Marine Actinobacteria Bioflocculant: A Storehouse of Unique Biotechnological Resources for Wastewater Treatment and Other Applications
Abstract
1. Introduction
2. The Phylum Actinobacteria
3. Bioactive Compounds from Marine Actinomycetes
3.1. Isolation and Maintenance of Cultivable Actinobacteria for Bioflocculant Production
3.1.1. Plackett–Burman (PB) Design for the Screening of Bioflocculant Production
3.1.2. Bioflocculation Process Optimization Using Central Composite Design (CCD)
4. Factors Affecting Bioflocculant Production
4.1. Effect of Inoculum Size
4.2. Effect of Cations
4.3. Effect of pH
4.4. Effect of Carbon and Nitrogen Sources
5. Characterization of Purified Bioflocculants
6. Applications of Actinobacteria in Biotechnology
6.1. Antimicrobials
6.2. Enzymes
6.3. Biofuels
6.4. Synthesis of Nanoparticles
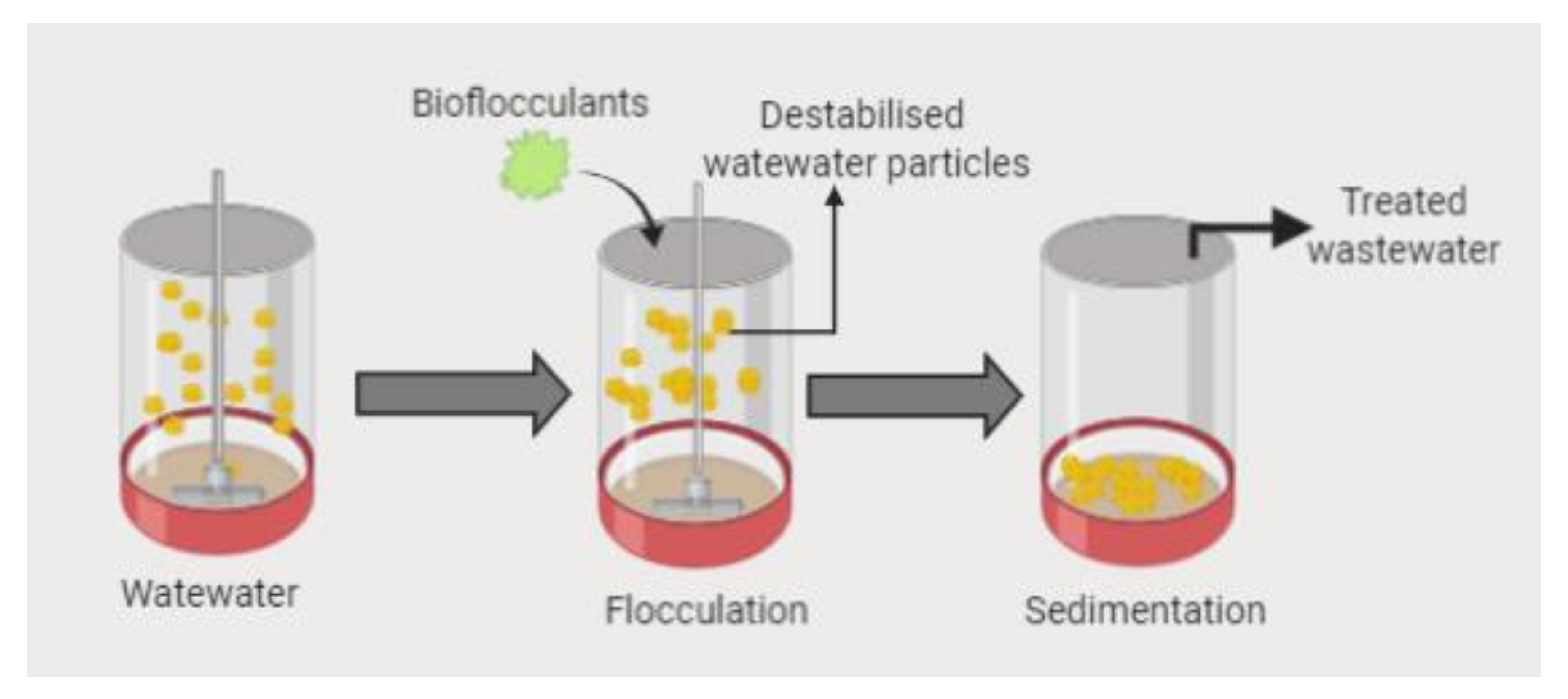
6.5. Bioremediation and Bioflocculation
7. Conclusions and Recommendations
Author Contributions
Funding
Conflicts of Interest
References
- Passari, A.K.; Chandra, P.; Mishra, V.K.; Leo, V.V.; Gupta, V.K.; Kumar, B.; Singh, B.P. Detection of biosynthetic gene and phytohormone production by endophytic actinobacteria associated with Solanum lycopersicum and their plant-growth-promoting effect. Res. Microbiol. 2016, 167, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Passari, A.K.; Chandra, P.; Leo, V.V.; Mishra, V.K.; Kumar, B.; Singh, B.P. Production of Potent Antimicrobial Compounds from Streptomyces cyaneofuscatus Associated with Fresh Water Sediment. Front. Microbiol. 2017, 8, 68. [Google Scholar] [CrossRef]
- Chaudhary, H.S.; Soni, B.; Shrivastava, A.R.; Shrivastava, S. Diversity and Versatility of Actinomycetes and its Role in Antibiotic Production. J. Appl. Pharm. Sci. 2013, 3, 83–94. [Google Scholar] [CrossRef]
- Yuan, M.; Yu, Y.; Li, H.R.; Dong, N.; Zhang, X.H. Phylogenetic diversity and biological activity of actinobacteria isolated from the chukchi shelf marine sediments in the arctic ocean. Mar. Drugs 2014, 12, 1281–1297. [Google Scholar] [CrossRef]
- Aljuboori, A.H.R.; Uemura, Y.; Osman, N.B.; Yusup, S. Production of a bioflocculant from Aspergillus niger using palm oil mill effluent as carbon source. Bioresour. Technol. 2014, 171, 66–70. [Google Scholar] [CrossRef]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol. Res. 2014, 169, 262–278. [Google Scholar] [CrossRef]
- Ajao, V.; Fokkink, R.; Leermakers, F.; Bruning, H.; Rijnaarts, H.; Temmink, H. Bioflocculants from wastewater: Insights into adsorption affinity, flocculation mechanisms and mixed particle flocculation based on biopolymer size-fractionation. J. Colloid Interface Sci. 2020, 581, 533–544. [Google Scholar] [CrossRef]
- Wang, T.; Tang, X.; Zhang, S.; Zheng, J.; Zheng, H.; Fang, L. Roles of functional microbial flocculant in dyeing wastewater treatment: Bridging and adsorption. J. Hazard. Mater. 2020, 384. [Google Scholar] [CrossRef]
- Wang, S.G.; Gong, W.X.; Liu, X.W.; Tian, L.; Yue, Q.Y.; Gao, B.Y. Production of a novel bioflocculant by culture of Klebsiella mobilis using dairy wastewater. Biochem. Eng. J. 2007, 36, 81–86. [Google Scholar] [CrossRef]
- Zhong, C.; Xu, A.; Chen, L.; Yang, X.; Yang, B.; Hong, W.; Mao, K.; Wang, B.; Zhou, J. Production of a bioflocculant from chromotropic acid waste water and its application in steroid estrogen removal. Colloids Surf. B Biointerfaces 2014, 122, 729–737. [Google Scholar] [CrossRef]
- Guo, J.; Chen, C.; Jiang, S.; Zhou, Y. Feasibility and Mechanism of Combined Conditioning with Coagulant and Flocculant to Enhance Sludge Dewatering. ACS Sustain. Chem. Eng. 2018, 6, 10758–10765. [Google Scholar] [CrossRef]
- Agunbiade, M.O.; Van Heerden, E.; Pohl, C.H.; Ashafa, A.T. Flocculating performance of a bioflocculant produced by Arthrobacter humicola in sewage waste water treatment. BMC Biotechnol. 2017, 17, 1. [Google Scholar] [CrossRef]
- Wan, C.; Zhao, X.Q.; Guo, S.L.; Asraful Alam, M.; Bai, F.W. Bioflocculant production from Solibacillus silvestris W01 and its application in cost-effective harvest of marine microalga Nannochloropsis oceanica by flocculation. Bioresour. Technol. 2013, 135, 207–212. [Google Scholar] [CrossRef]
- Ananda, R.; Dharumadurai, D.; Manogaran, G.P. An Introduction to Actinobacteria. In Actinobacteria-Basics and Biotechnological Applications; IntechOpen: London, UK, 2016. [Google Scholar]
- Gao, B.; Gupta, R.S. Phylogenetic Framework and Molecular Signatures for the Main Clades of the Phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 66–112. [Google Scholar] [CrossRef]
