Tree2C: A Flexible Tool for Enabling Model Deployment with Special Focus on Cheminformatics Applications
Abstract
:1. Introduction
2. Materials and Methods
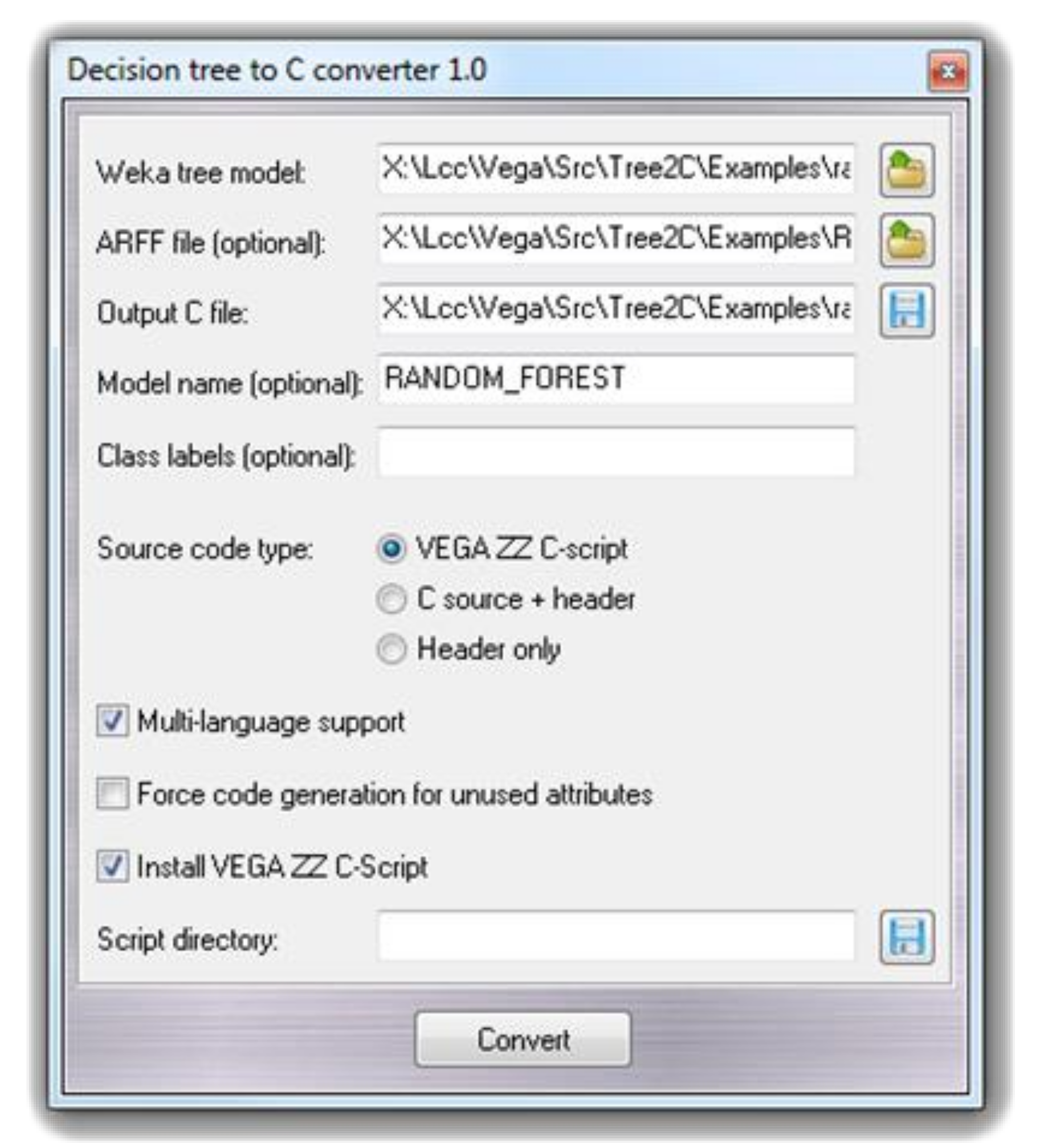
2.1. The Tree2C Implementation
2.1.1. Tree2C: The Input Model
2.1.2. Tree2C: How It Works
2.1.3. Tree2C: The Output Source Code
2.2. Computational Details
3. Results
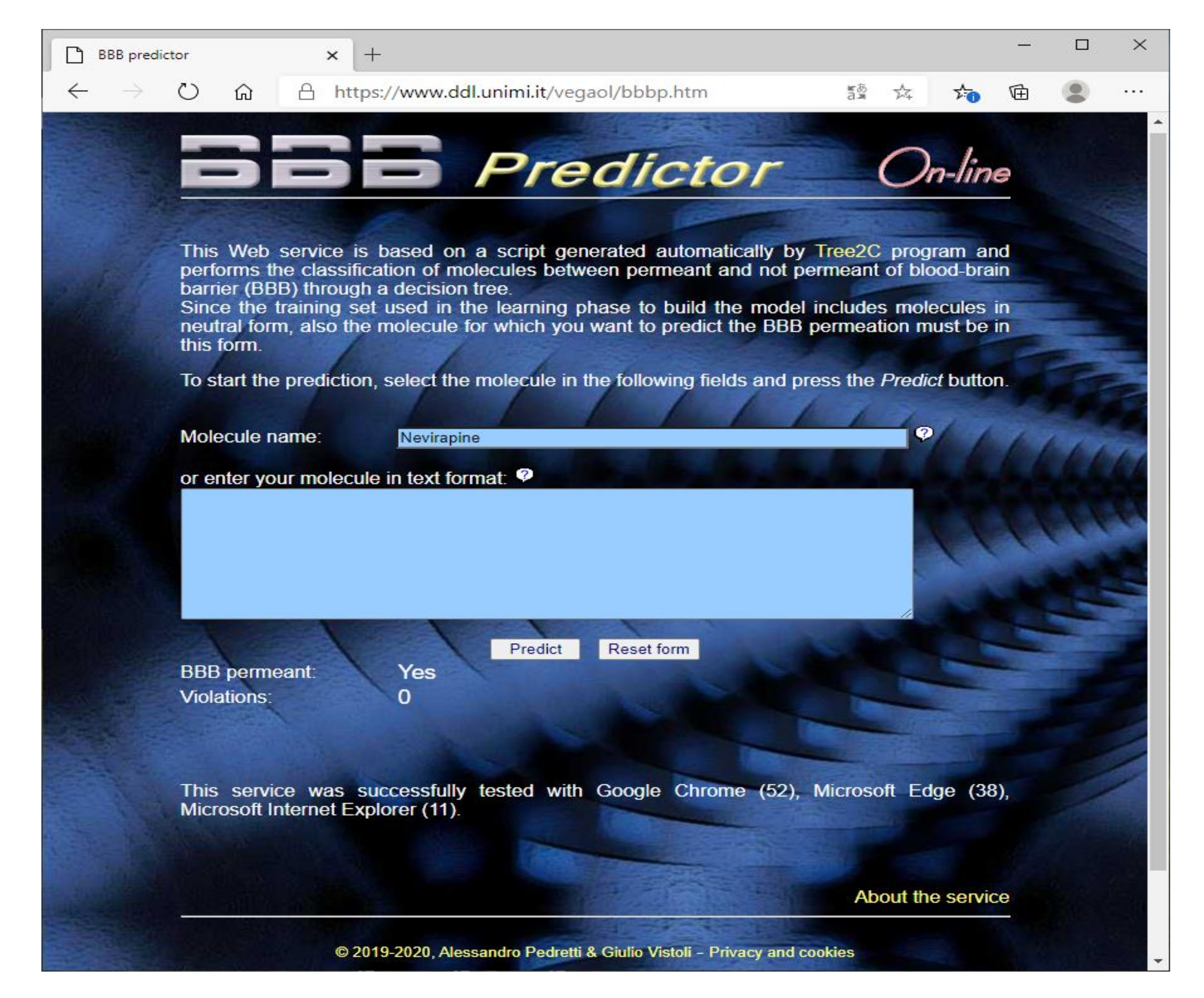
3.1. Prediction of the BBB Permeation
3.2. Prediction of Mutagenicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Abduljabbar, R.; Dia, H.; Liyanage, S.; Bagloee, S.A. Applications of Artificial Intelligence in Transport: An Overview. Sustainability 2019, 11, 189. [Google Scholar]
- Nemitz, P. Constitutional democracy and technology in the age of artificial intelligence. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2018, 376, 20180089. [Google Scholar]
- Smith, T.C.; Frank, E. Introducing Machine Learning Concepts with WEKA. Methods Mol. Biol. 2016, 1418, 353–378. [Google Scholar]
- Rampasek, L.; Goldenberg, A. TensorFlow: Biology’s Gateway to Deep Learning? Cell Syst. 2016, 27, 12–14. [Google Scholar]
- Schneider, P.; Walters, W.P.; Plowright, A.T.; Sieroka, N.; Listgarten, J.; Goodnow, R.A., Jr.; Fisher, J.; Jansen, J.M.; Duca, J.S.; Rush, T.S.; et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2020, 19, 353–364. [Google Scholar]
- Yang, X.; Wang, Y.; Byrne, R.; Schneider, G.; Yang, S. Concepts of Artificial Intelligence for Computer-Assisted Drug Discovery. Chem. Rev. 2019, 119, 10520–10594. [Google Scholar]
