Encapsulation of mRNA into Artificial Viral Capsids via Hybridization of a β-Annulus-dT20 Conjugate and the Poly(A) Tail of mRNA
Abstract
1. Introduction
2. Materials and Methods
2.1. General
2.2. Preparation of dT20-SS-β-Annulus and β-Annulus Peptides
2.3. Preparation and Characterization of Artificial Viral Capsids
2.4. Preparation of Complex of dT20-SS-β-Annulus:β-Annulus Peptide Hybridized with mCherry mRNA
2.5. Electrophoretic Mobility Shift Assay and Nuclease Resistance Assay
2.6. Confocal Laser Scanning Microscopy (CLSM) Measurements of In-Cell Expression of mCherry mRNA
3. Results and Discussion
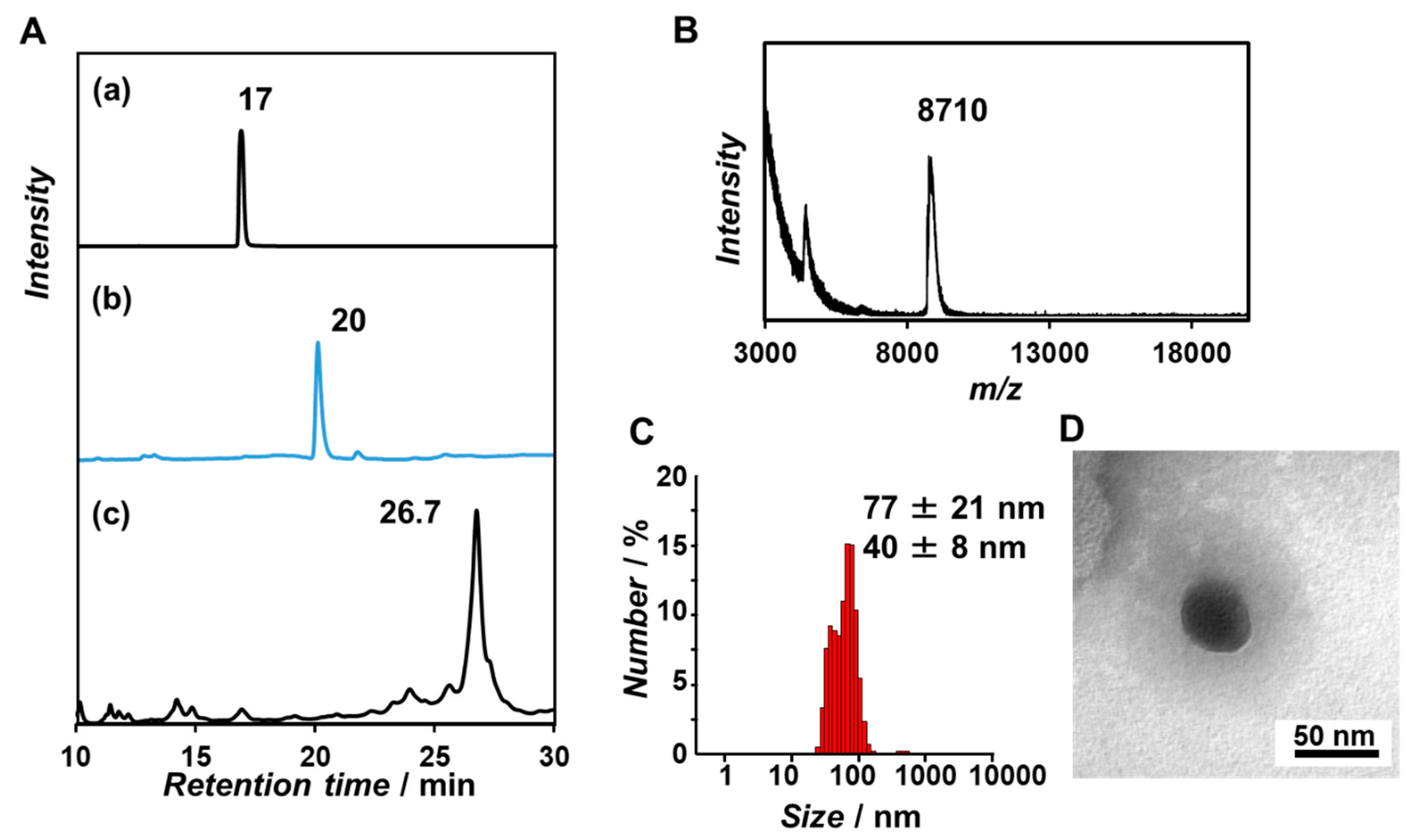
3.1. Synthesis and Self-Assembling Behavior of β-Annulus Peptide Bearing dT20 at the N-Terminus
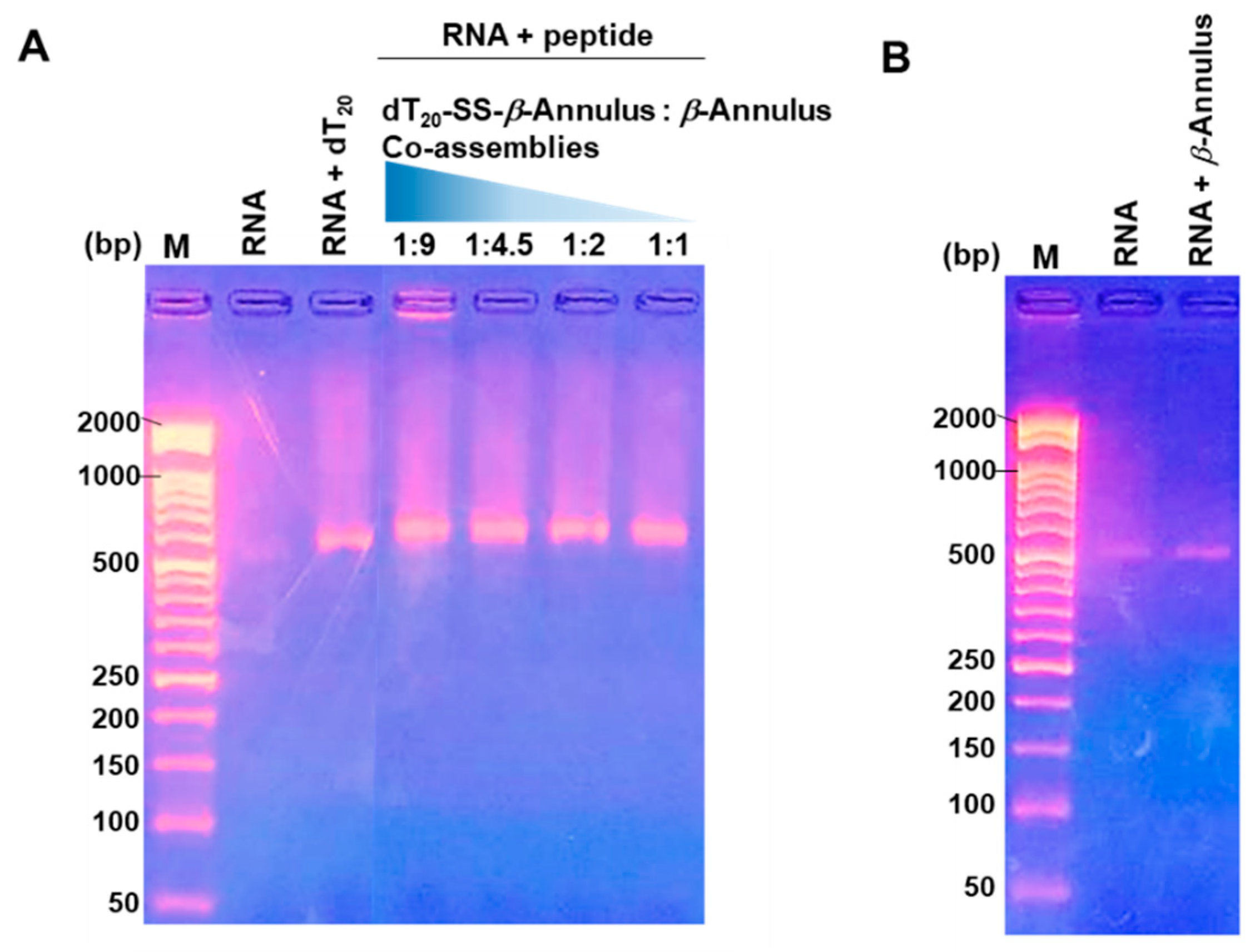
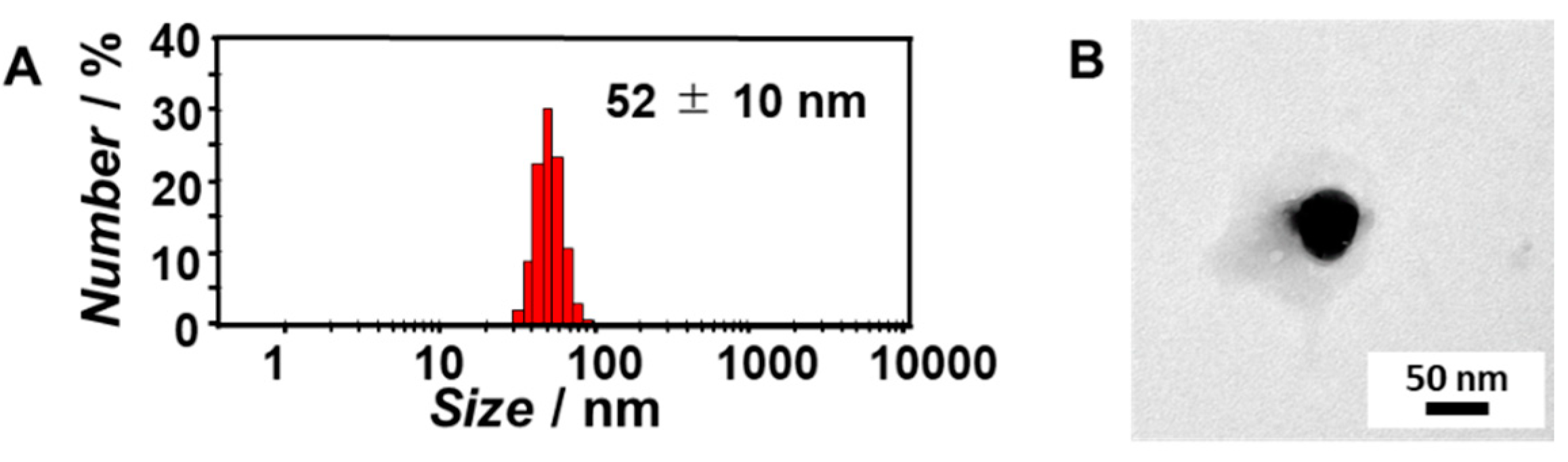
3.2. Complexation of mCherry mRNA and Artificial Viral Capsid Bearing dT20
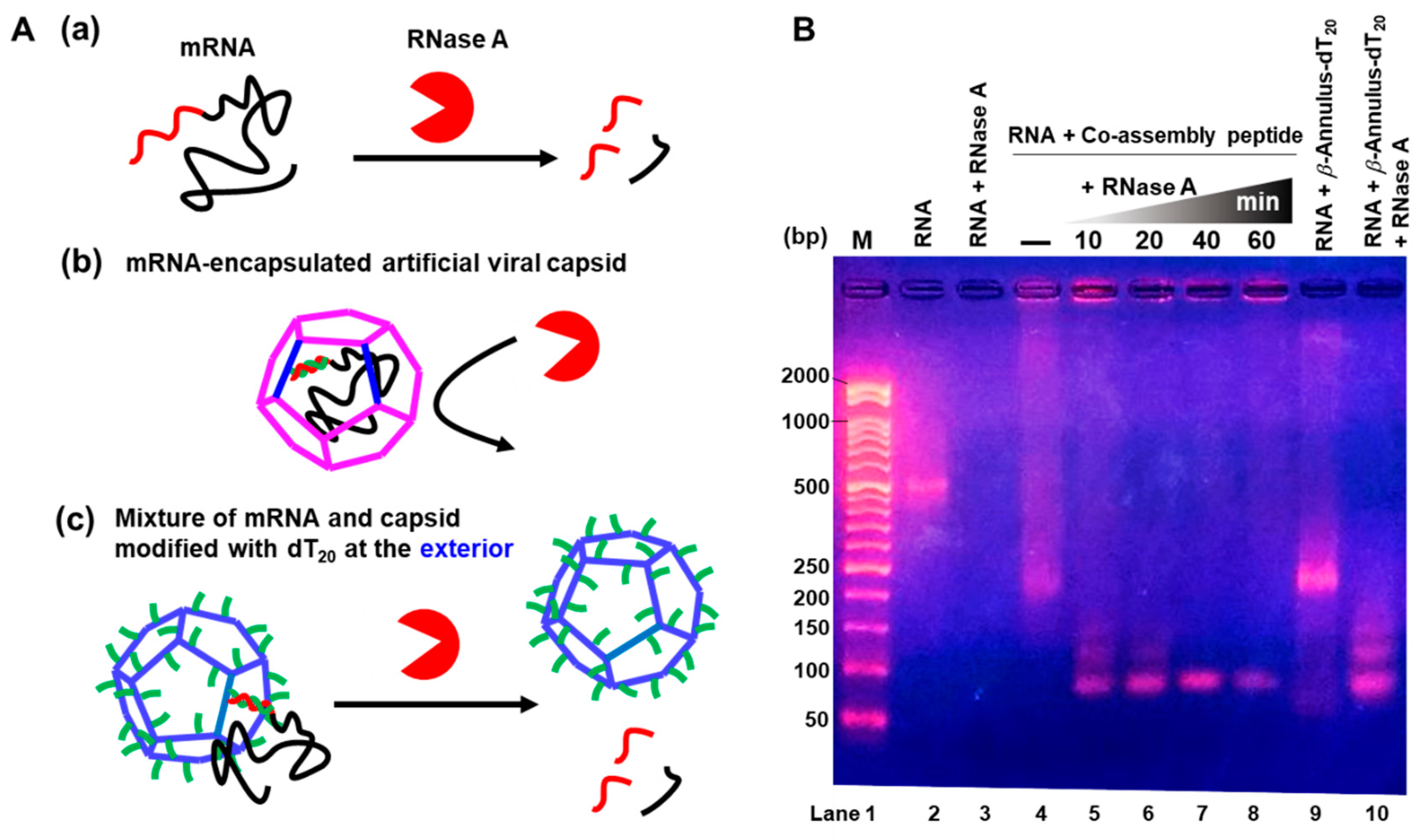
3.3. Nuclease Resistance of mRNA-Encapsulated Artificial Viral Capsid
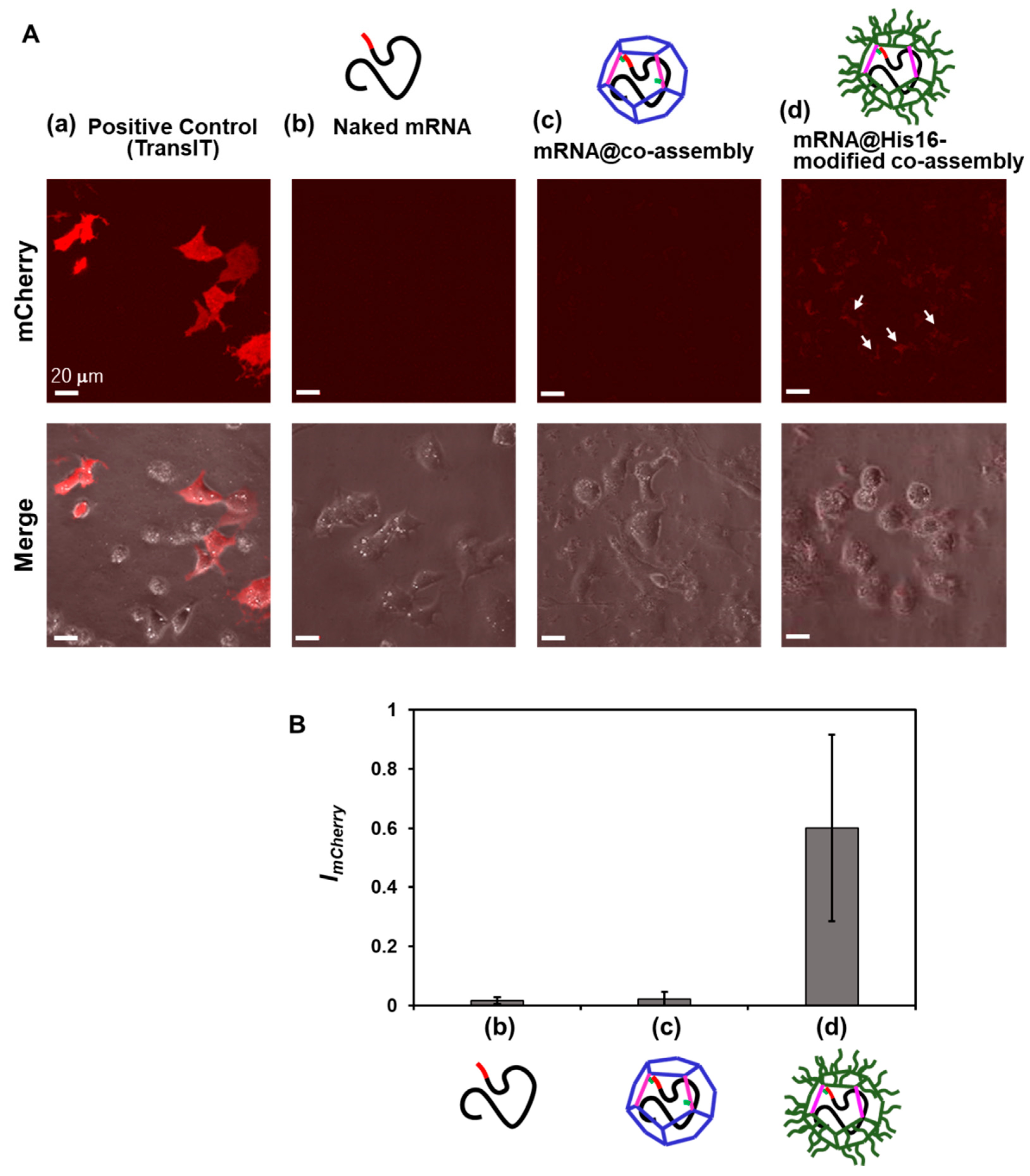
3.4. In-Cell Expression of mCherry mRNA Encapsulated in Artificial Viral Capsid
