CIMI: Classify and Itemize Medical Image System for PFT Big Data Based on Deep Learning
Abstract
1. Introduction
2. Related Work
2.1. Related Technology
2.2. Related Research
3. System Architecture
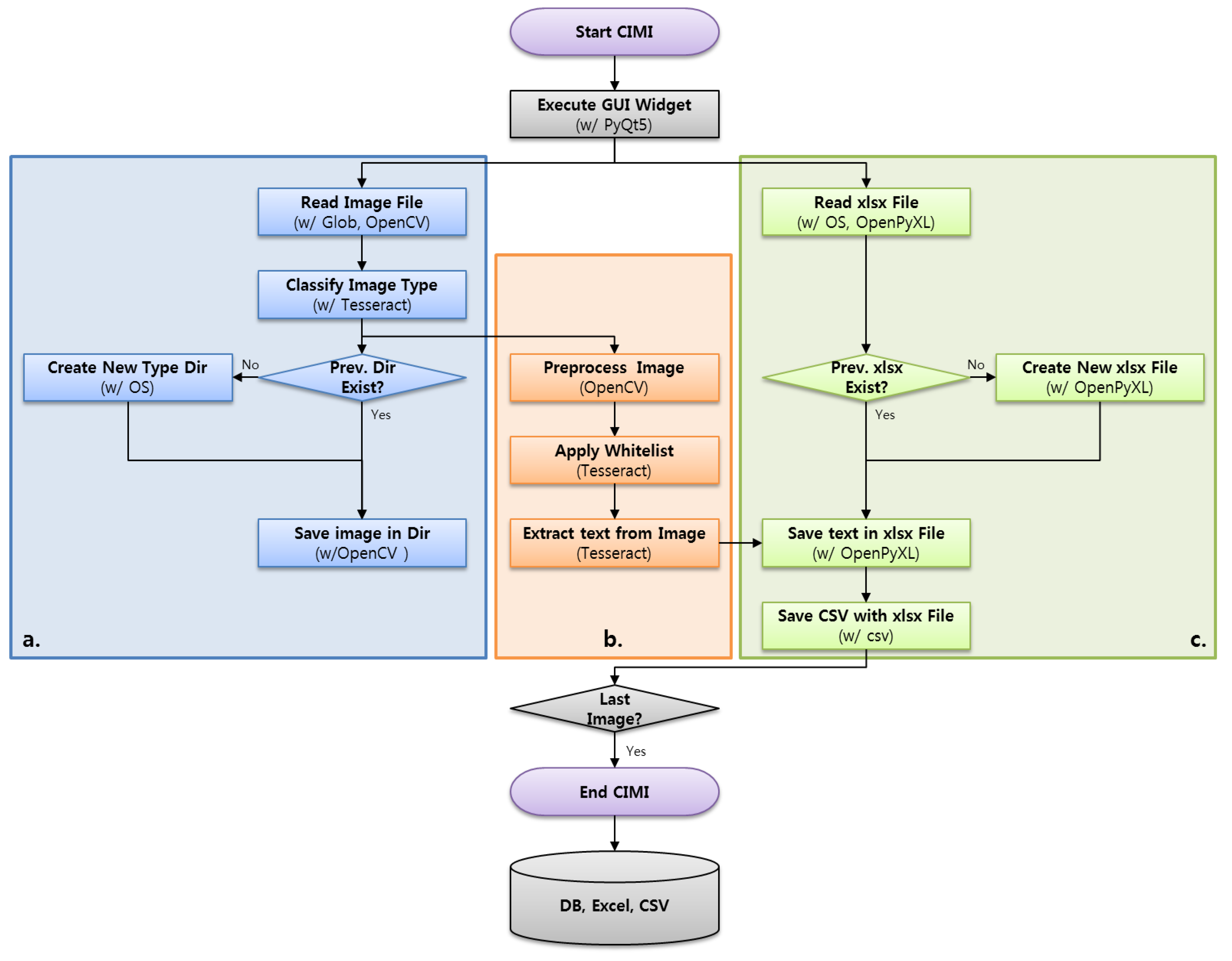
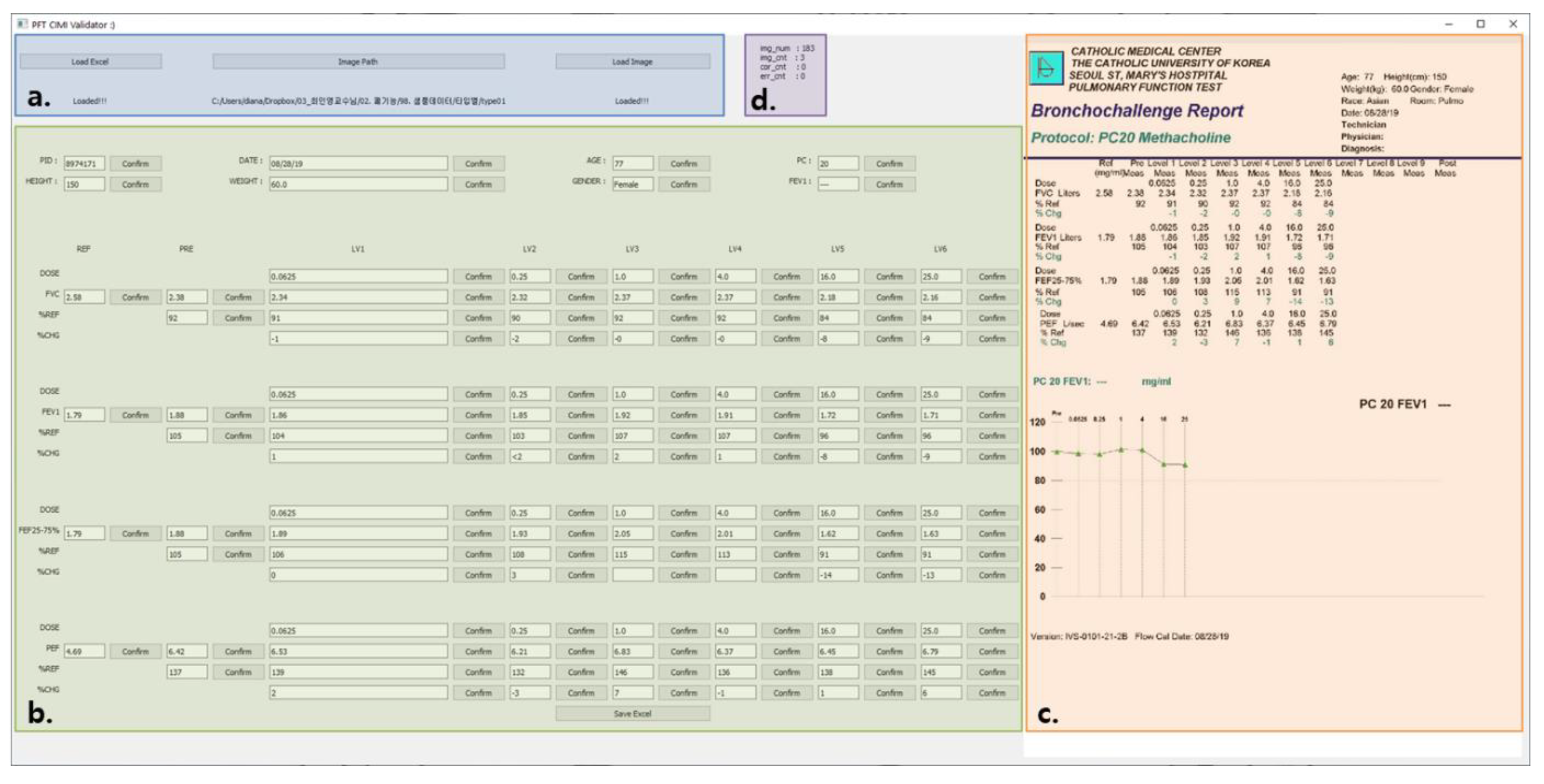
3.1. Overall System
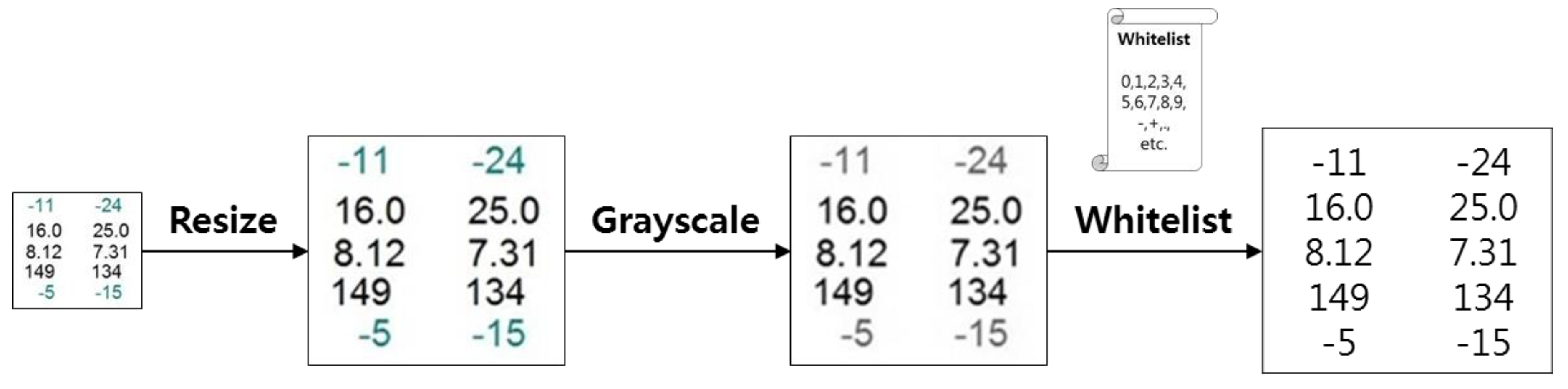
3.2. Major Function Specifications
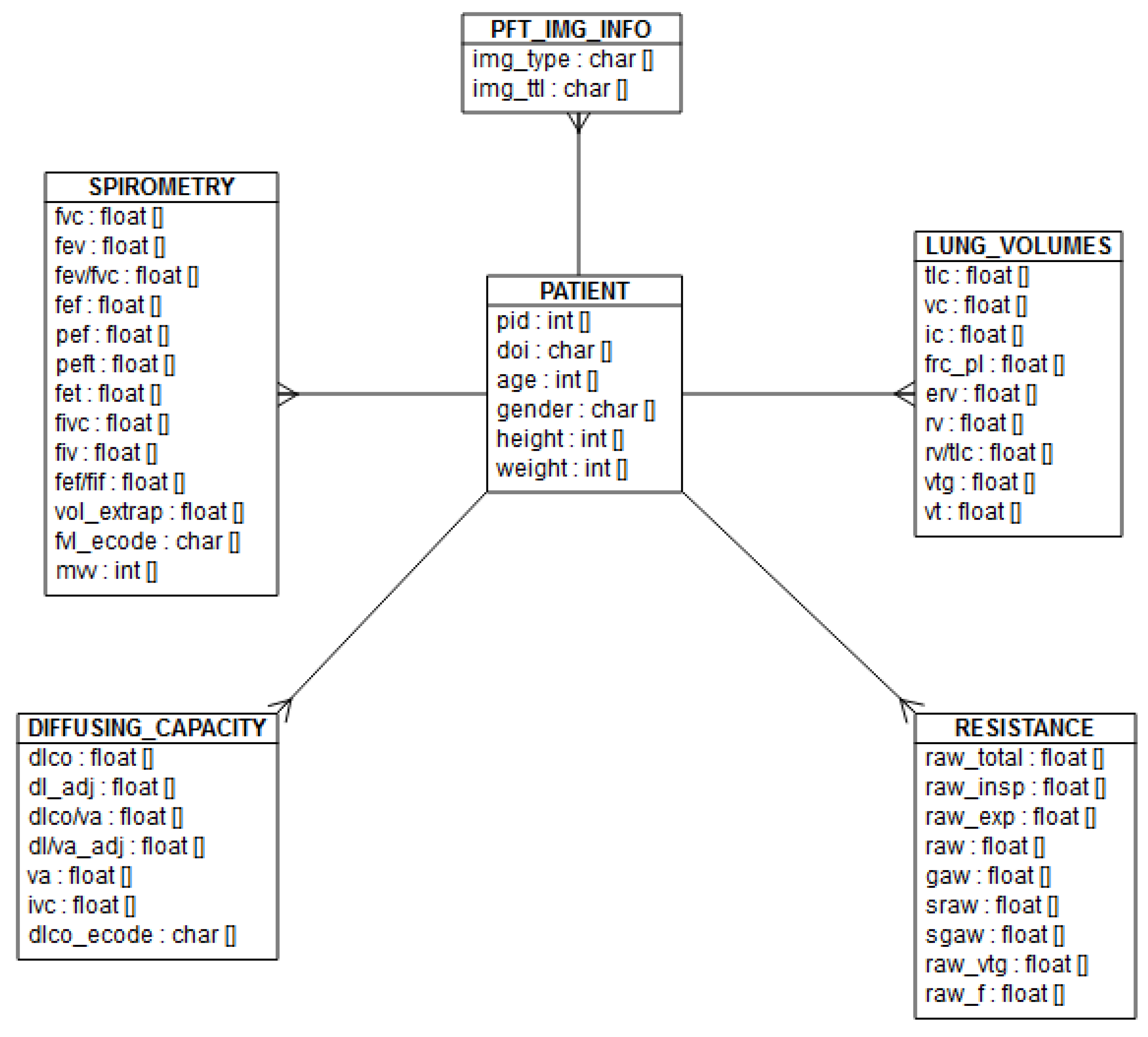
3.3. Database Architecture
4. Evaluation
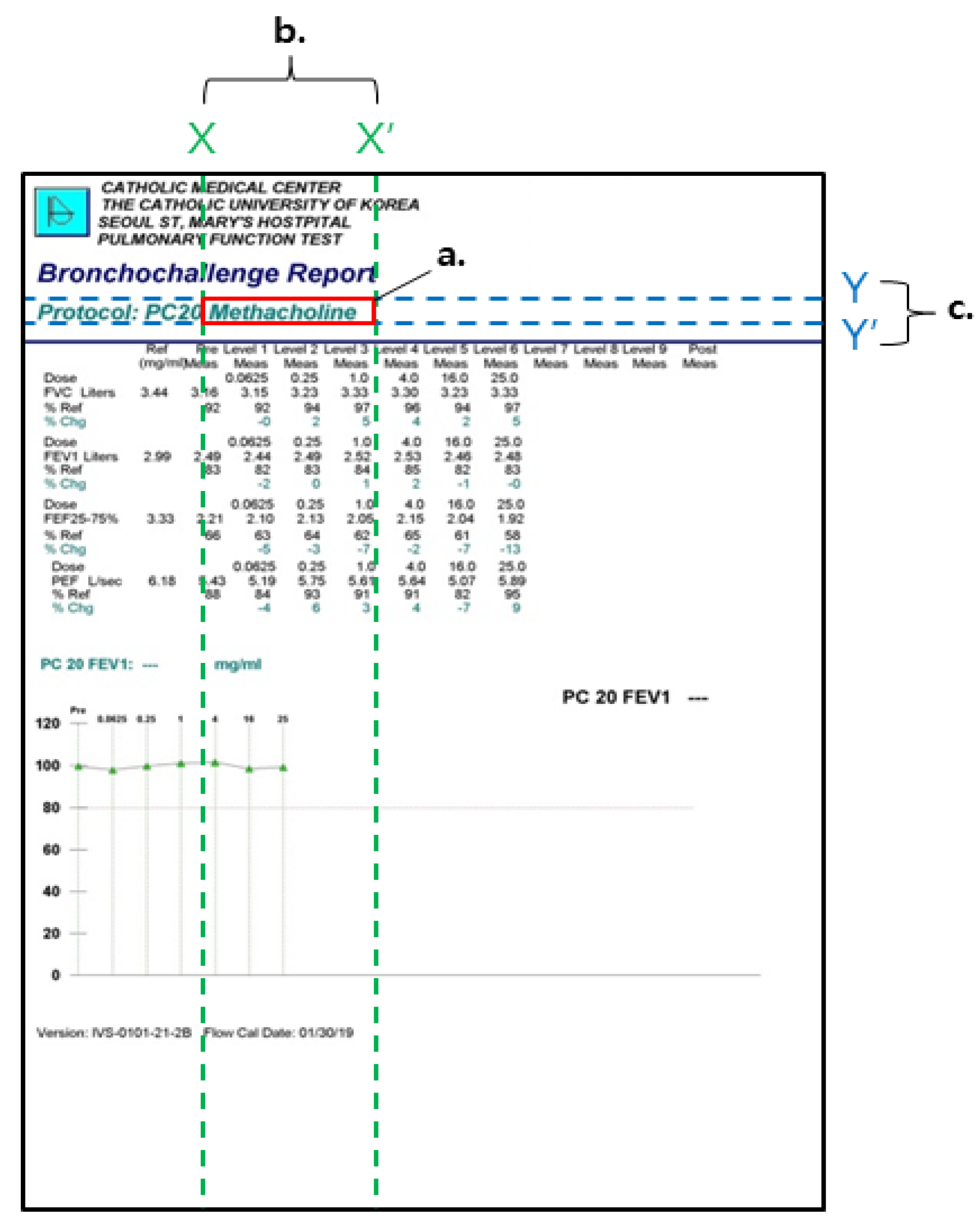
4.1. Materials and Methods
4.2. Performance Evaluation
4.2.1. Type Classification Results
4.2.2. Accuracy Performance Results
5. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gibson, G.J.; Loddenkemper, R.; Lundbäck, B.; Sibille, Y. Respiratory health and disease in Europe: The new European Lung White Book. Eur. Respir. Soc. 