- Goodfellow, M.; Kämpfer, P.; Busse, H.; Trujillo, M.; Suzuki, K.; Ludwig, W.; Whitman, W. No Title Bergey’s manual of systematic bacteriology. In The Actinobacteria, Part A and B; Springer: New York, NY, USA, 2012; Volume 5. [Google Scholar]
- Puttaswamygowda, G.H.; Olakkaran, S.; Antony, A.; Purayil, A.K. Present status and future perspectives of marine actinobacterial metabolites. In Recent Developments in Applied Microbiology and Biochemistry; Academic Press: Cambridge, MA, USA, 2019; p. 319. [Google Scholar]
- Veena, S.; Swetha, D.; Karthik, L.; Gaurav, K.; Bhaskara Rao, K. Antibiofouling activity of marine actinobacterial mediated titanium dioxide nanoparticles. Indian J. Geo-Marine Sci. 2016, 45, 583–590. [Google Scholar]
- Shao, J.W.; Fotso, S.; Li, F.; Qin, S.; Laatsch, H. Amorphane sesquiterpenes from a marine Streptomyces sp. J. Nat. Prod. 2007, 70, 304–306. [Google Scholar] [CrossRef]
- Dharmaraj, S. Marine Streptomyces as a novel source of bioactive substances. World J. Microbiol. Biotechnol. 2010, 26, 2123–2139. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 43. [Google Scholar] [CrossRef]
- Bérdy, J. Bioactive microbial metabolites: A personal view. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Sreedharan, V.; Bhaskara Rao, K.V. Efficacy of protease inhibitor from marine Streptomyces sp. VITBVK2 against Leishmania donovani—An In Vitro study. Exp. Parasitol. 2017, 174, 45–51. [Google Scholar] [CrossRef]
- Anwesha, S.; Isha, A.; Purbasha, S.; Sathish, K.S.R.; Bhaskara Rao, K. Antioxidant, haemolytic activity of l-glutaminase producing marine actinobacteria isolated from salt pan soil of coastal Andhra Pradesh. Res. J. Pharm. Tech. 2014, 7, 544–549. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep sea actinomycetes and their secondary metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef] [PubMed]
- Riedlinger, J.; Reicke, A.; Zähner, H.; Krismer, B.; Bull, A.T.; Maldonado, L.A.; Ward, A.C.; Goodfellow, M.; Bister, B.; Bischoff, D.; et al. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot. 2004, 57, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, R.W.; Talmage, S.C.; Miller, S.A.; Sarris, K.E.; Davidson, B.S.; Goldberg, A. Isolation and structure determination of an antimicrobial ester from a marine sediment-derived bacterium. J. Nat. Prod. 2003, 66, 1291–1293. [Google Scholar] [CrossRef] [PubMed]
- Soria-Mercado, I.E.; Prieto-Davo, A.; Jensen, P.R.; Fenical, W. Antibiotic terpenoid chloro-dihydroquinones from a new marine actinomycete. J. Nat. Prod. 2005, 68, 904–910. [Google Scholar] [CrossRef]
- Charan, R.D.; Schlingmann, G.; Janso, J.; Bernan, V.; Feng, X.; Carter, G.T. Diazepinomicin, a new antimicrobial alkaloid from a marine Micromonospora sp. J. Nat. Prod. 2004, 67, 1431–1433. [Google Scholar] [CrossRef]
- Bruntner, C.; Binder, T.; Pathom-Aree, W.; Goodfellow, M.; Bull, A.T.; Potterat, O.; Puder, C.; Hörer, S.; Schmid, A.; Bolek, W.; et al. Frigocyclinone, a novel angucyclinone antibiotic produced by a Streptomyces griseus strain from Antarctica. J. Antibiot. 2005, 58, 346–349. [Google Scholar] [CrossRef]
- El-Gendy, M.M.A.; Shaaban, M.; Shaaban, K.A.; El-Bondkly, A.M.; Laatsch, H. Essramycin: A first triazolopyrimidine antibiotic isolated from nature. J. Antibiot. 2008, 61, 149–157. [Google Scholar] [CrossRef]
- McArthur, K.A.; Mitchell, S.S.; Tsueng, G.; Rheingold, A.; White, D.J.; Grodberg, J.; Lam, K.S.; Potts, B.C.M. Lynamicins A-E, chlorinated bisindole pyrrole antibiotics from a novel marine actinomycete. J. Nat. Prod. 2008, 71, 1732–1737. [Google Scholar] [CrossRef]
- Hughes, C.C.; Prieto-Davo, A.; Jensen, P.R.; Fenical, W. The marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces sp. Org. Lett. 2008, 10, 629–631. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, C.; Schneider, K.; Bruntner, C.; Irran, E.; Nicholson, G.; Bull, A.T.; Jones, A.L.; Brown, R.; Stach, J.E.M.; Goodfellow, M.; et al. Caboxamycin, a new antibiotic of the benzoxazole family produced by the deep-sea strain Streptomyces sp. NTK 937. J. Antibiot. 2009, 62, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Maskey, R.P.; Helmke, E.; Laatsch, H. Himalomycin A and B: Isolation and structure elucidation of new fridamycin type antibiotics from a marine Streptomyces isolate. J. Antibiot. 2003, 56, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Maskey, R.P.; Li, F.C.; Qin, S.; Fiebig, H.H.; Laatsch, H. Chandrananimycins A~C: Production of novel anticancer antibiotics from a marine Actinomadura sp. isolate M048 by variation of medium composition and growth conditions. J. Antibiot. 2003, 56, 622–629. [Google Scholar] [CrossRef]
