 10908–10913. [Google Scholar] [CrossRef] [PubMed]
- Protozanova, E.; Demidov, V.V.; Nielsen, P.E.; Frank-Kamenetskii, M.D. Pseudocomplementary PNAs as selective modifiers of protein activity on duplex DNA: The case of type IIs restriction enzymes. Nucleic Acids Res. 2003, 31, 3929–3935. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, M.; Aiba, Y.; Yamamoto, Y.; Sumaoka, J. Artificial restriction DNA cutter for site-selective scission of double-stranded DNA with tunable scission site and specificity. Nat. Protoc. 2008, 3, 655–662. [Google Scholar] [CrossRef]
- Tomac, S.; Sarkar, M.; Ratilainen, T.; Wittung, P.; Nielsen, P.E.; Norden, B.; Graslund, A. Ionic effects on the stability and conformation of peptide nucleic acid complexes. J. Am. Chem. Soc. 1996, 118, 5544–5552. [Google Scholar] [CrossRef]
- Aiba, Y.; Honda, Y.; Komiyama, M. Promotion of Double-Duplex Invasion of Peptide Nucleic Acids through Conjugation with Nuclear Localization Signal Peptide. Chem. Eur. J. 2015, 21, 4021–4026. [Google Scholar] [CrossRef]
- Aiba, Y.; Hamano, Y.; Kameshima, W.; Araki, Y.; Wada, T.; Accetta, A.; Sforza, S.; Corradini, R.; Marchelli, R.; Komiyama, M. PNA-NLS conjugates as single-molecular activators of target sites in double-stranded DNA for site-selective scission. Org. Biomol. Chem. 2013, 11, 5233–5238. [Google Scholar] [CrossRef]
- Aiba, Y.; Ohyama, J.; Komiyama, M. Transfection of PNA-NLS Conjugates into Human Cells Using Partially Complementary Oligonucleotides. Chem. Lett. 2015, 44, 1547–1549. [Google Scholar] [CrossRef]
- Hibino, M.; Aiba, Y.; Watanabe, Y.; Shoji, O. Peptide Nucleic Acid Conjugated with Ruthenium-Complex Stabilizing Double-Duplex Invasion Complex Even under Physiological Conditions. ChemBioChem 2018, 19, 1601–1604. [Google Scholar] [CrossRef]
- Hibino, M.; Aiba, Y.; Shoji, O. Cationic guanine: Positively charged nucleobase with improved DNA affinity inhibits self-duplex formation. Chem. Commun. 2020, 56, 2546–2549. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, M.; Aiba, Y.; Ishizuka, T.; Sumaoka, J. Solid-phase synthesis of pseudo-complementary peptide nucleic acids. Nat. Protoc. 2008, 3, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, H.; Cherny, D.I.; Demidov, V.V.; Frank-Kamenetskii, M.D. Inducing and modulating anisotropic DNA bends by pseudocomplementary peptide nucleic acids. Proc. Natl. Acad. Sci. USA 2004, 101, 7548–7553. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, T.; Yoshida, J.; Yamamoto, Y.; Sumaoka, J.; Tedeschi, T.; Corradini, R.; Sforza, S.; Komiyama, M. Chiral introduction of positive charges to PNA for double-duplex invasion to versatile sequences. Nucleic Acids Res. 2008, 36, 1464–1471. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. P = proline inserted between the PNA and valine of the NLS. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).