Abstract
β-mannanase is one of the key enzymes to hydrolyze hemicellulose. At present, most β-mannanases are not widely applied because of their low enzyme activity and unsuitable enzymatic properties. In this work, a new β-mannanase from Enterobacter aerogenes was studied, which laid the foundation for its further application. Additionally, we will further perform directed evolution of the enzyme to increase its activity, improve its temperature and pH properties to allow it more applications in industry. A new β-mannanase (Man26E) from Enterobacter aerogenes was successfully expressed in Escherichia coli. Man26E showed about 40 kDa on SDS-PAGE gel. The SWISS-MODEL program was used to model the tertiary structure of Man26E, which presented a core (α/β)8-barrel catalytic module. Based on the binding pattern of CjMan26 C, Man26E docking Gal1Man4 was investigated. The catalytic region consisted of a surface containing four solvent-exposed aromatic rings, many hydrophilic and charged residues. Man26E displayed the highest activity at pH 6.0 and 55 °C, and high acid and alkali stability in a wide pH range (pH 4–10) and thermostability from 40 to 50 °C. The enzyme showed the highest activity on locust bean gum, and the Km and Vmax were 7.16 mg mL−1 and 508 U mg−1, respectively. This is the second β-mannanase reported from Enterobacter aerogenes B19. The β-mannanase displayed high enzyme activity, a relatively high catalytic temperature and a broad range of catalytic pH values. The enzyme catalyzed both polysaccharides and manno-oligosaccharides.
1. Introduction
Mannan is a component of hemicellulose, which is second only to xylan and is widespread in wood, tubers, beans and plant seeds [1,2]. According to the way that the sugar residues are connected, Mannan is composed of four subfamilies: linear mannan, glucomannan, galactomannan and galactoglucomannan. These glycans are made up of β-1,4 linked mannose or α-1,6-linked mannose residues [3]. Because most mannans are highly branched heterogeneous polysaccharides with many different substituents, the biodegradation of mannans require a complex enzyme system in which many components degrade mannans by synergistic action. The enzyme system of mannan biodegradation involves many kinds of hydrolases which hydrolyze the main chain and side chains of mannan, such as exo-β-mannosidase (EC 3.2.1.25), endo-β-mannanase (EC 3.2.1.78), α-galactosidase (EC 3.2.1.22), β-glucosidase (EC 3.2.1.21) and acetyl mannan esterases (EC 3.1.1.6) [1,4]. Mannanases can be divided into several glycosyl hydrolase families (GH families) and their primary structures are quite different, whereas possess high similarity in their spatial arrangements which contain (β/α)8-barrel folding structures and are classified as GH-A clan [5]. They usually present modular structures which are composed of catalytic domain(s), carbohydrate binding module(s) (CBM), and additional functional domain(s) [6]. The hydrolysis of mannan-based polysaccharides involves two crucial enzymes, namely, β-mannanase (endo-β-mannose) and β-mannosidase (exo-β-mannose). β-mannanase cleaves glucosidic bonds inside mannan to produce smaller manno-oligosaccharides, and then, β-mannosidase hydrolyzes these smaller manno-oligosaccharides from their non-reducing end into fermentable mannose sugars. β-mannanase catalyzes the degradation of mannan, glucomannan, and galactomannan by incising the β-1,4-mannoside bonds in the main chain in a random way, thereby degrades the abundant heterologous mannans to manno-oligosaccharides [7,8]. β- mannanase has shown potential application values in many industrial fields, for instance, pulp bleaching, oligosaccharides producing, food quality improving [9,10,11,12], oil drilling, feed additives and detergents [4,13]. Up to now, a variety of β-mannanases and their corresponding genes have been isolated, purified and identified from microbial strains, for example, Bacillus subtilis and other Bacillus [14], Aspergillus niger [15], Caldocellum saccharolyticum [16], Caldibacillus cellulovorans [17], Thermoanaerobacterium spp. [18], and Penicillium purpurogenum [19]. β-mannanases of the GH26 family are found to originate primarily from bacteria, for example, Bacillus subtilis [20], Clostridium cellulovorans [21], Paenibacillus polymyxa [22], Pseudomonas fluorescens, Cellulomonas fimi [23].
It is widely known that Enterobacter aerogenes is a producing strain for 2,3-butanediol and 1,3-propanediol [24], and it is also regarded as a very useful strain for isobutanol production [25], ethanol production [26] and hydrogen production [27,28]. In our previous work, we isolated a β-mannanase producing strain from forest soil. After physiological, biochemical and 16S rDNA identification, the strain belongs to Enterobacter aerogenes, named as Enterobacter aerogenes B19. From this strain, we have cloned and characterized a CH1 β-mannanase with molecular weight of 82.5 kDa [29]. In this work, another new GH26 β-mannanase from Enterobacter aerogenes B19 was investigated. We have successfully cloned and expressed Man26E gene in Escherichia coli, determined biochemical characteristics and substrate specificity of the enzyme. Three-dimensional structure of the β-mannanase was homologously modeled and its active domain was analyzed. The β-mannanase from Enterobacter aerogenes B19 may have special potential applications in different industries.
2. Materials and Methods
2.1. Strains, Medium, Plasmids and Reagents
Enterobacter aerogenes B19 was isolated and preserved by our laboratory. Escherichia coli DH5α and pMD18-T simple vector were bought from Takara. Escherichia coli BL21(DE3) and pET-28a (+) vector were provided by Vazyme Biotech Co. (Nanjing, China). Locust bean gum, konjac powder and p-nitrophenyl-β-d-mannopyranoside were purchased from Sigma (St. Louis, MO, USA). The 6× His-Tagged Protein Purification kit was provided by Cwbiotech (Beijing, China). The DNA purification kits, restriction endonuclease, 2× premix LA Taq and T4 DNA ligase were offered by Takara (Dalian, China). All other reagents were analytical grade. Escherichia coli strains grew at 37 °C and 200 rev min−1 in Luria-Bertani (LB) medium which was composed of 0.5% NaCl, 0.5% yeast extract, 1% tryptone, 1% glucose. LB agar medium was prepared by adding 2 % (w/v) agar in LB medium. 50 µg mL−1 of ampicillin and 40 µg mL−1 of X-Gal were added in LB agar medium for screening positive clones. 50 µg mL−1 of kalamycin was added in LB agar medium for screening the transformants, and 24 µg mL−1 of IPTG (isopropyl-beta-D-thiogalactopyranoside) was mixed in LB medium to improve the expression of recombinant proteins. Congo-red dyeing medium consisted of LB agar medium and additional 1% konjac powder.
2.2. Cloning of the β-Mannanase Gene and Sequence Analysis
The cloning of β-mannanase gene was carried out according to the standard method. The genomic DNA extracted from Enterobacter aerogenes B19 was used as the template DNA and amplification of β-mannanase gene was performed by PCR method. In the light of the gene sequence of a published β-mannanase from Pantoea agglomerans A021 (GenBank accession no. ACN30272), we input the amino acid sequence of β-mannanase (published) from Pantoea agglomerans A021 into NCBI to search for homologous sequences, and highly homologous sequences from Enterobacter ludwigii strains and Enterobacter cloacae complex strains were obtained. We analyzed 5′-terminal sequence and 3′-terminal sequence of these homologous proteins and designed a pair of degenerate primers. The forward primer was 5′-CGCGGATCCATGAGTACTTTTACTGTAGTACCCG-3′ and the reverse primer was 5′-CCCAAGCTTTTAACTCAGAACGCTGCCGTTCCAT-3′. The PCR reaction system was composed of 1 µL DNA template (1 ng µL−1), 2 µL each primer (10 µmol L−1), 20 µL double-distilled H2O and 25 µL 2× premix LA Taq. The PCR amplification was carried out as follows: initial template denaturation at 94 °C for 5 min, then 35 cycles, each containing 30 s at 94 °C, 30 s at 58 °C, 90 s at 72 °C, and finally an extension for 10 min at 72 °C. The PCR products were assayed by agarose gel electrophoresis and purified by the DNA Purification Kit. The target gene was ligated to plasmid pMD19-T (Takara) by T4 ligase to construct the recombinant vectors. Then, the recombinant vectors were transferred into Escherichia coli DH5α. The recombinant positive clones were cultured overnight, and a plasmid extraction kit was used to extract the recombinant vectors and further verified by DNA sequencing. The gene sequence of the β-mannanase was analyzed by the NCBI ORF Finder tool (https://www.ncbi.nlm.nih.gov/orffinder/). The corresponding amino acid sequence of the β-mannanase was translated by EMBOSS Transeq (http://www.ebi.ac.uk/Tools/st/emboss_transeq/), and the isoelectric point and molecular weight were predicted by ProtParam (http://web.expasy.org/protparam/). Signal peptide was examined by SignalP 4.0 [30] (http://www.cbs.dtu.dk/services/SignalP/). The homologous sequences of the amino acid sequence of Man26E were searched out by BLASTp tool (http://www.ncbi.nlm.nih.gov/BLAST/). Alignment of multiple sequences were carried out by Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/).
2.3. Structural Simulation and Analysis for the Active Region
The analysis of Man26E secondary structure was fulfilled using PredictProtein program (https://www.predictprotein.org/). The tertiary structure of Man26E was simulated by the method of homologous modeling with the crystallographic structure of a β-mannanase from Bacillus subtilis str. 168 (PDB ID. 2WHK-A, 47.97% identity) as a template and the SWISS-MODEL tool (http://swissmodel.expasy.org/). The structural model was evaluated with Molprobity program (http://molprobity.biochem.duke.edu/index.php) and SAVES v5.0 tool (https://servicesn.mbi.ucla.edu/SAVES/). The structure was visualized and analyzed using PyMol software. On the basis of the crystallographic structure of a homologous GH26 β-mannanase CjMan26 C (PDB ID. 2VX7) from Cellvibrio japonicus in complex with mannose residues, the modeled structure of Man26E was superimposed with that of CjMan26 C in complex with Gal1Man4, the modeled structure of Man26E in complex with Gal1Man4 was obtained and the active region of Man26E was analyzed using PyMol 2.3.2. Schematic diagram of the interactions of Man26E with manno-oligosaccharide Gal1Man4 was plotted with ChemDraw Prime 19 software.