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, K.J.; Webber, M.J.; Anderson, D.G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release 2016, 240, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Rosenecker, J. Nanotechnologies in delivery of mRNA therapeutics using non-viral vector-based delivery systems. Gene Ther. 2017, 24, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef]
- Meng, C.; Chen, Z.; Li, G.; Welte, T.; Shen, H. Nanoplatforms for mRNA Therapeutics. Adv. Ther. 2020. [Google Scholar] [CrossRef]
- Uchida, H.; Itaka, K.; Nomoto, T.; Ishii, T.; Suma, T.; Ikegami, M.; Miyata, K.; Oba, M.; Nishiyama, N.; Kataoka, K. Modulated Protonation of Side Chain Aminoethylene Repeats in N-Substituted Polyaspartamides Promotes mRNA Transfection. J. Am. Chem. Soc. 2014, 136, 12396–12405. [Google Scholar] [CrossRef]
- Baba, M.; Itaka, K.; Kondo, K.; Yamasoba, T.; Kataoka, K. Treatment of neurological disorders by introducing mRNA In Vivo using polyplex nanomicelles. J. Control. Release 2015, 201, 41–48. [Google Scholar] [CrossRef]
- Matsui, A.; Uchida, S.; Ishii, T.; Itaka, K.; Kataoka, K. Messenger RNA-based therapeutics for the treatment of apoptosis-associated diseases. Sci. Rep. 2015, 5, 15810. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; He, Y.; Li, Y.; Yan, E.Z.; Zhang, K.; Irvine, D.J.; Hammond, P.T. Structurally Programmed Assembly of Translation Initiation Nanoplex for Superior mRNA Delivery. ACS Nano 2017, 11, 2531–2544. [Google Scholar] [CrossRef]