- Gao, X.; Lu, Y.; Xing, Y.; Ma, Y.; Lu, J.; Bao, W.; Wang, Y.; Xi, T. A novel anticancer and antifungus phenazine derivative from a marine actinomycete BM-17. Microbiol. Res. 2012, 167, 616–622. [Google Scholar] [CrossRef]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef]
- Stritzke, K.; Schulz, S.; Laatsch, H.; Helmke, E.; Beil, W. Novel Caprolactones from a Marine Streptomycete. J. Nat. Prod. 2004, 67, 395–401. [Google Scholar] [CrossRef]
- Sánchez López, J.M.; Martinez Insua, M.; Pérez Baz, J.; Fernández Puentes, J.L.; Cañedo Hernández, L.M. New cytotoxic indolic metabolites from a marine Streptomyces. J. Nat. Prod. 2003, 66, 863–864. [Google Scholar] [CrossRef]
- Rodriguez, J.C.; Fernandez, P.I.L.; Perez, B.J.; Canedo, L.M. IB-00208, a New Cytotoxic Polycyclic Xanthone Produced by a Marine-derived Actinomadura II. Isolation, Physico-chemical Properties and Structure Determination. J. Antibiot. 2003, 3, 318–321. [Google Scholar] [CrossRef]
- Halaouli, S.; Record, E.; Casalot, L.; Hamdi, M.; Sigoillot, J.C.; Asther, M.; Lomascolo, A. Cloning and characterization of a tyrosinase gene from the white-rot fungus Pycnoporus sanguineus, and overproduction of the recombinant protein in Aspergillus niger. Appl. Microbiol. Biotechnol. 2006, 70, 580–589. [Google Scholar] [CrossRef]
- Kanoh, K.; Matsuo, Y.; Adachi, K.; Imagawa, H.; Nishizawa, M.; Shizuri, Y. Mechercharmycins A and B, cytotoxic substances from marine-derived Thermoactinomyces sp. YM3-251. J. Antibiot. 2005, 58, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.S.; Nicholson, B.; Teisan, S.; Lam, K.S.; Potts, B.C.M. Aureoverticillactam, a novel 22-atom macrocyclic lactam from the marine actinomycete Streptomyces aureoverticillatus. J. Nat. Prod. 2004, 67, 1400–1402. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.G.; Miller, E.D.; Asolkar, R.N.; Jensen, P.R.; Fenical, W. Arenicolides A-C, 26-membered ring macrolides from the marine actinomycete Salinispora arenicola. J. Org. Chem. 2007, 72, 5025–5034. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wu, S.; Zhang, R.; Wang, D.; Chen, J.; Zhao, J. Diversity and antimicrobial potential of Actinobacteria isolated from diverse marine sponges along the Beibu Gulf of the South China Sea. FEMS Microbiol. Ecol. 2019, 95, 89. [Google Scholar] [CrossRef]
- Baig, U.; Dahanukar, N.; Shintre, N.; Holkar, K.; Pund, A.; Lele, U.; Gujarathi, T.; Patel, K.; Jakati, A.; Singh, R.; et al. Phylogenetic diversity and activity screening of cultivable actinobacteria isolated 1 from marine sponges and associated environments from the western coast of India 2 3. bioRxiv 2020. [Google Scholar] [CrossRef]
- Abdelfattah, M.S.; Elmallah, M.I.Y.; Ebrahim, H.Y.; Almeer, R.S.; Eltanany, R.M.A.; Abdel Moneim, A.E. Prodigiosins from a marine sponge-associated actinomycete attenuate HCl/ethanol-induced gastric lesion via antioxidant and anti-inflammatory mechanisms. PLoS ONE 2019, 14, e0216737. [Google Scholar] [CrossRef]
- Undabarrena, A.; Ugalde, J.A.; Seeger, M.; Cámara, B. Genomic data mining of the marine actinobacteria Streptomyces sp. H-KF8 unveils insights into multi-stress related genes and metabolic pathways involved in antimicrobial synthesis. PeerJ 2017, 2017, e2912. [Google Scholar] [CrossRef]
- Myronovskyi, M.; Rosenkränzer, B.; Stierhof, M.; Petzke, L.; Seiser, T.; Luzhetskyy, A. Identification and heterologous expression of the albucidin gene cluster from the marine strain streptomyces albus subsp. Chlorinus NRRL B-24108. Microorganisms 2020, 8, 237. [Google Scholar] [CrossRef]
- Thongphrom, C.; Kim, J.H.; Bora, N.; Kim, W. Tessaracoccus arenae sp. nov., isolated from sea sand. Int. J. Syst. Evol. Microbiol. 2017, 67, 2008–2013. [Google Scholar] [CrossRef]
- He, J.; Zhen, Q.; Qiu, N.; Liu, Z.; Wang, B.; Shao, Z.; Yu, Z. Medium optimization for the production of a novel bioflocculant from Halomonas sp. V3a′ using response surface methodology. Bioresour. Technol. 2009, 100, 5922–5927. [Google Scholar] [CrossRef]
- Khaouane, L.; Si-Moussa, C.; Hanini, S.; Benkortbi, O. Optimization of culture conditions for the production of pleuromutilin from pleurotus mutilus using a hybrid method based on central composite design, neural network, and particle swarm optimization. Biotechnol. Bioprocess Eng. 2012, 17, 1048–1054. [Google Scholar] [CrossRef]
- Baumann, P.; Hubbuch, J. Downstream process development strategies for effective bioprocesses: Trends, progress, and combinatorial approaches. Eng. Life Sci. 2017, 17, 1142–1158. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhong, S.; Lei, H.Y.; Chen, R.W.; Yu, Q.; Li, H.L. Production of a novel bioflocculant by Bacillus licheniformis X14 and its application to low temperature drinking water treatment. Bioresour. Technol. 2009, 100, 3650–3656. [Google Scholar] [CrossRef] [PubMed]