2.4. Expression of Man26E in Escherichia coli
The recombinant pMD19-T –Man26E plasmids and pET-28a (+) vectors were cleaved by Bam HI and Hind III, and then the products were ligated by DNA ligase to form the recombinant expression pET-28a (+) –Man26E vectors. The vectors were transformed into competent Escherichia coli BL21 (DE3). Positive transformants were selected on the LB resistant plates with 50 μg mL−1 kalamycin. Moreover, the positive transformants were further confirmed by DNA sequencing. Then, the screened positive transformants were applied for Congo-red dyeing assay established by Teather and Wood [31]. A positive transformant with maximum hydrolytic circle was picked out and used for further culture and analysis. The transformant grew in 50 mL of LB liquid medium with 50 µg mL−1 kanamycin at 37 °C and 200 rev min−1. When the biomass (represented by OD600) achieved 0.8, IPTG was supplemented into the medium at a final concentration of 1 mmol L−1. After IPTG treatment for 8 h at 32 °C, the cells and the supernatant were gathered respectively. Then, the cells were washed twice with stroke-physiological saline solution and resuspended in 10 mL of 100 mmol L−1 phosphate buffer (pH 7.0). The cell suspension was processed by ultrasonication for 20 min in an ice-water bath. The cellular debris was detached by centrifugation for 30 min at 11,000× g and 4 °C, and the resulting supernate was preserved at −20 °C for β-mannanase assays.
2.5. Purification. of Man26E
Affinity chromatography of the stored supernatant was carried out on a nickel-chelate column with a 6× His-Tagged Protein Purification kit in accordance with the operation manuals. After equilibration with binding buffer containing 10 mmol L−1 imidazole, the column was eluted with elution buffer containing 500 mmol L−1 imidazole. The active components were combined and imidazoles were removed by dialysis with 100 mmol L−1 phosphate buffer (pH 7.0), and the enzyme liquid was stored at −20 °C. The purified enzyme was used for SDS-PAGE and enzyme activity assays.
2.6. Activity Measurement and Protein Examination
The activity of β mannanase was calculated by determining the amount of reducing sugar produced from the substrate, which was quantified by reacting with 3,5-Dinitrosalicylic acid (DNS) reagent. The enzyme reaction system included 0.1 mL of enzyme solution with appropriate dilution and 0.9 mL of 0.5% (w/v) locust bean gum prepared by100 mmol L−1 citric acid buffer (pH 6.0). After incubation at 50 °C for 10 min, the enzyme reaction was stopped with 1.0 mL DNS reagent and the solution after reaction was boiled for 20 min. The mannose (Sigma) taken as the standard was treated under the same condition to make a standard curve. After the reaction solution cooled, its absorbance at 540 nm was determined colorimetrically. And the quantities of reducing sugars were computed from the standard curve. One unit (U) of β-mannanase activity meant the quantities of enzyme required to produce 1 μmol of reducing sugar (calculated as D-mannose) from the substrate per minute. The protein was quantified according to the Bradford’s method [32] with bovine serum albumin as a standard. The homogeneity and molecular weight of Man26E was detected by 12% SDS-PAGE in light of Laemmli’s description [33]. The protein bands were visualized by staining with Coomassie brilliant blue R-250. The low molecular weight calibration kit (TaKaRa, China) was used as the protein marker, which was composed of phosphorylase b (97.4 kDa), albumin (66.2 kDa), ovalbumin (43 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa).
2.7. Analysis of Enzymatic Characteristics
The characterization of Man26E was investigated with 0.5% locust bean gum as the substrate. Three replicates were set in all experiments independently. So as to obtain the optimal pH value, the enzyme reaction solution was incubated for 10 min at different pH (pH 3–10) conditions and 50 °C, and the enzyme activity was quantified according to the standard method. To test pH stability, the enzyme solutions were preincubated in the range of pH 4 to 10 for 1 h at 37 °C, and then the residual activities were measured. The buffers with different pH values included 100 mmol L−1 sodium citrate (pH 3–6), 100 mmol L−1 sodium phosphate (pH 7–8) and 100 mmol L−1 glycine-NaOH (9–10). To examine the optimal temperature, the enzyme activities were determined after the reaction mixtures were incubated at 40, 45, 50, 55, 60, 65, and 70 °C for 10 min, respectively. The thermostability of Man26E was investigated by preincubating the enzyme in 100 mmol L−1 citric acid buffer (pH 6.0) at 40, 50 and 60 °C respectively for different time (15, 30, 45 and 60 min), then the residual enzyme activities were determined. For investigating the action of metal cations on the purified β-mannanase activity, the enzyme solutions were preincubated in 100 mmol L−1 citric acid buffer (pH 6.0) containing 1, 5 and 10 mmol L−1 various ions for 5 min at 40 °C, separately, the enzyme activities were assayed at 50 °C and pH 6.0. The reaction group without adding any ions was used as a control. In this experiment, the metal ions included Ni2+, Zn2+, Mg2+, Co2+, Mn2+, Cu2+, K+, Fe2+, and Ca2+.
2.8. Substrate Specificity and Kinetic Parameters
For investigating the substrate specificity, the reaction solutions containing 5 mg mL−1 various substrates were incubated for 10 min, and the enzyme activities were quantified according to the dinitrosalicylic acid (DNS) method as described above. The tested substrates included locust bean gum, konjac powder, starch, manno-oligosaccharides, sodium carboxymethyl cellulose (CMC-Na), xylan and p-nitrophenyl-β-d-mannopyranoside (pNPM). Relative activities against the various substrates were determined using locust bean gum as the control. The enzyme reactions were performed at 50 °C for 10 min with 0.1 mg mL−1 to 20 mg mL−1 of LBG as the substrate, and then the dynamic parameters (Km and Vmax) of Man26E were computed from Lineweaver–Burk double-reciprocal chart [34].
3. Results
3.1. Gene Cloning and Sequence Assay
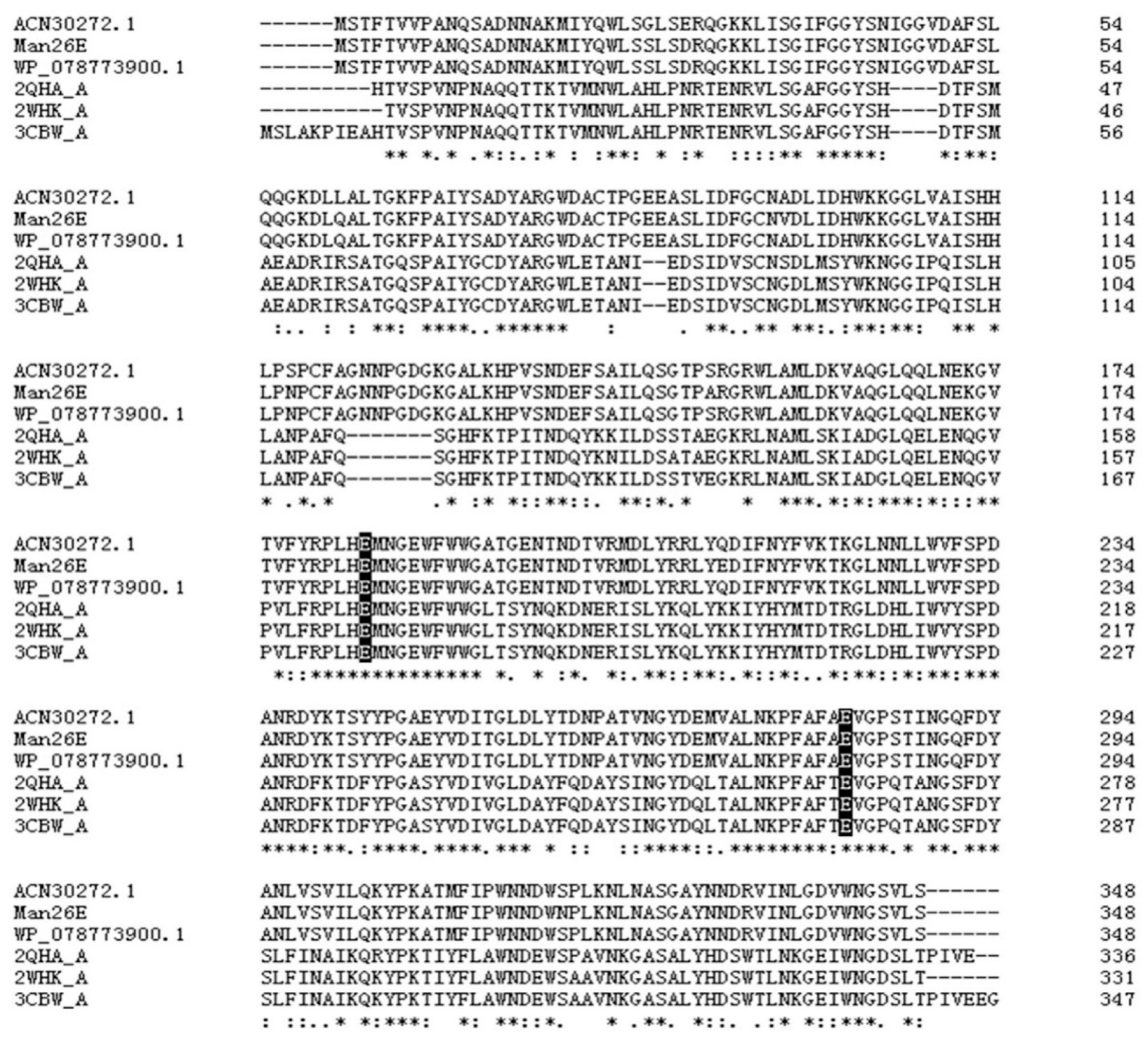
A β-mannanase gene was successfully amplified with the PCR method from Enterobacter aerogenes B19. The ORF of the β-mannanase gene contained 1047 bp encoding 348 amino acids. The molecular mass (Mw) and isoelectric point (pI) of the enzyme were predicted by ProtParam to be 38.61 kDa and 5.15, respectively. SignalP predicted that no signal peptide existed in the amino acid sequence. BLASTp result showed that Man26E shared high identity in the amino acid sequence (above 97%, Query cover 100%) with many Enterobacter bacteria strains, which included a Pantoea agglomerans A021 strain, Enterobacter ludwigii strains and Enterobacter cloacae strains. However, up to now, there are few reports on the identification, structure analysis, purification and heterologous expression of these β-mannanases. Multiple sequence alignment of Man26E (GenBank accession no. ANA84055.1) and other bacterial β-mannanases from Enterobacter ludwigii (GenBank accession no. WP_078773900.1), Bacillus subtilis strain 168 (PDB ID: 3CBW_A), Bacillus subtilis strain 168 (PDB ID. 2WHK_A), Pantoea agglomerans (GenBank accession no. ACN30272.1) and Bacillus subtilis Z-2 (PDB ID. 2QHA_A) were done by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The result showed that Man26E had 98.85% sequence identity with WP_078773900.1, 98.28% with WP_108648449.1, 47.97% with 2WHK_A, 97.70% with ACN30272.1, 47.97% with 2QHA _A and 47.41% with 3CBW_A. Although the similarity between three Enterobacter strains and three Bacillus subtilis strains was less than 50%, a large number of highly conserved amino acid residues were found in their sequences. The residues which have been considered to be involved in the catalysis of GH26 enzymes were conserved, and they were Glu183 (putative catalytic acid-base) and Glu282 (putative catalytic nucleophile) in Man26E (Figure 1).