- Mazzolari, A.; Afzal, A.M.; Pedretti, A.; Testa, B.; Vistoli, G.; Bender, A. Prediction of UGT-mediated Metabolism Using the Manually Curated MetaQSAR Database. ACS Med. Chem. Lett. 2019, 10, 633–638. [Google Scholar]
- Šícho, M.; Stork, C.; Mazzolari, A.; de Bruyn Kops, C.; Pedretti, A.; Testa, B.; Vistoli, G.; Svozil, D.; Kirchmair, J. FAME 3: Predicting the Sites of Metabolism in Synthetic Compounds and Natural Products for Phase 1 and Phase 2 Metabolic Enzymes. J. Chem. Inf. Model. 2019, 59, 3400–3412. [Google Scholar]
- Coiera, E. The Last Mile: Where Artificial Intelligence Meets Reality. J. Med. Internet Res. 2019, 21, e16323. [Google Scholar]
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA—An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Aided Mol. Des. 2004, 18, 167–173. [Google Scholar]
- Li, H.; Yap, C.W.; Ung, C.Y.; Xue, Y.; Cao, Z.W.; Chen, Y.Z. Effect of selection of molecular descriptors on the prediction of blood-brain barrier penetrating and nonpenetrating agents by statistical learning methods. J. Chem. Inf. Model. 2005, 45, 1376–1384. [Google Scholar]
- Kazius, J.; McGuire, R.; Bursi, R. Derivation and validation of toxicophores for mutagenicity prediction. J. Med. Chem. 2005, 48, 312–320. [Google Scholar]
- Morales, J.F.; Montoto, S.S.; Fagiolino, P.; Ruiz, M.E. Current State and Future Perspectives in QSAR Models to Predict Blood-Brain Barrier Penetration in Central Nervous System Drug R&D. Mini Rev. Med. Chem. 2017, 17, 247–257. [Google Scholar]