- Agunbiade, M.; Pohl, C.; Ashafa, O. Bioflocculant production from Streptomyces platensis and its potential for river and waste water treatment. Braz. J. Microbiol. 2018, 49, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Wang, Y.; Yu, Y.; Li, Q.; Wang, H.; Chen, R.; He, N. Production and characterization of a novel bioflocculant from Bacillus Licheniformis. Appl. Environ. Microbiol. 2010, 76, 2778–2782. [Google Scholar] [CrossRef]
- Zulkeflee, Z.; Aris, A.Z.; Shamsuddin, Z.H.; Yusoff, M.K. Cation dependence, pH tolerance, and dosage requirement of a bioflocculant produced by bacillus spp. UPMB13: Flocculation performance optimization through kaolin assays. Sci. World J. 2012, 2012. [Google Scholar] [CrossRef]
- Zhong, C.; Sun, S.; Zhang, D.; Liu, L.; Zhou, S.; Zhou, J. Production of a bioflocculant from ramie biodegumming wastewater using a biomass-degrading strain and its application in the treatment of pulping wastewater. Chemosphere 2020, 253. [Google Scholar] [CrossRef]
- Wu, J.Y.; Ye, H.F. Characterization and flocculating properties of an extracellular biopolymer produced from a Bacillus subtilis DYU1 isolate. Process Biochem. 2007, 42, 1114–1123. [Google Scholar] [CrossRef]
- Li, X.M.; Yang, Q.; Huang, K.; Zeng, G.M.; Liao, D.X.; Liu, J.J.; Long, W.F. Screening and characterization of a bioflocculant produced by Aeromonas sp. Biomed. Environ. Sci. 2007, 20, 274–278. [Google Scholar]
- He, J.; Zou, J.; Shao, Z.; Zhang, J.; Liu, Z.; Yu, Z. Characteristics and flocculating mechanism of a novel bioflocculant HBF-3 produced by deep-sea bacterium mutant Halomonas sp. V3a’. World J. Microbiol. Biotechnol. 2010, 26, 1135–1141. [Google Scholar] [CrossRef]
- Prasertsan, P.; Dermlim, W.; Doelle, H.; Kennedy, J.F. Screening, characterization and flocculating property of carbohydrate polymer from newly isolated Enterobacter cloacae WD7. Carbohydr. Polym. 2006, 66, 289–297. [Google Scholar] [CrossRef]
- Feng, D.L.; Xu, S.H. Characterization of bioflocculant MBF3-3 produced by an isolated Bacillus sp. World J. Microbiol. Biotechnol. 2008, 24, 1627–1632. [Google Scholar] [CrossRef]
- Guo, J.; Yang, C.; Peng, L. Preparation and characteristics of bacterial polymer using pre-treated sludge from swine wastewater treatment plant. Bioresour. Technol. 2014, 152, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Qian, H.; Li, X.; Cheng, Y.; He, H.; Zeng, G.; Xi, J. Simultaneous Removal of Multicomponent VOCs in Biofilters. Trends Biotechnol. 2018, 36, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Agunbiade, M.O.; Pohl, C.; Van Heerden, E.; Oyekola, O.; Ashafa, A. Evaluation of fresh water actinomycete bioflocculant and its biotechnological applications in wastewaters treatment and removal of heavy metals. Int. J. Environ. Res. Public Health 2019, 16, 3337. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, Z.; Liu, P.; Liu, Y.; Wang, Y.; Li, Q.; He, N. Characterization of a novel bioflocculant from a marine bacterium and its application in dye wastewater treatment. BMC Biotechnol. 2017, 17, 1. [Google Scholar] [CrossRef]
- Tawila, Z.M.A.; Ismail, S.; Dadrasnia, A.; Usman, M.M. Production and characterization of a bioflocculant produced by bacillus salmalaya 139si-7 and its applications in wastewater treatment. Molecules 2018, 23, 2689. [Google Scholar] [CrossRef]
- Rasulov, B.A.; Li, L.; Liu, Y.H.; Mohamad, O.A.; Xiao, M.; Ma, J.B.; Li, W.J. Production, characterization and structural modification of exopolysaccharide-based bioflocculant by Rhizobium radiobacter SZ4S7S14 and media optimization. 3 Biotech 2017, 7. [Google Scholar] [CrossRef]
- Salehizadeh, H.; Yan, N. Recent advances in extracellular biopolymer flocculants. Biotechnol. Adv. 2014, 32, 1506–1522. [Google Scholar] [CrossRef]
- Xia, S.; Zhang, Z.; Wang, X.; Yang, A.; Chen, L.; Zhao, J.; Leonard, D.; Jaffrezic-Renault, N. Production and characterization of a bioflocculant by Proteus mirabilis TJ-1. Bioresour. Technol. 2008, 99, 6520–6527. [Google Scholar] [CrossRef]
- Deng, S.; Yu, G.; Ting, Y.P. Production of a bioflocculant by Aspergillus parasiticus and its application in dye removal. Colloids Surf. B Biointerfaces 2005, 44, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, C.; Jiang, J.; Lu, Q.; Hao, Y.; Wang, L.; Liu, C. Bioflocculant production from untreated corn stover using Cellulosimicrobium cellulans L804 isolate and its application to harvesting microalgae. Biotechnol. Biofuels 2015, 8, 170. [Google Scholar] [CrossRef] [PubMed]