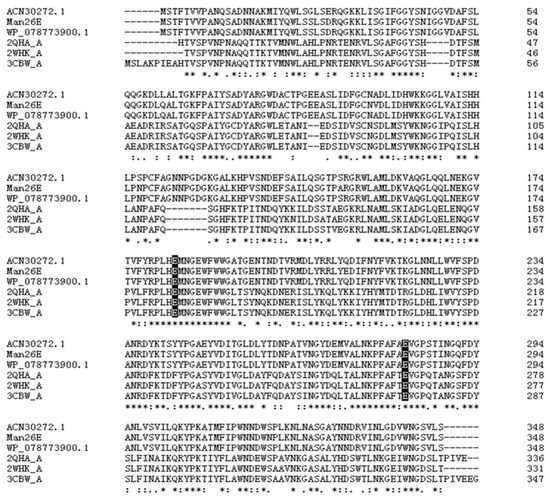
Figure 1.
Multiple analysis of amino acid sequences of Man26E (GenBank accession no. ANA84055.1) with other mannan hydrolases. Man26E and the Genbank accession numbers and PDB ID of these hydrolases were listed on the left side of the figure. Identical, highly conserved (residues with strong similar properties), and conserved residues (residues with weak similar properties) among sequences are marked by asterisk (*), colon (:), and dot (.), respectively. The residues participate in catalytic action are marked as dark shade.
3.2. The Structural Prediction and the Catalytic Region Analysis
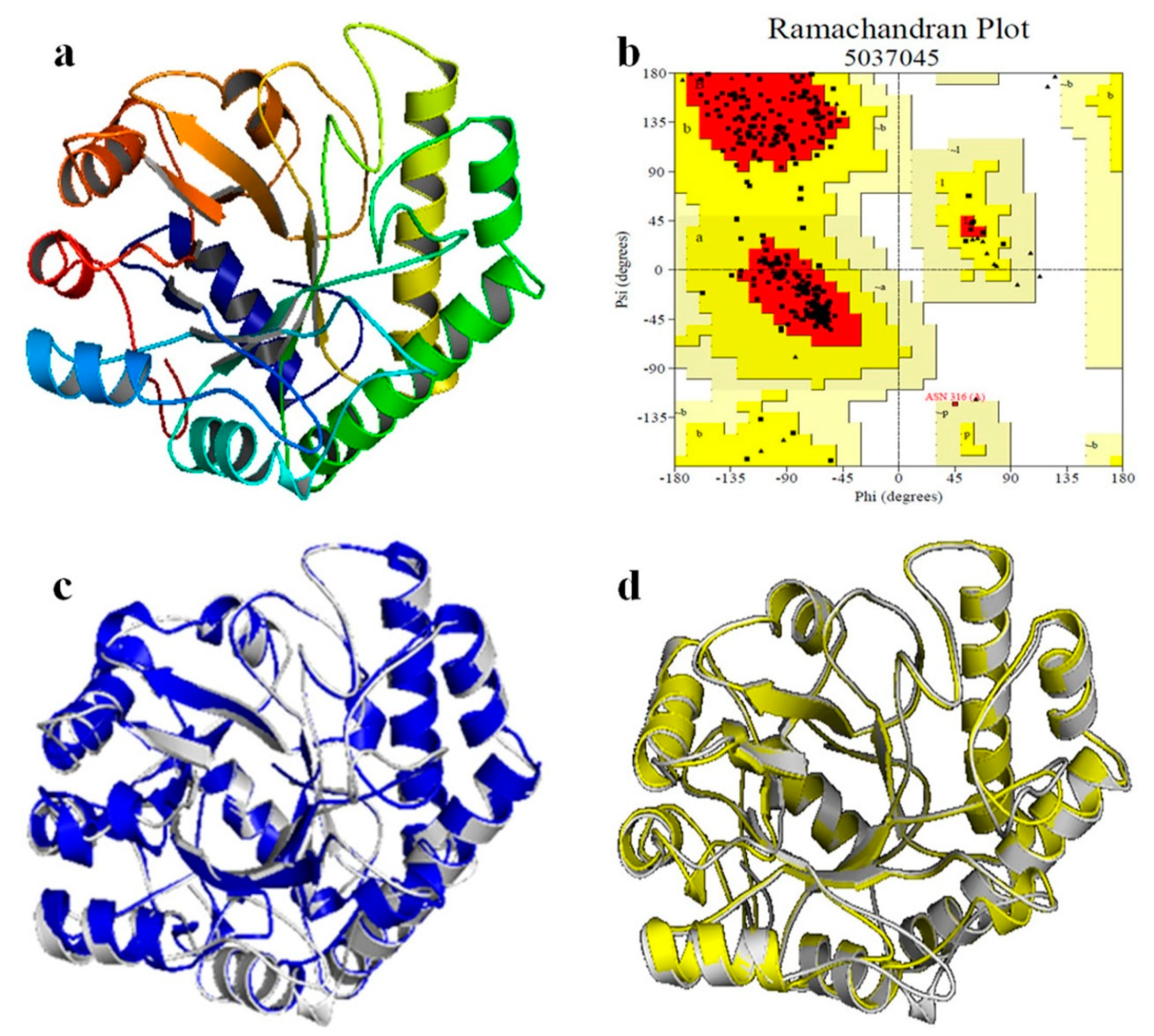
The amino acid sequence of Man26E was assayed with NCBI CDD program (https://www.ncbi.nlm.nih.gov/cdd) and only one CDD matches obtained in Man26E was Glycosyl hydrolase family 26 (9 aa-340 aa). The result showed that Man26E belonged to GH26 family with a (β/α)8 TIM barrel domain. The secondary structure, which was analyzed with SOPMA tool (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html), included 29.89% alpha-helix (104 aa), 17.53% extended strand (61 aa), 45.40% random coil (158 aa) and 7.18% beta-turn (25 aa). β-mannanase from Bacillus subtilis str. 168 (PDB ID. 2WHK-A) has 47.97% similarity (>30%) in the amino acid sequence with Man26E, so it can be used as the template for homology modelling. The final structural model covering residue 5 to 348 of Man26E was constructed with SWISS-MODEL tool, based on the structure of B. subtilis β-mannanase (PDB ID. 2WHK-A). The rationality of the structure model of Man26E was evaluated with SAVES v5.0 tool, the Ramachandran plot was drawn using the PROCHECK program (Figure 2b).
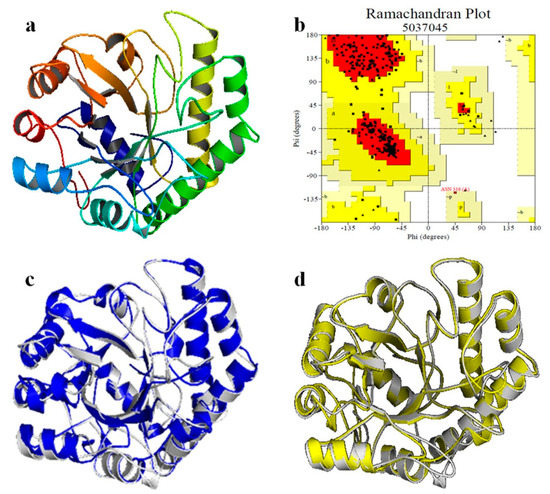
Figure 2.
Homology modeling of β-mannanase of Man26E. (a) The predicted 3D-structure of Man26E based on B. subtilis β-mannanase (2WHK_A). The figure was generated using the program SWISS-MODEL and the software PyMol 2.3.2; C-terminus (bright blue), N-terminus (red); (b) Ramachandran plot of the homology-modeled structure of Man1E based on B. subtilis β-mannanase; (c) the overlap between Man26E (represented in gray) and BCman (represented in blue); (d) the overlap between Man26E (represented in gray) and BsMan26A (represented in yellow).
The result evaluated with SAVES v5.0 tool revealed that there were 91.1% of total residues (266 aa) in most favoured regions, 8.6% residues in additional allowed regions (25 aa), 0.3% residues in generously allowed regions (1 aa) and no residues in disallowed regions. MolProbity analysis indicated that 95.3% of all residues (326/342) were in favored regions. 99.4% of all residues (340/342) were in allowed regions and there were two outliers. The modeled three-dimensional structure of Man26E (Figure 2a) contained nine α-helixes, nine β-strands and a few of loops. These loops that connected β2 and α3, β3 and α4, β4 and α5, and β5 and β6 usually consisted of 20–30 amino acid residues. This structural characteristic implies that the catalytic domain of Man26E allows for much more flexibility, which may affect its function. The modeled structure of Man26E did not contain disulfide bond. Although the similarity of amino acid sequences are less than 50%, the modeled three-dimensional structure of Man26P almost completely overlapped those of BsMan26A or BCman (Figure 2c,d).