2013, 42, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Bowen, T.S.; Aakerøy, L.; Eisenkolb, S.; Kunth, P.; Bakkerud, F.; Wohlwend, M.; Ormbostad, A.M.; Fischer, T.; Wisloff, U.; Schuler, G.; et al. Exercise Training Reverses Extrapulmonary Impairments in Smoke-exposed Mice. Med. Sci. Sports Exerc. 2017, 49, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Hadi, A.G.; Kadhom, M.; Hairunisa, N.; Yousif, E.; Mohammed, S.A. A Review on COVID-19: Origin, Spread, Symptoms, Treatment, and Prevention. Biointerface Res. Appl. Chem. 2020, 10, 7234–7242. [Google Scholar]
- World Health Organization. The Global Impact of Respiratory Disease, 2nd ed.; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- World Health Organization. Global Status Report on Noncommunicable Diseases 2014; no. WHO/NMH/NVI/15.1; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Galode, F.; Dournes, G.; Chateil, J.-F.; Fayon, M.; Collet, C.; Bui, S. Impact at school age of early chronic methicillin-sensitiveStaphylococcus aureusinfection in children with cystic fibrosis. Pediatr. Pulmonol. 2020, 55, 2641–2645. [Google Scholar] [CrossRef]
- Wang, C.; Qi, Y.; Zhu, G. Deep learning for predicting the occurrence of cardiopulmonary diseases in Nanjing, China. Chemosphere 2020, 257, 127176. [Google Scholar] [CrossRef]
- Vergeer, M. Artificial Intelligence in the Dutch Press: An Analysis of Topics and Trends. Commun. Stud. 2020, 71, 1–20. [Google Scholar] [CrossRef]
- Zhavoronkov, A.; Vanhaelen, Q.; Oprea, T.I. Will Artificial Intelligence for Drug Discovery Impact Clinical Pharmacology? Clin. Pharmacol. Ther. 2020, 107, 780–785. [Google Scholar] [CrossRef]