- Taokaew, S.; Phisalaphong, M.; Newby, B. min Z. Modification of bacterial cellulose with organosilanes to improve attachment and spreading of human fibroblasts. Cellulose 2015, 22, 2311–2324. [Google Scholar] [CrossRef]
- Tang, W.; Song, L.; Li, D.; Qiao, J.; Zhao, T.; Zhao, H. Production, Characterization, and Flocculation Mechanism of Cation Independent, pH Tolerant, and Thermally Stable Bioflocculant from Enterobacter sp. ETH-2. PLoS ONE 2014, 9, e114591. [Google Scholar] [CrossRef]
- Pathak, M.; Sarma, H.K.; Bhattacharyya, K.G.; Subudhi, S.; Bisht, V.; Lal, B.; Devi, A. Characterization of a Novel Polymeric Bioflocculant Produced from Bacterial Utilization of n-Hexadecane and Its Application in Removal of Heavy Metals. Front. Microbiol. 2017, 8, 170. [Google Scholar] [CrossRef]
- Peng, L.; Yang, C.; Zeng, G.; Wang, L.; Dai, C.; Long, Z.; Liu, H.; Zhong, Y. Characterization and application of bioflocculant prepared by Rhodococcus erythropolis using sludge and livestock wastewater as cheap culture media. Appl. Microbiol. Biotechnol. 2014, 98, 6847–6858. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.A.; Azim, E.-B.A.; Hassan, A.A.; El-Barawy, A.M.; Mokhtar, E.; Ahed, M. Evaluation of Biological Compounds of Streptomyces Species for Control of some Fungal Diseases. J. Am. Sci. 2011, 7, 752–760. [Google Scholar]
- Jensen, P.R.; Dwight, R.; Fenical, W. Distribution of actinomycetes in near-shore tropical marine sediments. Appl. Environ. Microbiol. 1991, 57, 1102–1108. [Google Scholar] [CrossRef]
- Pathalam, G.; Rajendran, H.A.D.; Appadurai, D.R.; Gandhi, M.R.; Michael, G.P.; Savarimuthu, I.; Naif, A.A.-D. Isolation and molecular characterization of actinomycetes with antimicrobial and mosquito larvicidal properties. Beni-Suef Univ. J. Basic Appl. Sci. 2017, 6, 209–217. [Google Scholar] [CrossRef]
- Shaik, M.; Girija Sankar, G.; Iswarya, M.; Rajitha, P. Isolation and characterization of bioactive metabolites producing marine Streptomyces parvulus strain sankarensis-A10. J. Genet. Eng. Biotechnol. 2017, 15, 87–94. [Google Scholar] [CrossRef]
- Prakash, D.; Nawani, N.; Prakash, M.; Bodas, M.; Mandal, A.; Khetmalas, M.; Kapadnis, B. Actinomycetes: A Repertory of Green Catalysts with a Potential Revenue Resource. Biomed Res. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Nawani, N.; Aigle, B.; Mandal, A.; Bodas, M.; Ghorbel, S.; Prakash, D. Actinomycetes: Role in Biotechnology and Medicine. Biomed Res. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, S.; Zaheer, A.; Aiysha, D.; Abdulla Malik, K.; Mehnaz, S. Actinomycetes: A Source of Industrially Important Enzymes. J. Proteom. Bioinform. 2017, 10. [Google Scholar] [CrossRef]
- Pandey, A.; Nigam, P.; Soccol, C.R.; Soccol, V.T.; Singh, D.; Mohan, R. Advances in microbial amylases. Biotechnol. Appl. Biochem. 2000, 31, 135. [Google Scholar] [CrossRef]
- Lee, N.; Lee, S.H.; Baek, K.; Kim, B.G. Heterologous expression of tyrosinase (MelC2) from Streptomyces avermitilis MA4680 in E. coli and its application for ortho-hydroxylation of resveratrol to produce piceatannol. Appl. Microbiol. Biotechnol. 2015, 99, 7915–7924. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Williams, S.T.; Mordarski, M. Actinomycetes in Biotechnology; Academic Press: Cambridge, MA, USA, 1988. [Google Scholar]
- Fulton, L.M.; Lynd, L.R.; Körner, A.; Greene, N.; Tonachel, L.R. The need for biofuels as part of a low carbon energy future. Biofuels Bioprod. Biorefin. 2015, 9, 476–483. [Google Scholar] [CrossRef]
- Lombard, F.; Guidi, L.; Kiørboe, T. Effect of Type and Concentration of Ballasting Particles on Sinking Rate of Marine Snow Produced by the Appendicularian Oikopleura dioica. PLoS ONE 2013, 8, e75676. [Google Scholar] [CrossRef]
- Mason, M.G.; Ball, A.S.; Reeder, B.J.; Silkstone, G.; Nicholls, P.; Wilson, M.T. Extracellular Heme Peroxidases in Actinomycetes: A Case of Mistaken Identity. Appl. Environ. Microbiol. 2001, 67, 4512–4519. [Google Scholar] [CrossRef]
- Saha, S.; Dhanasekaran, D.; Shanmugapriya, S.; Latha, S. Nocardiopsis sp. SD5: A potent feather degrading rare actinobacterium isolated from feather waste in Tamil Nadu, India. J. Basic Microbiol. 2012, 53. [Google Scholar] [CrossRef]
- Liu, W.; Yuan, H.; Yang, J.; Li, B. Characterization of bioflocculants from biologically aerated filter backwashed sludge and its application in dying wastewater treatment. Bioresour. Technol. 2009, 100, 2629–2632. [Google Scholar] [CrossRef]