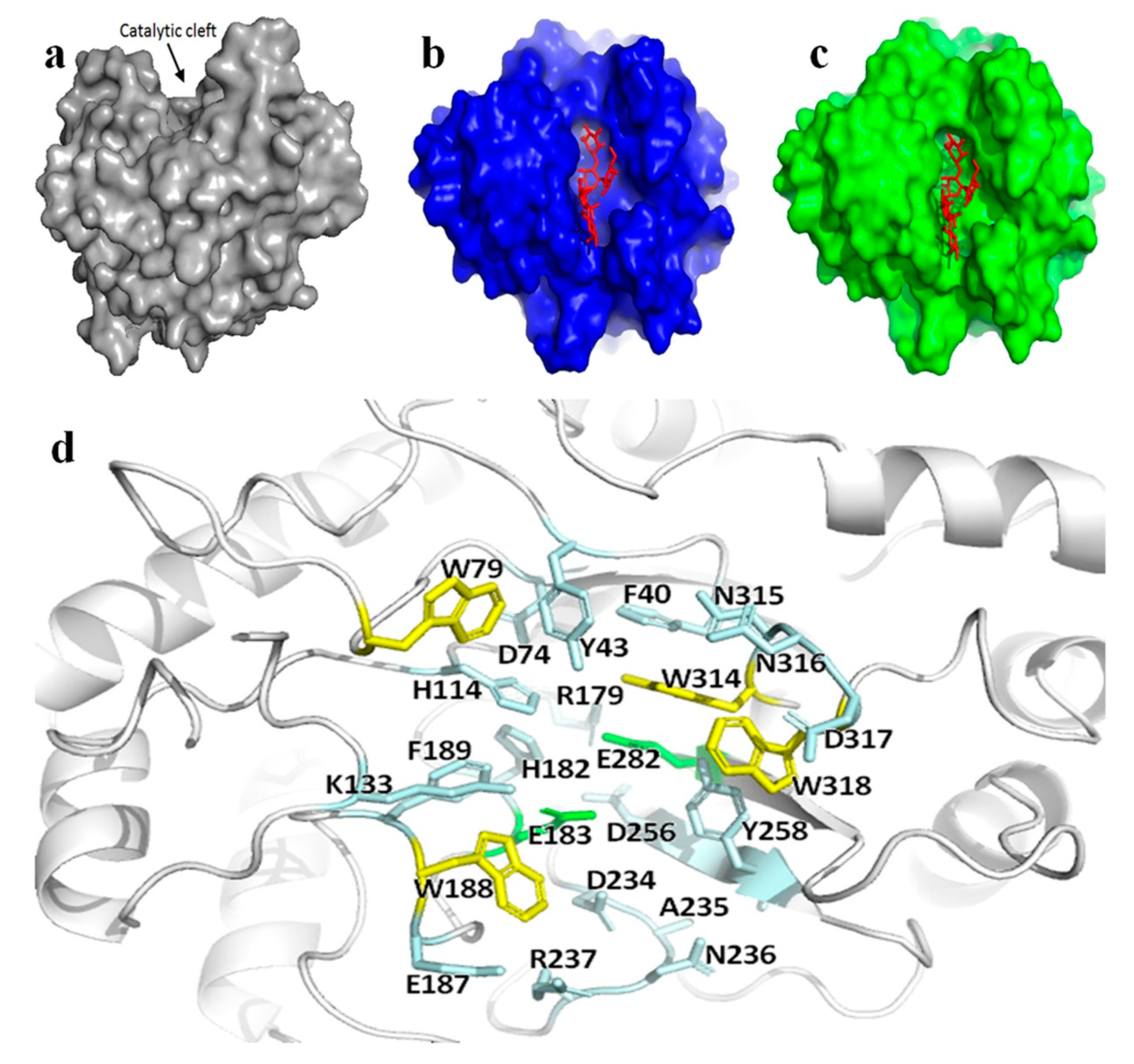
Three-dimensional structure of Man26E indicated that it had a saddle-shaped active center (Figure 3a).
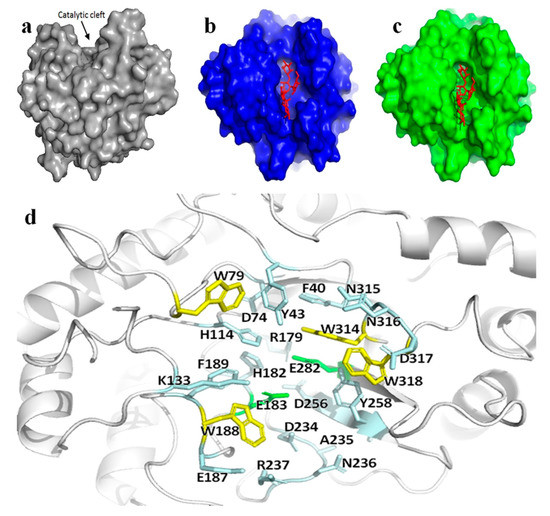
Figure 3.
The active apparatus of Man26E. (a) Schematic representation of surface electrostatic potential distribution for Man26E. The catalytic cleft on the upper part of the molecule was indicated by an arrow. The figure was made using the software PyMol 2.3.2; (b) sketch map of surface electrostatic potential distribution for Man26E docking Gal1Man4; (c) sketch map of surface electrostatic potential distribution for CjMan26C docking Gal1Man4. Gal1Man4 represented in red; (d) the structural elements participating in the Man26E active center. Residues were shown in ball-and-stick model. The conserved catalytic residues (Glu183, Glu282) and other hydrogen-bonding polar residues that surrounded the exposed tryptophan residues (containing Trp79, Trp188, Trp314 and Trp318) jointly constituted the active center of Man26E. The catalytic residues were shown in green, the hydrophilic and charged residues in light blue, and the exposed tryptophan residues in yellow.
In terms of the three-dimensional map (Figure 2a), six key loops in the active region of Man26E participated in binding in the saddle -shaped active site. CjMan26C (PDB ID. 2VX7) from Cellvibrio japonicus, a homologous β-mannanase (GH26) crystallographic structure in complex with mannose residues in PDB, was chosen as the template of substrate docking for Man26E. Man26E had 30.45% sequence homology (comparatively high in PDB) with CjMan26C, and the conserved active center of Man26E showed high similarity to that of CjMan26C. The substrate docking for Man26E and CjMan26C were shown in Figure 3b,c. The result indicated that all the residues located in the loops around the active region, except for Glu282 and Asp256, could form potential hydrogen bonds with the substrate. These residues included Trp318, Asp317, Asn316, Asn315, Trp314, Glu282, Tyr258, Asp256, Arg237, Asn236, Ala235, Asp234, Phe189, Trp188, Glu187, Glu183, His182, Arg179, Lys133, His114, Trp79, Asp74, Tyr43 and Phe40 (Figure 3d). The acid–base catalytic glutamate (Glu183) was at the loop between β-strand 4 and α- helix 6, and the nucleophilic glutamate (Glu282) was at the N-terminal end of β-strand 8. The binding of Man26E to mannan was mainly mediated by a hydrophobic plane composed of a strip of solvent-exposed aromatic rings of tryptophans, which included Trp318, Trp314, Trp188, and Trp79. Moreover, many hydrophilic and charged residues, such as Asp317, Asn316, Asn315, Tyr258, Asp256, Arg237, Asn236, Asp234, Glu187, His182, Arg179, Lys133, His114, Asp74 and Tyr43, surrounded the four aromatic rings in the binding site. All these residues were so oriented that their side chain groups could form hydrogen bonds with a polysaccharide, thus exercising certain functions in the binding of Man26E with mannan, and as a result, affecting the catalytic function.
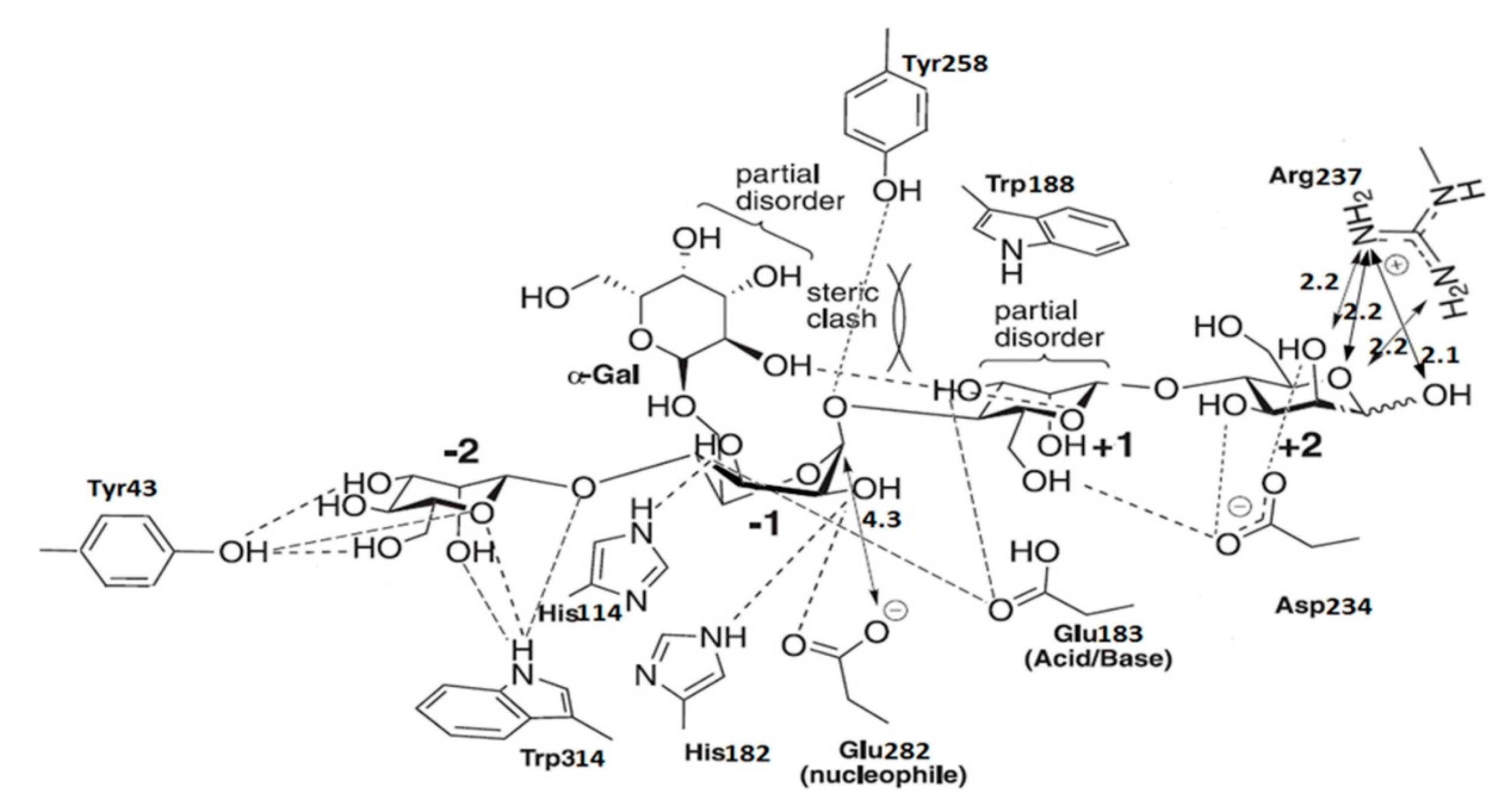
On the basis of the binding mode of CjMan26 C, Gal1Man4 was docked into the Man26E modeled structure (Figure 4).