- Ahmed, Z.; Mohamed, K.; Zeeshan, S.; Dong, X. Artificial intelligence with multi-functional machine learning platform development for better healthcare and precision medicine. Database J. Biol. Databases Curation 2020, 2020. [Google Scholar] [CrossRef]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing Medical Imaging Data for Machine Learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef]
- Tappert, C.C.; Suen, C.Y.; Wakahara, T. The state of the art in online handwriting recognition. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 787–808. [Google Scholar] [CrossRef]
- Zanibbi, R.; Blostein, D. Recognition and retrieval of mathematical expressions. Int. J. Doc. Anal. Recognit. 2012, 15, 331–357. [Google Scholar] [CrossRef]
- Thompson, P.; Batista-Navarro, R.T.; Kontonatsios, G.; Carter, J.; Toon, E.; McNaught, J.; Timmermann, C.; Worboys, M.; Ananiadou, S. Text Mining the History of Medicine. PLoS ONE 2016, 11, e0144717. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ashley, K.D.; Bridewell, W. Emerging AI & Law approaches to automating analysis and retrieval of electronically stored information in discovery proceedings. Artif. Intell. Law 2010, 18, 311–320. [Google Scholar]
- Memon, J.; Sami, M.; Khan, R.A.; Uddin, M. Handwritten optical character recognition (OCR): A comprehensive systematic literature review (SLR). IEEE Access 2020, 8, 142642–142668. [Google Scholar] [CrossRef]
- Smith, R. An overview of the Tesseract OCR engine. In Proceedings of the Ninth International Conference on Document Analysis and Recognition (ICDAR 2007), Curitiba, Parana, 23–26 September 2007; Volume 2, pp. 629–633. [Google Scholar]
- Socher, R.; Lin, C.C.-Y.; Ng, A.Y.; Manning, C.D. Parsing natural scenes and natural language with recursive neural networks. In Proceedings of the ICML—28th International Conference on Machine Learning, Bellevue, WA, USA, 28 June–2 July 2011. [Google Scholar]
- Fabbri, M.; Moro, G. Dow Jones Trading with Deep Learning: The Unreasonable Effectiveness of Recurrent Neural Networks. In Proceedings of the 7th International Conference on Data Science, Technologies and Applications (DATA 2018), Porto, Portugal, 26–28 July 2018; pp. 142–153. [Google Scholar]
- Bengio, Y.; Simard, P.; Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Netw. 1994, 5, 157–166. [Google Scholar] [CrossRef]
- Olah, C. Understanding lstm Networks. 2015. Available online: http//colah.github.io/posts/2015-08-Understanding-LSTMs (accessed on 30 November 2020).