- Benner, N.L.; McClellan, R.L.; Turlington, C.R.; Haabeth, O.A.W.; Waymouth, R.M.; Wender, P.A. Oligo(serine ester) Charge-Altering Releasable Transporters: Organocatalytic Ring-Opening Polymerization and their Use for In Vitro and In Vivo mRNA Delivery. J. Am. Chem. Soc. 2019, 141, 8416–8421. [Google Scholar] [CrossRef] [PubMed]
- Rein, A. Retroviral RNA packaging: A review. In Positive-Strand RNA Viruses; Brinton, M.A., Calisher, C.H., Rueckert, R., Eds.; Springer: Vienna, Austria, 1994; Volume 9, pp. 513–522. [Google Scholar]
- Perlmutter, J.D.; Hagan, M.F. Mechanisms of Virus Assembly. Annu. Rev. Phys. Chem. 2015, 66, 217–239. [Google Scholar] [CrossRef] [PubMed]
- Azuma, Y.; Edwardson, T.G.W.; Terasaka, N.; Hilvert, D. Modular Protein Cages for Size-Selective RNA Packaging in Vivo. J. Am. Chem. Soc. 2018, 140, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Terasaka, N.; Azuma, Y.; Hilvert, D. Laboratory evolution of virus-like nucleocapsids from non-viral protein cages. Proc. Natl. Acad. Sci. USA 2018, 115, 5432–5437. [Google Scholar] [CrossRef]
- Munroe, D.; Jacobson, A. Tales of poly(A): A review. Gene 1990, 91, 151–158. [Google Scholar] [CrossRef]