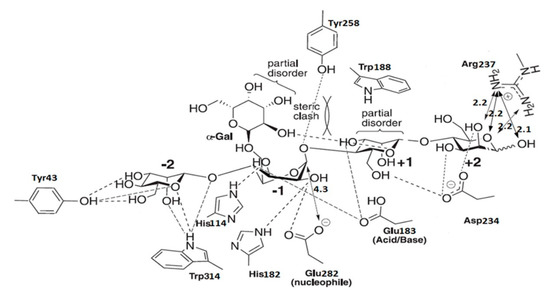
Figure 4.
Diagrammatic sketch of the interactions of Man26E and manno-oligosaccharide Gal1Man4.
The catalytic device, which was located at the −1 subsite, consisted of the catalytic acid/base Glu183, and nucleophile Glu282. The mannose at the −2 subsite made several interactions with Man26E. O2 produced a polar connection with the side chains of Trp314. The exocyclic oxygen made polar interactions with Trp314 and Tyr43, O3 and O6 also formed hydrogen bonds with Tyr43. The amino acid residues interacting with the substrate were less conserved a at the −2 subsite of CjMan26C and Man26E, and only the residue Trp314 in Man26E was structurally equivalent to Trp373 in CjMan26C. The catalytic nucleophile was likely to stabilize its position by interacting with Arg179 and Tyr258. His182 and Glu282 made hydrogen bonds with O2 of the −1 subsite sugar in its distorted conformation. Other two polar interactions between the −1 subsite sugar and Man26E occurred at O3 which formed polar links with the imidazole ring of His114 and carbonyl oxygen in the side chain of Glu183. Unusually, no more polar interactions were found between the −1 sugar and Man26E. The sugar at the +1 subsite made limited polar interactions with Man26E. O3 hydrogen bonded to carbonyl oxygen in the side chain of Glu183, whereas O6 produced a polar link to the side chain of Asp234 (Figure 3d). Man26E had a hydrophobic platform extending from the-1 subsite, in which Trp314 stacked against the sugar ring, while Trp188 took on the same planar orientation as the pyranose sugar at the +1 subsite (Figure 3d). At the +2 subsite, Asp234 interacted with O2 and O3, whereas Arg237 made polar connections with O1, O2 and the endocyclic oxygen at almost equal distances.
3.3. Gene Expression and Protein Purification
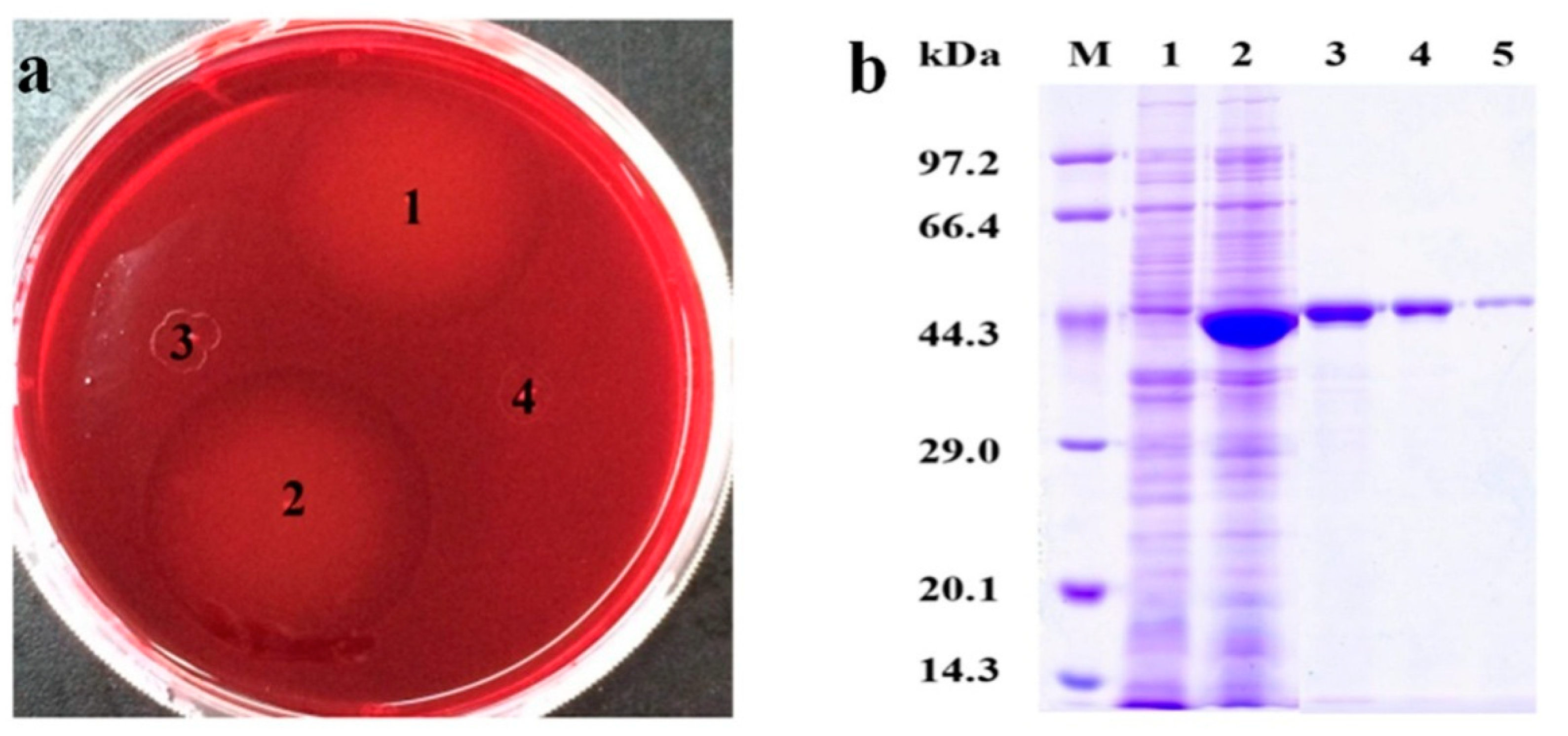
The screened positive transformants from antibiotic resistant plates were applied for Congo-red dyeing assay. Obvious hydrolytic circles presented around Escherichia coli BL21(DE3) harboring pET-28a (+) −Man26E colonies, but no hydrolytic circles around Escherichia coli BL21(DE3) harboring pET-28a (+) and Escherichia coli BL21(DE3) colonies were found, indicating that the positive transformants could produce β-mannanase and some enzymes were secreted out of the cells (Figure 5a).
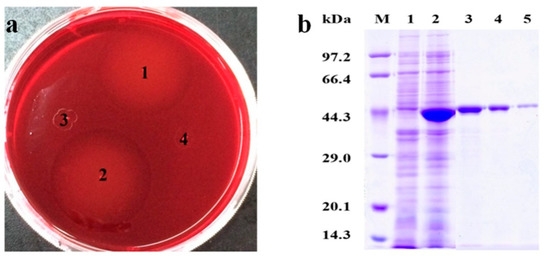
Figure 5.
The activity analysis of positive colonies dyed by Congo-red and SDS-PAGE analysis. (a) 1 and 2: Escherichia coli BL21(DE3) harboring pET-28a (+)-Man26E; 3: Escherichia coli BL21(DE3) harboring pET-28a (+); 4: Escherichia coli BL21(DE3). (b) SDS-PAGE analysis. Lane M: Marker; Lane 1: Escherichia coli BL21(DE3) control; Lane 2: Escherichia coli BL21(DE3) harboring pET-28a (+)-Man26E Lane 3, Lane 4 and Lane 5: The purified Man26E.
The transformant with a maximal hydrolytic circle was picked out and cultured in LB medium with 1 mmol L−1 IPTG at 32 °C, 200 rev min−1 for 8 h. The cell and the supernate were collected respectively for β-mannanase assay. The results showed that total β-mannanase activity reached 497.9 U/mg in the crude extract. SDS-PAGE assays revealed that the molecular mass of the recombinant protein was about 40 kDa (Figure 5b). The crude enzyme liquid was purified with a 6× His-Tagged Protein Purification kit, and The purified enzyme appeared a single band of approximately 40 kDa on SDS-PAGE gel (Figure 5b), which contained a 6× His-Tag (the Mw about 822.85 Da).