- Chen, X.; Stewart, P.S. Role of electrostatic interactions in cohesion of bacterial biofilms. Appl. Microbiol. Biotechnol. 2002, 59, 718–720. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhu, X.H.; Mu, J.; Zhang, Y.; Dong, X.W. Using Ruditapes philippinarum conglutination mud to produce bioflocculant and its applications in wastewater treatment. Bioresour. Technol. 2009, 100, 4996–5001. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Salam, A.E.; Abd-El-Haleem, D.; Youssef, A.S.; Zaki, S.; Abu-Elreesh, G.; El-Assar, S.A. Isolation, characterization, optimization, immobilization and batch fermentation of bioflocculant produced by Bacillus aryabhattai strain PSK1. J. Genet. Eng. Biotechnol. 2017, 15, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Zhou, H.; Chen, Y.; Yang, G.; Cui, X. Revealing a novel natural bioflocculant resource from Ruditapes philippinarum: Effective polysaccharides and synergistic flocculation. Carbohydr. Polym. 2018, 186, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Sajayan, A.; Seghal Kiran, G.; Priyadharshini, S.; Poulose, N.; Selvin, J. Revealing the ability of a novel polysaccharide bioflocculant in bioremediation of heavy metals sensed in a Vibrio bioluminescence reporter assay. Environ. Pollut. 2017, 228, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liang, Y.; Dou, Y.; Jia, X.; Che, H. Isolation and identification of a bioflocculant-producing strain and optimisation of cultural conditions via a response surface model. Chem. Ecol. 2015, 31, 650–660. [Google Scholar] [CrossRef]
- Guo, J.; Chen, C. Removal of arsenite by a microbial bioflocculant produced from swine wastewater. Chemosphere 2017, 181, 759–766. [Google Scholar] [CrossRef]
- Sivasankar, P.; Poongodi, S.; Lobo, A.O.; Pugazhendhi, A. Characterization of a novel polymeric bioflocculant from marine actinobacterium Streptomyces sp. and its application in recovery of microalgae. Int. Biodeterior. Biodegrad. 2020, 148, 104883. [Google Scholar] [CrossRef]
- Sarang, M.C.; Nerurkar, A.S. Amyloid protein produced by B. cereus CR4 possesses bioflocculant activity and has potential application in microalgae harvest. Biotechnol. Lett. 2020, 42, 79–91. [Google Scholar] [CrossRef]
- Guo, J.; Yu, J. Sorption characteristics and mechanisms of Pb(II) from aqueous solution by using bioflocculant MBFR10543. Appl. Microbiol. Biotechnol. 2014, 98, 6431–6441. [Google Scholar] [CrossRef]
- Aljuboori, A.H.R.; Uemura, Y.; Thanh, N.T. Flocculation and mechanism of self-flocculating lipid producer microalga Scenedesmus quadricauda for biomass harvesting. Biomass Bioenergy 2016, 93, 38–42. [Google Scholar] [CrossRef]
- Tang, J.; Qi, S.; Li, Z.; An, Q.; Xie, M.; Yang, B.; Wang, Y. Production, purification and application of polysaccharide-based bioflocculant by Paenibacillus mucilaginosus. Carbohydr. Polym. 2014, 113, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, G.; Wang, G. Characteristics for production of hydrogen and bioflocculant by Bacillus sp. XF-56 from marine intertidal sludge. Int. J. Hydrog. Energy 2015, 40, 1414–1419. [Google Scholar] [CrossRef]
- Liu, C.; Wang, K.; Jiang, J.H.; Liu, W.J.; Wang, J.Y. A novel bioflocculant produced by a salt-tolerant, alkaliphilic and biofilm-forming strain Bacillus agaradhaerens C9 and its application in harvesting Chlorella minutissima UTEX2341. Biochem. Eng. J. 2015, 93, 166–172. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Y.; Liu, L.; Jiang, X.; Zhang, K.; Zheng, T.; Wang, H. First evidence of bioflocculant from Shinella albus with flocculation activity on harvesting of Chlorella vulgaris biomass. Bioresour. Technol. 2016, 218, 807–815. [Google Scholar] [CrossRef]
- Zhao, H.; Zhong, C.; Chen, H.; Yao, J.; Tan, L.; Zhang, Y.; Zhou, J. Production of bioflocculants prepared from formaldehyde wastewater for the potential removal of arsenic. J. Environ. Manage. 2016, 172, 71–76. [Google Scholar] [CrossRef]
- Yin, Y.-J.; Tian, Z.-M.; Tang, W.; Li, L.; Song, L.-Y.; McElmurry, S.P. Production and characterization of high efficiency bioflocculant isolated from Klebsiella sp. ZZ-3. Bioresour. Technol. 2014, 171, 336–342. [Google Scholar] [CrossRef]
- Sun, P.-F.; Lin, H.; Wang, G.; Lu, L.-L.; Zhao, Y.-H. Preparation of a new-style composite containing a key bioflocculant produced by Pseudomonas aeruginosa ZJU1 and its flocculating effect on harmful algal blooms. J. Hazard. Mater. 2015, 284, 215–221. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Hu, Z.Q.; Wang, T.; Chen, Y.Y.; Zhang, J.; Yu, J.R.; Zhang, T.; Zhang, Y.F.; Li, Y.L. Production of novel microbial flocculants by Klebsiella sp. TG-1 using waste residue from the food industry and its use in defecating the trona suspension. Bioresour. Technol. 2013, 139, 265–271. [Google Scholar] [CrossRef]