- Sachs, A. The role of poly(A) in the translation and stability of mRNA. Curr. Opin. Cell Biol. 1990, 2, 1092–1098. [Google Scholar] [CrossRef]
- Eckmann, C.R.; Rammelt, C.; Wahle, E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA 2011, 2, 348–361. [Google Scholar] [CrossRef]
- Weill, L.; Belloc, E.; Bava, F.-A.; Méndez, R. Translational control by changes in poly(A) tail length: Recycling mRNAs. Nat. Struct. Mol. Biol. 2012, 19, 577–585. [Google Scholar] [CrossRef]
- Gruber, A.J.; Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 2019, 20, 599–614. [Google Scholar] [CrossRef]
- Nicholson, A.L.; Pasquinelli, A.E. Tales of Detailed Poly(A) Tails. Trends Cell Biol. 2019, 29, 191–200. [Google Scholar] [CrossRef]
- Matsuura, K.; Watanabe, K.; Matsuzaki, T.; Sakurai, K.; Kimizuka, N. Self-Assembled Synthetic Viral Capsids from a 24-mer Viral Peptide Fragment. Angew. Chem. Int. Ed. 2010, 49, 9662–9665. [Google Scholar] [CrossRef]
- Matsuurua, K. Rational design of self-assembled proteins and peptides for nano and micro-sized architectures. RSC Adv. 2014, 4, 2942–2953. [Google Scholar] [CrossRef]
- Matsuura, K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem. Commun. 2018, 54, 8944–8959. [Google Scholar] [CrossRef]