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Sundermeyer, M.; Schlüter, R.; Ney, H. LSTM neural networks for language modeling. In Proceedings of the Thirteenth Annual Conference of the International Speech Communication Association, Portland, OR, USA, 9–13 September 2012. [Google Scholar]
- Gers, F.A.; Schmidhuber, J.; Cummins, F. Learning to forget: Continual prediction with LSTM. In Proceedings of the 9th International Conference on Artificial Neural Networks—ICANN ’991999, Edinburgh, UK, 7–10 September 1999. [Google Scholar]
- Gers, F.A.; Schraudolph, N.N.; Schmidhuber, J. Learning precise timing with LSTM recurrent networks. J. Mach. Learn. Res. 2002, 3, 115–143. [Google Scholar]
- Culjak, I.; Abram, D.; Pribanic, T.; Dzapo, H.; Cifrek, M. A brief introduction to OpenCV. In Proceedings of the 35th International Convention MIPRO, Opatija, Croatia, 21–25 May 2012; pp. 1725–1730. [Google Scholar]
- Laique, S.N.; Hayat, U.; Sarvepalli, S.; Vaughn, B.; Ibrahim, M.; McMichael, J.; Qaiser, K.N.; Burke, C.; Bhatt, A.; Rhodes, C.; et al. Application of optical character recognition with natural language processing for large-scale quality metric data extraction in colonoscopy reports. Gastrointest. Endosc. 2020. [Google Scholar] [CrossRef]
- Park, M.Y.; Park, R.W. Construction of an PFT database with various clinical information using optical character recognition and regular expression technique. J. Internet Comput. Serv. 2017, 18, 55–60. [Google Scholar]
- Hinchcliff, M.; Just, E.; Podlusky, S.; Varga, J.; Chang, R.W.; Kibbe, W.A. Text data extraction for a prospective, research-focused data mart: Implementation and validation. BMC Med. Inform. Decis. Mak. 2012, 12, 1–7. [Google Scholar] [CrossRef]
- González, D.R.; Carpenter, T.; van Hemert, J.I.; Wardlaw, J. An open source toolkit for medical imaging de-identification. Eur. Radiol. 2010, 20, 1896–1904. [Google Scholar] [CrossRef] [PubMed]
- Rybalkin, V.; Wehn, N.; Yousefi, M.R.; Stricker, D. Hardware architecture of bidirectional long short-term memory neural network for optical character recognition. In Proceedings of the Design, Automation & Test in Europe Conference & Exhibition, Lausanne, Switzerland, 27–31 March 2017; pp. 1390–1395. [Google Scholar]
- Koistinen, M.; Kettunen, K.; Kervinen, J. How to Improve Optical Character Recognition of Historical Finnish Newspapers Using Open Source Tesseract OCR Engine. In Proceedings of the 8th Language and Technology Conference LTC, Poznan, Poland, 17–19 November 2017; pp. 279–283. [Google Scholar]


| Type | 01 | 02 | 03 | 04 | 05 | 06 | 07 | Unknown | Total |
|---|---|---|---|---|---|---|---|---|---|
| PFT Sheets | 213 | 12 | 20 | 5728 | 1956 | 697 | 139 | 5955 | 14,720 |
| Texts in Each Sheet | 116 | 164 | 21 | 130 | 37 | 73 | 130 | 0 | 671 |
| Extracted Texts | 24,708 | 1968 | 420 | 744,640 | 72,372 | 50,881 | 18,070 | 0 | 913,059 |