- Saxena, D.; Sharma, A.; Siddiqui, M.H.; Kumar, R. Blood Brain Barrier Permeability Prediction Using Machine Learning Techniques: An Update. Curr. Pharm. Biotechnol. 2019, 20, 1163–1171. [Google Scholar]
- Gupta, M.; Lee, H.J.; Barden, C.J.; Weaver, D.F. The Blood-Brain Barrier (BBB) Score. J. Med. Chem. 2019, 62, 9824–9836. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar]
- Wu, F.; Zhou, Y.; Li, L.; Shen, X.; Chen, G.; Wang, X.; Liang, X.; Tan, M.; Huang, Z. Computational Approaches in Preclinical Studies on Drug Discovery and Development. Front. Chem. 2020, 8, 726. [Google Scholar]
- Rim, K.T. In silico prediction of toxicity and its applications for chemicals at work. Toxicol. Environ. Health Sci. 2020, 12, 191–202. [Google Scholar]
- Honma, M. An assessment of mutagenicity of chemical substances by (quantitative) structure-activity relationship. Genes Environ. 2020, 42, 23. [Google Scholar]
- Benigni, R.; Bossa, C. Data-based review of QSARs for predicting genotoxicity: The state of the art. Mutagenesis 2019, 34, 17–23. [Google Scholar]
- Frank, E.; Hall, M.A.; Witten, I.H. The WEKA Workbench. Online Appendix for Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann: Burlington, MA, USA, 2016; Available online: https://www.cs.waikato.ac.nz/ml/weka/Witten_et_al_2016_appendix.pdf (accessed on 30 October 2020).