- Matsuura, K.; Watanabe, K.; Matsushita, Y.; Kimizuka, N. Guest-binding behavior of peptide nanocapsules self-assembled from viral peptide fragments. Polym. J. 2013, 45, 529–534. [Google Scholar] [CrossRef]
- Fujita, S.; Matsuura, K. Inclusion of zinc oxide nanoparticles into virus-like peptide nanocapsules self-assembled from viral β-annulus peptide. Nanomaterials 2014, 4, 778–791. [Google Scholar] [CrossRef]
- Fujita, S.; Matsuura, K. Encapsulation of CdTe Quantum Dots into Synthetic Viral Capsids. Chem. Lett. 2016, 45, 922–924. [Google Scholar] [CrossRef]
- Matsuura, K.; Nakamura, T.; Watanabe, K.; Noguchi, T.; Minamihata, K.; Kamiya, N.; Kimizuka, N. Self-assembly of Ni-NTA-modified β-annulus peptides into artificial viral capsids and encapsulation of His-tagged proteins. Org. Biomol. Chem. 2016, 14, 7869–7874. [Google Scholar] [CrossRef]
- Matsuura, K.; Ueno, G.; Fujita, S. Self-assembled artificial viral capsid decorated with gold nanoparticles. Polym. J. 2014, 47, 146–151. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yamada, S.; Nishikawa, S.; Matsuura, K. DNA-modified artificial viral capsids self-assembled from DNA-conjugated β-annulus peptide. J. Pept. Sci. 2017, 23, 636–643. [Google Scholar] [CrossRef]