3.4. Enzymatic Properties of Recombinant Man26E
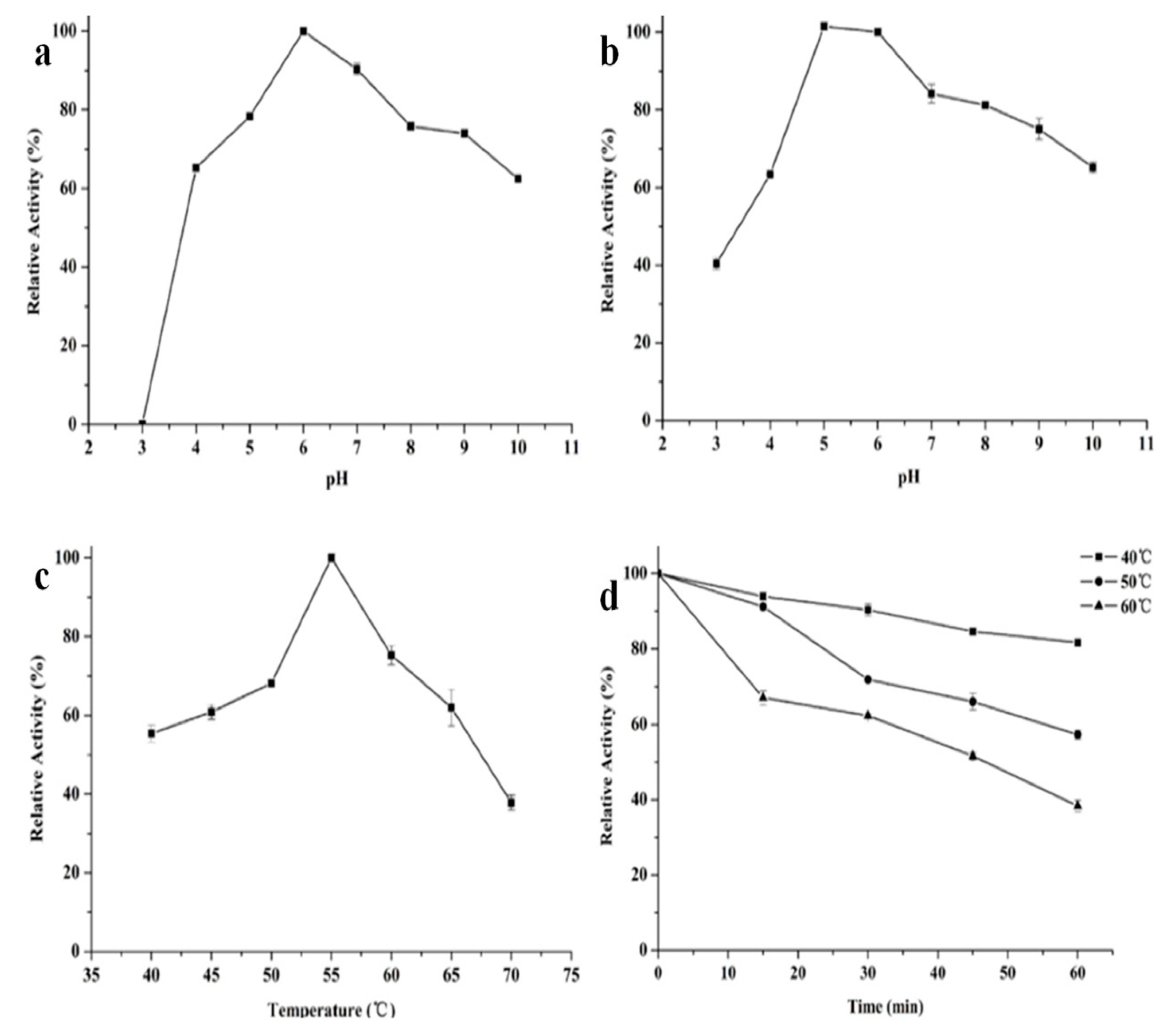
The properties of recombinant β-mannanase were investigated under different pHs (3–10) and temperature (40–70 °C) conditions. The recombinant Man26E had the maximal activity at pH 6.0 and kept over 75% of the highest activity at 5–9. The recombinant Man26E still showed more than 60% relative activity at pH 4.0 and pH 10.0, but no activity at pH 3.0 (Figure 6a). The enzyme activities at other pH values were significantly different from that at pH 6.0 (p < 0.01).
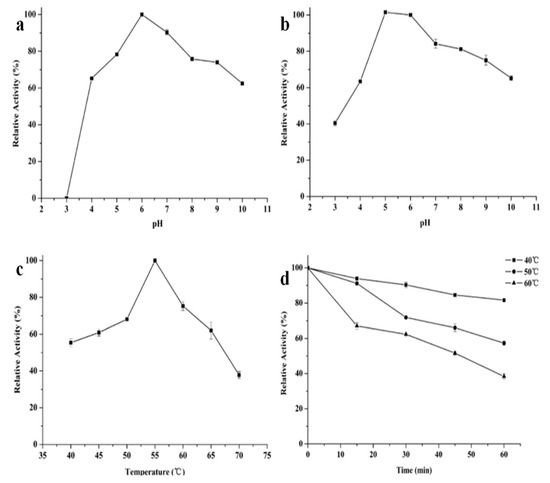
Figure 6.
Influence of pH and temperature on the enzyme. (a) enzyme activity at different pH values; (b) pH stability of the enzyme; (c) enzyme activity at different temperatures; (d) thermostability of the enzyme.
In addition, the recombinant Man26E exhibited a good pH stability and over 80% of the optimal activity was maintained after incubation at 50 °C and pH 5–8 for 60 min. Its stability rapidly decreased when pH values was below 5.0 and gradually declined in the range of pH 8.0–10.0, but remained about 40% residual activity at pH 3.0 and 65% residual activity at pH 10.0 after incubation at 50 °C for 60 min (Figure 6b). The optimal temperature for most β- mannanases is between 40 and 75 °C. As shown in Figure 6c, Man26E had the optimal temperature at 55 °C. The recombinant Man26E was active at 40–70 °C, and kept more than 60% relative activity in the temperature range of 45 to 65 °C. Man26E still remained about 40% relative activity at 70 °C. The enzyme activities at other temperatures were significantly different from that at 55 °C (p < 0.01). For testing thermostability, the purified enzyme was first pretreated at different temperatures for 60 minutes, and then the enzyme activity was determined. The enzyme was stable up to 40 °C for 60 min, and maintained more than 80% of its activity. At temperatures above 50 °C, its stability decreased gradually, but remained about 60% residual activity at 50 °C and nearly 40% residual activity at 60 °C for 60 min (Figure 6d).
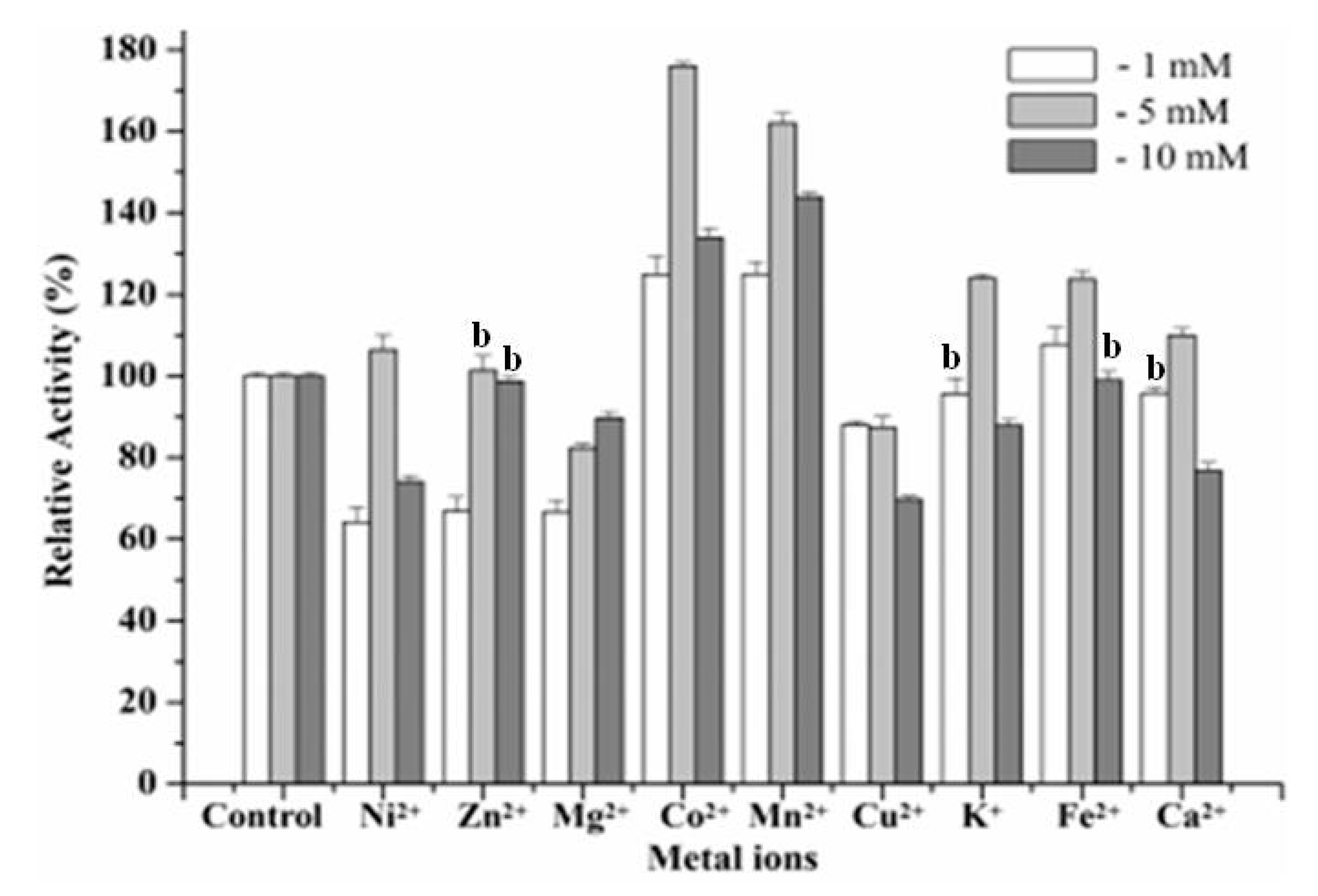
Different kinds of metal ions (1, 5 and 10 mmol L−1) were respectively added into the solution of the purified enzyme and preincubated at 55 °C for 10 min, and the residual activity was measured according to the standard method. Among these additives, the action of metal cations on the activity of the recombinant Man26E was quite interesting, as shown in Figure 7.
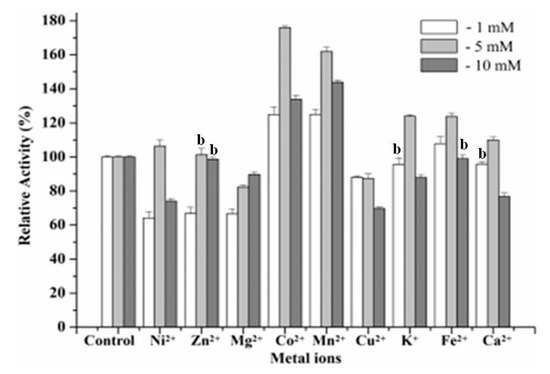
Figure 7.