- Karthiga Devi, K.; Natarajan, K.A. Production and characterization of bioflocculants for mineral processing applications. Int. J. Miner. Process. 2015, 137, 15–25. [Google Scholar] [CrossRef]
- Mabrouk, M.E.M. Production of bioflocculant by the marine actinomycete Nocardiopsis aegyptia sp. nov. Life Sci. J. 2014, 11, 27–35. [Google Scholar]
| Inorganic Flocculants | Organic Flocculants | Natural Flocculants |
|---|---|---|
| Polyaluminum chloride | Polyacrylamide | Chitosan |
| Aluminum sulphate | Polyethylene amine | Cellulose |
| Aluminum chloride | Gum and mucilage | |
| Ferric chloride | Sodium alginate | |
| Alum | Tannin | |
| Ferrous sulphate | Microbial flocculants |
| Compound | Species | Other Biological Activity | References |
|---|---|---|---|
| Antibacterial activity | |||
| Abyssomicins | Verrucosispora sp. | – | [27] |
| Bonactin | Streptomyces sp. | Antifungal | [28] |
| Chloro-dihydroquino | Streptomyces sp. | Anticancer | [29] |
| Diazepinomicin | Micromonospora sp. | Anticancer; anti-inflammatory | [30] |
| Frigocyclinone | Streptomyces griseus | – | [31] |
| Essramycin | Streptomyces sp. | – | [32] |
| Lynamicins | Marinispora sp. | – | [33] |
| Marinopyrroles | Streptomyces sp. | Cytotoxic | [34] |
| Caboxamycin | Streptomyces sp. | Cytotoxic | [35] |
| Himalomycins | Streptomyces sp. | – | [36] |
| Chandrananimycin | Actinomadura sp. | Antialgal; antibacterial | [37] |
| N-(2-hydroxyphenyl)-2-phenazinamine(NHP) | Nocardia dassonvillei | Anticancer | [38] |
| Anticancer activity | |||
| Salinosporamide A | Salinispora tropica | – | [39] |
| Caprolactones | Streptomyces sp. | – | [40] |
| 3, 6-Disubstituted indoles | Streptomyces sp. | – | [41] |
| IB-00208 | Actinomadura sp. | – | [42] |
| Antitumor activity | |||
| Chinikomycins | Streptomyces sp. | – | [43] |
| Glyciapyrroles | Streptomyces sp. | – | [35] |
| Mechercharmycin A | Thermoactinomyces sp | – | [44] |
| Aureoverticillactam | Streptomyces aureoverticillatus | – | [45] |
| Arenicolides | Salinispora arenicola | – | [46] |
| Chalcomycin | Streptomyces sp. | – | [9] |
| Use | Enzyme | Applications |
|---|---|---|
| Detergent (laundry and dish wash) | Protease | Protein stain removal |
| Amylase | Starch stain removal | |
| Lipase | Lipid stain removal | |
| Cellulase | Cleaning, color clarification, anti-redeposition (cotton) | |
| Mannanase | Mannanan stain removal (reappearing stains) | |
| Starch and fuel | Amylase | Starch liquefaction and saccharification |
| Amyloglucosidase | Saccharification | |
| Pullulanase | Saccharification | |
| Glucose isomerase | Glucose to fructose conversion | |
| Cyclodextrin-glycosyltransferase | Cyclodextrin production | |
| Xylanase | Viscosity reduction (fuel and starch) | |
| Food (including dairy) | Protease | Milk clotting, infant formulas (low allergenic), flavor |
| Lipase | Cheese flavor | |
| Lactase | Lactose removal (milk) | |
| Pectin methyl esterase | Firming fruit-based products | |
| Pectinase | Fruit-based products | |
| Transglutaminase | Modify visco-elastic properties | |
| Baking | Amylase | Bread softness and volume, flour adjustment dough conditioning |
| Xylanase | Dough stability and conditioning (in situ emulsifier) | |
| Lipase | Dough stability and conditioning (in situ emulsifier) | |
| Phospholipase | Dough strengthening | |
| Glucose oxidase | Dough strengthening | |
| Lipoxygenase | Bread whitening | |
| Protease | Biscuits, cookies | |
| Transglutaminase | Laminated dough strengths | |
| Animal feed | Phytase | Phytate digestibility–phosphorus release |
| Xylanase | Digestibility | |
| β-Glucanase | Digestibility | |
| Beverage | Pectinase | De-pectinization, mashing |
| Amylase | Juice treatment, low calorie beer | |
| β-Glucanase | Mashing | |
| Acetolactate decarboxylase | Maturation (beer) | |
| Laccase | Clarification (juice), flavor (beer), cork stopper treatment | |
| Textile | Cellulase | Denim finishing, cotton softening |
| Amylase | De-sizing | |
| Pectatelyase | Scouring | |
| Catalase | Bleach termination | |
| Laccase | Bleaching | |
| Peroxidase | Excess dye removal | |
| Pulp and paper | Lipase | Pitch control, contaminant control |
| Protease | Biofilm removal | |
| Amylase | Starch-coating, de-inking, drainage improvement | |
| Xylanase | Bleach boosting | |
| Cellulase | De-inking, drainage improvement, fiber modification | |
| Fats and oils | Lipase | Transesterification |
| Phospholipase | De-gumming, lyso-lecithin production | |
| Organic synthesis | Lipase | Resolution of chiral alcohols and amides |
| Acylase | Synthesis of semisynthetic penicillin | |
| Nitrilase | Synthesis of enantiopure carboxylic acids | |
| Leather | Protease | Unhearing, bating |