- Matsuura, K.; Matsuura, K. Self-assembled artificial viral capsids bearing coiled-coils at the surface. Org. Biomol. Chem. 2017, 15, 5070–5077. [Google Scholar] [CrossRef]
- Matsuura, K.; Honjo, T. Artificial Viral Capsid Dressed Up with Human Serum Albumin. Bioconjug. Chem. 2019, 30, 1636–1641. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Ota, J.; Fujita, S.; Shiomi, Y.; Inaba, H. Construction of Ribonuclease-Decorated Artificial Virus-like Capsid by Peptide Self-assembly. J. Org. Chem. 2019, 85, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K. Dressing up artificial viral capsids self-assembled from C-terminal-modified β-annulus peptides. Polym. J. 2020, 52, 1035–1041. [Google Scholar] [CrossRef]
- Nakamura, Y.; Inaba, H.; Matsuura, K. Construction of Artificial Viral Capsids Encapsulating Short DNAs via Disulfide Bonds and Controlled Release of DNAs by Reduction. Chem. Lett. 2019, 48, 544–546. [Google Scholar] [CrossRef]
- Iwasaki, T.; Tokuda, Y.; Kotake, A.; Okada, H.; Takeda, S.; Kawano, T.; Nakayama, Y. Cellular uptake and In Vivo distribution of polyhistidine peptides. J. Control. Release 2015, 210, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Shinagawa, M.; Kawano, T.; Iwasaki, T. Drug delivery using polyhistidine peptide-modified liposomes that target endogenous lysosome. Biochem. Biophys. Res. Commun. 