- ARFF Format. Available online: https://waikato.github.io/weka-wiki/formats_and_processing/arff/ (accessed on 30 October 2020).
- James, J.P. MOPAC2016, Stewart, Stewart Computational Chemistry, Colorado Springs, CO, USA. Available online: http://OpenMOPAC.net (accessed on 30 October 2020).
- Tree2C. Classification Tree to Code Converter. Available online: https://www.ddl.unimi.it/manual/utilities/tree2c.htm (accessed on 30 October 2020).
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA: A versatile program to convert, handle and visualize molecular structure on Windows-based PCs. J. Mol. Graph. Model. 2002, 21, 47–49. [Google Scholar]
- Stewart, J.J. Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 2013, 19, 1–32. [Google Scholar]
- Gaillard, P.; Carrupt, P.A.; Testa, B.; Boudon, A. Molecular lipophilicity potential, a tool in 3D QSAR: Method and applications. J. Comput. Aided Mol. Des. 1994, 8, 83–96. [Google Scholar]
- Hall, L.H.; Mohney, B.; Kier, L.B. The electrotopological state: Structure information at the atomic level for molecular graphs. J. Chem. Inf. Comput. Sci. 1991, 31, 76–82. [Google Scholar]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar]
- Parr, R.G.; Chattaraj, P.K. Principle of maximum hardness. J. Am. Chem. Soc. 1991, 113, 1854–1855. [Google Scholar]
| Class | TP Rate | FP Rate | Precision | F-measure | PRC Area |
|---|---|---|---|---|---|
| 0 | 0.705 | 0.101 | 0.778 | 0.740 | 0.787 |
| 1 | 0.899 | 0.295 | 0.858 | 0.878 | 0.912 |
| mean | 0.834 | 0.230 | 0.831 | 0.832 | 0.870 |
| Class | TP Rate | FP Rate | Precision | F-measure | PRC Area |
|---|---|---|---|---|---|
| 0 | 0.798 | 0.155 | 0.806 | 0.802 | 0.867 |
| 1 | 0.845 | 0.202 | 0.838 | 0.842 | 0.910 |
| mean | 0.824 | 0.181 | 0.824 | 0.824 | 0.891 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedretti, A.; Mazzolari, A.; Gervasoni, S.; Vistoli, G. Tree2C: A Flexible Tool for Enabling Model Deployment with Special Focus on Cheminformatics Applications. Appl. Sci. 2020, 10, 7704. https://doi.org/10.3390/app10217704
Pedretti A, Mazzolari A, Gervasoni S, Vistoli G. Tree2C: A Flexible Tool for Enabling Model Deployment with Special Focus on Cheminformatics Applications. Applied Sciences. 2020; 10(21):7704. https://doi.org/10.3390/app10217704
Chicago/Turabian StylePedretti, Alessandro, Angelica Mazzolari, Silvia Gervasoni, and Giulio Vistoli. 2020. "Tree2C: A Flexible Tool for Enabling Model Deployment with Special Focus on Cheminformatics Applications" Applied Sciences 10, no. 21: 7704. https://doi.org/10.3390/app10217704
APA StylePedretti, A., Mazzolari, A., Gervasoni, S., & Vistoli, G. (2020). Tree2C: A Flexible Tool for Enabling Model Deployment with Special Focus on Cheminformatics Applications. Applied Sciences, 10(21), 7704. https://doi.org/10.3390/app10217704