| Type01 (texts = 116) | Type02 (texts = 164) | Type03 (texts = 21) | Type04 (texts = 130) | Type05 (texts = 37) | Type06 (texts = 73) | Type07 (texts = 130) | Average Accuracy | ||
|---|---|---|---|---|---|---|---|---|---|
| CIMI | Correct (Average) | 100.3 | 142.9 | 20.7 | 127.4 | 36.9 | 72.2 | 128.1 | |
| Omitted (Average) | 9.7 | 19.0 | 0.1 | 1.2 | 0.0 | 0.0 | 0.1 | ||
| Error (Average) | 6.0 | 2.3 | 0.2 | 1.4 | 0.1 | 0.8 | 1.7 | ||
| Accuracy (%) | 86.5 | 87.1 | 98.6 | 98.0 | 99.7 | 98.9 | 98.5 | 95.3 | |
| Vanilla | Correct (Average) | 83.3 | 122.5 | 10.8 | 106.4 | 34.0 | 59.8 | 76.3 | |
| Omitted (Average) | 14.6 | 25.8 | 9.5 | 13.5 | 0.0 | 8.9 | 38.4 | ||
| Error (Average) | 18.7 | 15.3 | 0.7 | 10.1 | 3.0 | 4.3 | 15.2 | ||
| Accuracy (%) | 71.8 | 74.7 | 51.4 | 81.8 | 91.9 | 81.9 | 58.7 | 73.2 | |
| Resize Pre-processed | Correct (Average) | 98.4 | 142.5 | 19.7 | 126.2 | 36.2 | 64.0 | 101.1 | |
| Omitted (Average) | 7.8 | 18.4 | 1.0 | 1.8 | 0.5 | 9.0 | 16.8 | ||
| Error (Average) | 10.4 | 2.7 | 0.3 | 2.0 | 0.3 | 0.0 | 12.0 | ||
| Accuracy (%) | 84.8 | 86.9 | 93.8 | 97.1 | 97.8 | 87.7 | 77.8 | 89.4 | |
| Grayscale Pre-Processed | Correct (Average) | 82.8 | 122.2 | 10.8 | 106.4 | 34.1 | 59.9 | 77.7 | |
| Omitted (Average) | 14.0 | 25.2 | 9.5 | 13.5 | 0.0 | 8.9 | 37.1 | ||
| Error (Average) | 19.2 | 16.2 | 0.7 | 10.1 | 2.9 | 4.2 | 15.1 | ||
| Accuracy (%) | 71.4 | 74.5 | 51.4 | 81.8 | 92.2 | 82.1 | 59.8 | 73.3 | |
| Whitelist Applied | Correct (Average) | 93.1 | 145 | 19.8 | 117.9 | 34.8 | 65.5 | 111.0 | |
| Omitted (Average) | 4.0 | 0.9 | 0.1 | 3.4 | 0.0 | 0.8 | 1.9 | ||
| Error (Average) | 19.5 | 17.7 | 1.1 | 8.7 | 2.2 | 6.7 | 17.0 | ||
| Accuracy (%) | 80.3 | 88.4 | 94.3 | 90.7 | 94.1 | 89.7 | 85.4 | 89.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.M.; Lee, S.-J.; Lee, H.Y.; Chang, D.-J.; Yoon, C.I.; Choi, I.-Y.; Yoon, K.-H. CIMI: Classify and Itemize Medical Image System for PFT Big Data Based on Deep Learning. Appl. Sci. 2020, 10, 8575. https://doi.org/10.3390/app10238575
Kim TM, Lee S-J, Lee HY, Chang D-J, Yoon CI, Choi I-Y, Yoon K-H. CIMI: Classify and Itemize Medical Image System for PFT Big Data Based on Deep Learning. Applied Sciences. 2020; 10(23):8575. https://doi.org/10.3390/app10238575
Chicago/Turabian StyleKim, Tong Min, Seo-Joon Lee, Hwa Young Lee, Dong-Jin Chang, Chang Ii Yoon, In-Young Choi, and Kun-Ho Yoon. 2020. "CIMI: Classify and Itemize Medical Image System for PFT Big Data Based on Deep Learning" Applied Sciences 10, no. 23: 8575. https://doi.org/10.3390/app10238575
APA StyleKim, T. M., Lee, S.-J., Lee, H. Y., Chang, D.-J., Yoon, C. I., Choi, I.-Y., & Yoon, K.-H. (2020). CIMI: Classify and Itemize Medical Image System for PFT Big Data Based on Deep Learning. Applied Sciences, 10(23), 8575. https://doi.org/10.3390/app10238575