Action of metal cations on enzyme activity. b No significant difference between the group and the control group.
For example, Co2+ and Mn2+ enhanced remarkably the β-mannanase activity, especially 5 mmol L−1 of Co2+ and Mn2+ increased enzyme activity by 72% and 60%, respectively. Fe2+ at 1 mmol L−1 and 5 mmol L−1 showed activation on the recombinant Man26E, but at 10 mmol L−1 had no effect. Ni2+, K+ and Ca2+ at 5 mmol L−1 had activation on the recombinant Man26E, whereas at 1 and 10 mmol L−1 displayed varying degrees of inhibition effect. All three concentrations of Mg2+ and Cu2+ showed inhibition on the recombinant Man26E. Zn2+ at 1 mmol L−1 reduced the enzyme activity of the recombinant Man26E, but at 5 and 10 mmol L−1 had little impact. Except for no significant difference (p > 0.05) between 1 mmol L−1 of K+ and the control group, between 1 mmol L−1 of Ca2+ and the control group, between 5 mmol L−1, 10 mmol L−1 of Zn2+ and the control group, 10 mmol L−1 of Fe2+ and the control group, other experimental groups showed significant differences (p < 0.01) with the control group.
3.5. Substrate Specificity and Kinetic Parameters
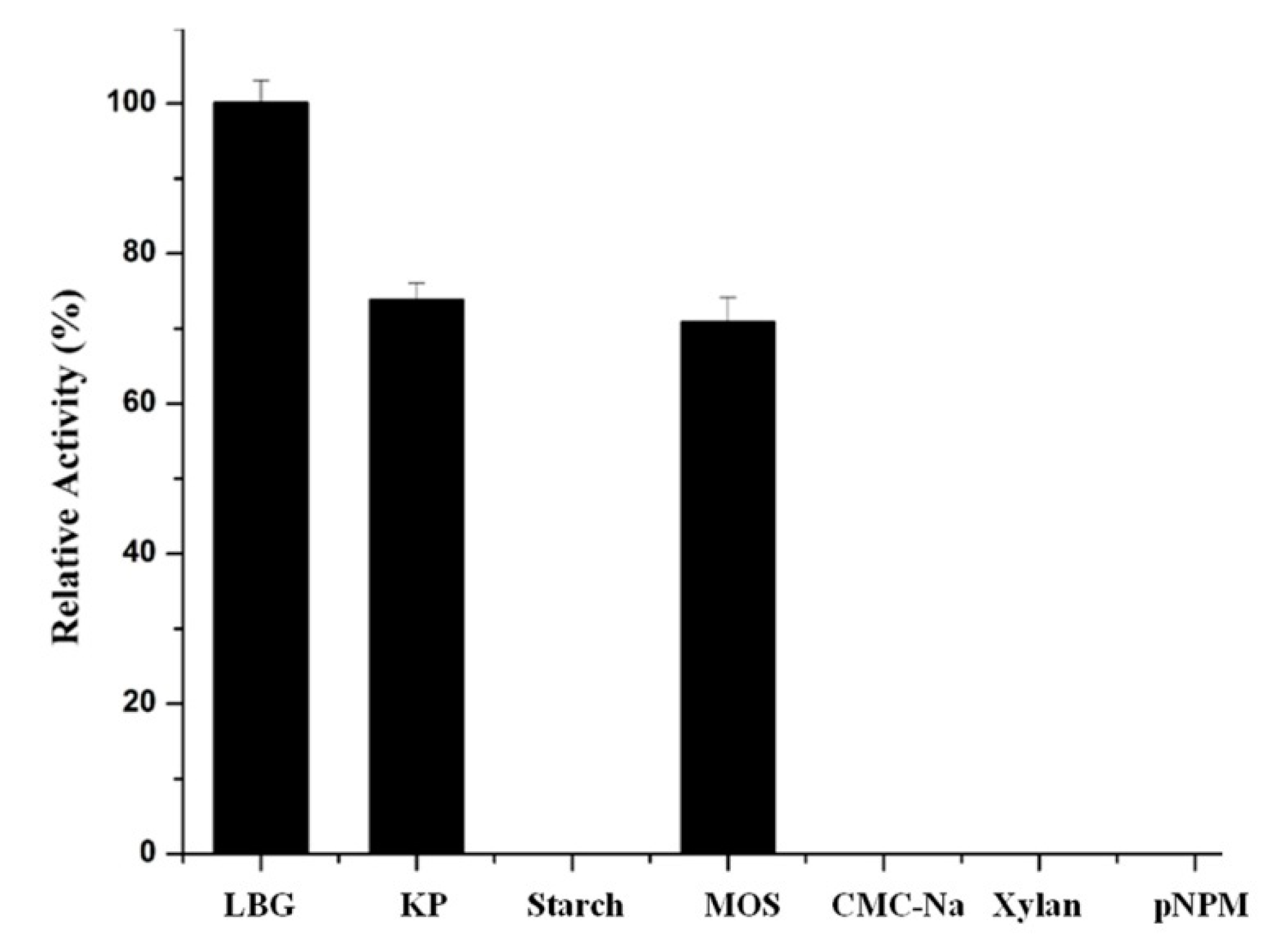
The ability for the recombinant Man26E to catalyze various substrates was examined (Figure 8). The enzyme had the highest activity on locust bean gum galactomannan (100%), followed by glucomannan from konjac powder (73.93%) and manno-oligosaccharides (70.89% relative activity). The LBG group displayed significant differences (p < 0.01) with the KP group and the MOS group, but the KP group and the MOS group showed no significant difference (p < 0.01). However, the recombinant Man26E had no activity on starch, CMC-Na, xylan and pNPM. Therefore, the enzyme acted specifically on the β-1,4-mannose glycoside bonds of mannopolysaccharides, and it suggested that the types of the branched chain in mannan might affect the enzymatic hydrolysis of polysaccharide. Locust bean gum and Konjacmannan are the most commonly applied substrates for assessing kinetic parameters. The kinetic parameters of the recombinant Man26E hydrolyzing locust bean gum were measured in 100 mmol L−1 citric acid buffer (pH 6.0) at 55 °C. Calculated according to the Lineweaver-Burk Plot, the values of Km and Vmax were 7.16 mg mL−1 and 508 U/mg, respectively.
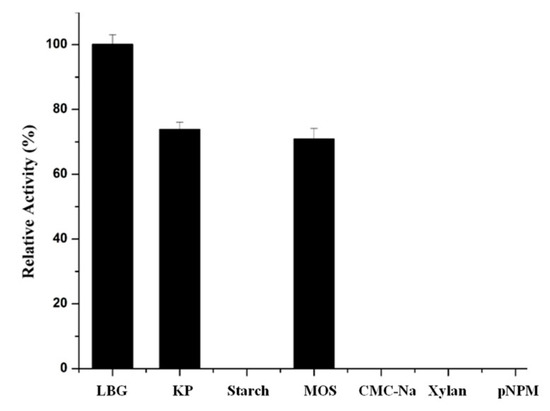
Figure 8.
Substrate specificity of the recombinant Man26E. LBG: locust bean gum; KP: konjac powder; MOS: manno-oligosaccharides; pNPM: p-nitrobenzene-d-pyranoside.
4. Discussion
Hemicellulose biomass is considered as a renewable source of sugar next to cellulose. In order to make better use of hemicellulose, more attention should be paid to hemicellulase producing bacteria. In recent years, culturable β-mannanase producing bacteria have been isolated from many sources. Enterobacter aerogenes, which is a gram-negative bacillary microorganism from Enterobacteriaceae, is widely found in water, soil and other environments. Enterobacter aerogenes is one of the members of the normal intestinal flora, but also an important conditional pathogenic bacteria. Enterobacter aerogenes can ferment a variety of sugars to produce clean and regenerated energy, such as ethanol [26], hydrogen [28], butanediol [35]. Here, we first reported a β-mannanase (Man26E) from Enterobacter aerogenes. Although a large number of highly homologous sequences with Man26E are found in NCBI library. However, at present, there are only a few reports about β-mannanase from Enterobacteriaceae strains, such as Klebsiella pneumoniae SS11 [36], Klebsiella oxytoca KUB-CW2-3 [37], Enterobacter sp. N18 [38], Enterobacter ludwigii MY271 [39], Enterobacter asburiae SD26 [40] and Pantoea agglomerans A021 [41]. Man26E belonged to GH26 family whose overall structure of domain possessed (β/α)8 TIM barrel domain [42]. The modeled 3D structure of Man26E was close to the standard for a good quality model which is 90% of total residues in the most favoured regions [43], indicating that it was reasonable and reliable. The modeled structure of Man26E is similar to those of BCman (PDB ID. 2QHA) [44] and BsMan26A (PDB ID. 3CBW) [45]. A major difference among the three structures lies in the number and the position of α- helix, β-strand and loop. In addition, no disulfide bond was found in Man26E, however, a disulfide bond exists between Cys75 at β-strand 2and Cys95 at α- helix 4 in BsMan26A, between Cys66 at β-strand 2and Cys 86 at α- helix 4 in BCman, respectively.
The conserved active center of Man26E showed high similarity to that of CjMan26C (PDB ID. 2VX7) from Cellvibrio japonicus, suggesting that the similar binding patterns of mannose are present in their active centers. The acid–base catalytic residue and the nucleophilic residue in GH26 family β-mannanases are highly conserved, and are all glutamate. For instance, they are Glu183and Glu282 in Man26E, Glu221 and Glu338 in CjMan26C [46], Glu176 and Glu275 in BsMan26A, respectively. The four enzymes contain a similar platform composed of four solvent-exposed tryptophan rings, which are Trp318, Trp314, Trp188 and Trp79 in Man26E, Trp302, Trp298, Trp172 and Trp72 in BCman, Trp373, Trp298, Trp226 and Trp167 in CjMan26C, Trp328, Trp324, Trp198, Trp98 in BsMan26A. Two of the four aromatic residues exhibit highly conserved properties in GH26 enzymes, they are Trp172 and Trp298 in BCman, Trp198 and Trp324 in BsMan26A, Trp373 and Trp226 in CjMan26C, which are equivalent to Trp188 and Trp314 in Man26E, respectively. The platform is involved in the binding of the active domain to the substrate. Yan et al. [43] have demonstrated that Trp298 and Trp172 are very important for binding and catalysis. We speculated that Trp188 and Trp314 may have similar functions.