| Lipase | De-pickling | |
| Personal care | Amyloglucosidase | Antimicrobial (combined with glucose oxidase) |
| Glucose oxidase | Bleaching, antimicrobial | |
| Peroxidase | Antimicrobial | |
| L-Asparagine | Antitumor | |
| Neuraminidase | Antiviral agents | |
| Aminoacylase | Regulation of urea cycle |
| Name | Source | Chemical Composition | Flocculating Activity (%) | Applications | Citations |
|---|---|---|---|---|---|
| Bacillus aryabhattai | Egyptian Agricultural soils | Glycoprotein | 92.8% at 50 °C and 94.6% at pH 2.0 | N/A | [97] |
| Ruditapes philippinarum | Aquaculture | Complex heteropolysaccharides | 86.7% in deionized water and 91.8% in sea water | N/A | [98] |
| Sphingomonas Yabuuchiae | Chromotrophic acid waste water | Polysaccharides | 0.4% (w/w) kaolin suspensions over pH 3.9 and 20–80 °C | Steroid estrogen removal | [10] |
| Alteromonas sp CGMCC 10612 | Surface Sea water | Polysaccharides | 2575.4 U/mL achieved in a 2-L fermenter | Dye decolorization | [69] |
| Bacillus cereus | Marine sponge | Polysaccharides | 94% F/A in kaolin suspension | synthesis of Ag nanoparticles and bioremediation of wastewater | [99] |
| Bacillus Mucilaginous | Mixed activated Sludge | Extracellular polysaccharides | 90% F/A in kaolin suspension | N/A | [100] |
| Bacillus Megaterium | Swine waste water treatment plant | Polysaccharides | 90.2% in 4 L kaolin suspension | Arsenite removal | [101] |
| Streptomyces sp | Mangrove sediments | Polysaccharides | 99.18% on Nannochloropsis culture medium | Recovery of microalgae | [102] |
| Bacillus cereus | Activated sludge Flocs | N/A | 86.87% | Microalgae harvest | [103] |
| Rhodococcus erythropolis | Alkaline thermal pretreated sludge | N/A | N/A | Removal of Pb (II) | [104] |
| Scendesmus quadricauda | Algaetech International Sdn Bhd | Glycoprotein | flocculate 86.7% of Scenedesmus quadricauda cells in presence of ZnCl2 | Harvesting of biomass | [105] |
| Paenibacillus mucilaginosus | Soil sample | Polysaccharides | 97% flocculation on kaolin clay suspension | Industrial waste water treatment | [106] |
| Bacillus sp. XF-56 | Marine intertidal Sludge | N/A | Up to 93.5% hydrogen and bioflocculant produced in marine culture condition and 96.8% in fresh ones | N/A | [107] |
| Bacillus agaradhaerens C9 | Alkaline lake sample | Polysaccharides Protein & nucleic Acids | 95.29% kaolin suspension | Biofilms formation and harvesting of Chlorella minuttssima | [108] |
| Shinella albus Xn-1 | Phycosphere of Microcyctis aeruginosa 7820 | Non proteins & carbohydrate | 86.65% | Harvest of Chlorella vulgaris biomass | [109] |
| Panebacillius polymyxa MBF-79 | Recycled activated sludge samples | Glycoproteins | 94.7% flocculation was achieved | Removal of arsenic acid | [110] |
| Klebsiella | Activated sludge | Polysaccharide | 93.9% flocculation was achieved | N/A | [111] |
| Pseudomonas aeruginosa ZJU1 | Water sample by a routine enrichment | Polysaccharide proteins & nucleic acids | N/A | Treatment of HABs caused by Microcystis aeruginosa | [112] |
| Klebsiella sp. TG-1 | Waste water of a starchy factory | Polysaccharides | 98% kaolin clay | Defecating Trona suspension | [113] |
| Bacillus firmus and Bacillus licheniformis | National collection of industrial microorganisms | Glycoproteins | N/A | Decolorization of dye and remediation of toxic metal solution | [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awolusi, O.O.; Ademakinwa, A.N.; Ojo, A.; Erasmus, M.; Bux, F.; Agunbiade, M.O. Marine Actinobacteria Bioflocculant: A Storehouse of Unique Biotechnological Resources for Wastewater Treatment and Other Applications. Appl. Sci. 2020, 10, 7671. https://doi.org/10.3390/app10217671
Awolusi OO, Ademakinwa AN, Ojo A, Erasmus M, Bux F, Agunbiade MO. Marine Actinobacteria Bioflocculant: A Storehouse of Unique Biotechnological Resources for Wastewater Treatment and Other Applications. Applied Sciences. 2020; 10(21):7671. https://doi.org/10.3390/app10217671
Chicago/Turabian StyleAwolusi, Oluyemi Olatunji, Adedeji Nelson Ademakinwa, Abidemi Ojo, Mariana Erasmus, Faizal Bux, and Mayowa Oladele Agunbiade. 2020. "Marine Actinobacteria Bioflocculant: A Storehouse of Unique Biotechnological Resources for Wastewater Treatment and Other Applications" Applied Sciences 10, no. 21: 7671. https://doi.org/10.3390/app10217671
APA StyleAwolusi, O. O., Ademakinwa, A. N., Ojo, A., Erasmus, M., Bux, F., & Agunbiade, M. O. (2020). Marine Actinobacteria Bioflocculant: A Storehouse of Unique Biotechnological Resources for Wastewater Treatment and Other Applications. Applied Sciences, 10(21), 7671. https://doi.org/10.3390/app10217671