2018, 501, 648–653. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamura, Y.; Sato, Y.; Inaba, H.; Iwasaki, T.; Matsuura, K. Encapsulation of mRNA into Artificial Viral Capsids via Hybridization of a β-Annulus-dT20 Conjugate and the Poly(A) Tail of mRNA. Appl. Sci. 2020, 10, 8004. https://doi.org/10.3390/app10228004
Nakamura Y, Sato Y, Inaba H, Iwasaki T, Matsuura K. Encapsulation of mRNA into Artificial Viral Capsids via Hybridization of a β-Annulus-dT20 Conjugate and the Poly(A) Tail of mRNA. Applied Sciences. 2020; 10(22):8004. https://doi.org/10.3390/app10228004
Chicago/Turabian StyleNakamura, Yoko, Yuki Sato, Hiroshi Inaba, Takashi Iwasaki, and Kazunori Matsuura. 2020. "Encapsulation of mRNA into Artificial Viral Capsids via Hybridization of a β-Annulus-dT20 Conjugate and the Poly(A) Tail of mRNA" Applied Sciences 10, no. 22: 8004. https://doi.org/10.3390/app10228004
APA StyleNakamura, Y., Sato, Y., Inaba, H., Iwasaki, T., & Matsuura, K. (2020). Encapsulation of mRNA into Artificial Viral Capsids via Hybridization of a β-Annulus-dT20 Conjugate and the Poly(A) Tail of mRNA. Applied Sciences, 10(22), 8004. https://doi.org/10.3390/app10228004