The Man26E- Gal1Man4 and CjMan26C- Gal1Man4 complexes display the interactions between Gal1Man4 and the −1, −2, +1 and +2 subsites in the active region. However, the positions and quantities of hydrogen bond between amino acid residues and sugars are different. For example, at the −2 subsite, only Trp314 and Tyr43 in Man26E made limited interactions with the mannose, by comparison, an amount of interactions are found, involving Arg374, Gln385, Trp373 and Asp130 in CjMan26C. In GH26 β-mannanases, the residues binding the substrate at the −1 subsite show strict conservative feature, such as His114, His182, Glu183 and Glu282 in Man26E have structural equivalents in CjMan26C and CjMan26A, which correspond to His156 and His143, His182 His220 and His211, Glu221 and Glu212, Glu338 and Glu320, respectively [46]. At the +1 subsite, Glu183 and Asp234 of Man26E formed limited polar interactions with the sugar, whereas in CjMan26C and CjMan26A, the equivalents to Asp234 of Man26E are Asp266 and Asn257, respectively. The +2 site is quite different among Man26E, CjMan26C and CjMan26A. In Man26E, Asp234 and Arg237 produced polar connections with the sugar. For CjMan26C, Ser268 and Arg269 generates interactions with the sugar, however, the equivalent loop in CjMan26A does not exist any residues near +2 subsite.
The activity (497.9 U/mg) of the recombinant Man26E was higher than those of β-mannanases from Bacillus circulans K-1 (481.55 U/mg) [47] and Bacillus sp. JAMB-750 (36.3 U/mg) [48], however, lower than those of β-mannanase from Clostridium thermocellum (520 U/mg) [49] and Pantoea agglomerans A021 (514 U/mg). The molecular mass of Man26E was predicted to be 38.61 kDa and the purified enzyme was about 40 kDa evaluated by SDS-PAGE. It has been reported that the molecular weights of β- mannanases from a variety of sources are in the range of 30–80 kDa [50]. Exceptionally, a GH26 β-mannanase from Clostridium thermocellum shows low molecular weight of 15 kDa [51], and the other β-mannanase from Thermoanaerobacterium polysaccharolyticum has a high molecular weight of 120 kDa [52]. The optimal pH value (6.5) of the recombinant Man26E is comparable to those of β-mannanases from Bacillus subtilis WL7 [53], Pantoea agglomerans A021, Bacillus subtilis WL-3 [54], Bacillus sp. SWU60 [55] and Bacillus subtilis (TBS2) [56]. Most bacterial β-mannanases have the highest activity from neutral to alkaline pH. For instance, β-mannanases (GH 26) from Bacillus sp. CFR1601 [57], Bacillus subtilis BCC41051 [58], Bacillus licheniformis [59] and Bacillus subtilis B23 [20] are optimally active at pH 7. β-mannanases (GH 26) from Bacillus N16-5 (GH 26) [60], Bacillus licheniformis THCM 3.1 [61] and Bacillus sp. strain JAMB-750 [62] are optimally active at pH 9.6, 9.0 and 10.0, respectively. Bacterial β-mannanases show remarkable stability in the range of pH 6–9 [50]. Exceptionally, the activity of β-mannanase from Acinetobacter sp. ST 1-1 is maintained over 80% in the range of pH 3–10 [63]. The optimal temperature (55 °C) of Man26E is similar to β-mannanases from Bacillus sp. CFR1601, Pantoea agglomerans A021, Erwinia carotovora CXJZ95-198 [64] and Bacillus subtilis WL7. However, β-mannanases from Bacillus N16-5 (GH 26) and Caldicellulosiruptor Rt 8B.4 (GH 26) [6] show the highest activity above 70 °C. Interestingly, β-mannanase from Thermotoga neapolitana 5068 exhibits maximum activity at 92 °C [65]. The recombinant Man26E is less stable than those of β-mannanases from Pantoea agglomerans A021, Bacillus subtilis B23, Bacillus sp. SWU60 and Enterobacter sp. N18. However, the thermostability of Man26E is better than those of β-mannanases from Cellulosimicrobium sp. strain HY-13 [66] and Sphingomonas sp. ERB1–3 [67]. Similar results to Man26E have been reported that Co2+ shows positive action on the activities of the recombinant β-mannanases from Enterobacter sp strain N18 and Bacillus subtilis WY34 [68], and Mn2+ performs positive effect on the β-mannanases from Bacillus sp. N16-5 and Bacillus subtilis WY34. The activation of Fe2+ on the recombinant Man26E is different from several published β-mannanases. For instance, Fe2+ inhibits the activity of β-mannanase from Paenibacillus cookie [69], however, Fe2+ makes no difference in β-mannanases from Bacillus subtilis WY34 and Cellulosimicrobium sp. strain HY-13. The inhibition of Mg2+ and Cu2+ on the recombinant Man26E is in consistent with several reported β-mannanases [38,70,71].
The substrate specificity of the recombinant Man26E is the same as those of Bacillus circulans NT 6.7 [72], Bacillus subtilis WY34, Bacillus sp. N16-5, Bacillus stearothermophilus [73], Bacillus circulans CGMCC 1416 [74] and Bacillus subtilis WL-3, which show higher activity toward LBG than toward KGM. By comparison, the substrate specificity of Man26E is different from those of β-mannanases from B. licheniformis mannanase [48], Bacillus sp. SWU60, Bacillus subtilis WD23 [75] and Paenibacillus cookie, which show more activity against glucomannan than against galactomannan. It has been reported that the Km values of β-mannanase are between 0.0095 and 27.4 mg mL−1, and the Vmax values are ranging from 29 to 16,666.7 μmol mL−1 or mg min−1 [50]. The Km value (7.16 mg mL−1) of the recombinant Man26E for LBG is similar to the values (7.22 mg mL−1) reported for a β-mannanase from Bacillus nealsonii PN-11 [76]. The Km value of the recombinant Man26E is significantly lower than those of BCman from Bacillus subtilis Z-2 (10.2 mg mL−1) and Bacillus subtilis YH12 (30 mg mL−1) [77]. Distinguishingly, the Km value of the β-mannanase from Paenibacillus cookie is 0.0095 mg mL−1 for LBG.
Man26E displayed many excellent features, for example, acid and alkali tolerance, high stability over a wide range of neutral-to-alkaline pH, good thermostability, high stability in the presence of various metal ions. These characteristics suggested that Man26E could have great potential for commercial application in the biofuels, the food/feed and paper/pulp and other industries. In addition, the simulation of the three-dimensional structure and the analysis of the active domain of this mannanase will help to understand the structure- function relationship, and lay a foundation for the molecular modification of the enzyme to improve its enzymatic properties.
Author Contributions
This paper was accomplished based on collaborative work of the authors. Conceptualization, T.C. and H.L.; methodology, T.C.; software, T.C.; validation, H.L., J.L. and T.C.; formal analysis, H.L.; investigation, H.L.; resources, J.L.; data curation, H.L. and J.L.; writing—original draft preparation, H.L.; writing—review and editing, T.C.; visualization, T.C.; supervision, T.C.; project administration, T.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
No grants in this work. The authors thank Lixin Luo from School of Biological Science and Bioengineering, South China University of Technology because he useful suggestions and language editing help us greatly improve the quality of our manuscript.
Conflicts of Interest
We declare that we have no conflict of interest.
References
- Danaher, B.; Dhanasobhon, S.; Smith, M.D.; Telang, R. Microbial mannanases: An overview of production and applications. Crit. Rev. Biotechnol. 2007, 27, 197–216. [Google Scholar]
- Timell, T.E. Recent progress in the chemistry of wood hemicelluloses. Wood Sci. Technol. 1967, 1, 45–70. [Google Scholar] [CrossRef]
- Petkowicz, C.D.O.; Reicher, F.; Chanzy, H.; Taravel, F.; Vuong, R. Linear mannan in the endosperm of Schizolobium amazonicum. Carbohydr. Polym. 2001, 44, 107–112. [Google Scholar] [CrossRef]
- Malgas, S.; Van Dyk, J.S.; Pletschke, B.I. A review of the enzymatic hydrolysis of mannans and synergistic interactions between β-mannanase, β-mannosidase and α-galactosidase. World J. Microbiol. Biotechnol. 2015, 31, 1167–1175. [Google Scholar] [CrossRef]
- Hilge, M.; Gloor, S.M.; Rypniewski, W.; Sauer, O.; Heightman, T.D.; Zimmermann, W.; Winterhalter, K.; Piontek, K. High-resolution native and complex structures of thermostable β-mannanase from Thermomonospora fusca—Substrate specificity in glycosyl hydrolase family 5. Structure 1998, 6, 1433–1444. [Google Scholar] [CrossRef]
- Sunna, A. Modular organisation and functional analysis of dissected modular β-mannanase CsMan26 from Caldicellulosiruptor Rt8B.4. Appl. Microbiol. Biotechnol. 2010, 86, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Gübitz, G.M.; Sachslehner, A.; Haltrich, D. Microbial Mannanases: Substrates, Production, and Applications. ACS Symp. Ser. 2001, 45, 523–538. [Google Scholar]
- Sánchez, C. Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnol. Adv. 2009, 27, 185–194. [Google Scholar] [CrossRef]
- Khanongnuch, C.; Lumyong, S.; Ooi, T.; Kinoshita, S. A non-cellulase producing strain of Bacillus subtilis and its potential use in pulp biobleaching. Biotechnol. Lett. 1999, 21, 61–63. [Google Scholar] [CrossRef]
- Bettiol, J.L.P.; Boutique, J.P.; Gualco, L.M.P.; Johnston, J.P. Non-Aqueous Liquid Detergent Compositions Comprising a Borate-Releasing Compound and a Mannanase. EP Patent EP1059351A1, 13 December 2000. [Google Scholar]
- Buchert, J.; Salminen, J.; Siika-Aho, M.; Ranua, M.; Viikari, L. The Role ofTrichoderma reeseiXylanase and Mannanase in the Treatment of Softwood Kraft Pulp Prior to Bleaching. Holzforschung 1993, 47, 473–478. [Google Scholar] [CrossRef]
- Smith, D.L., Jr.; Nagy, T.R.; Wilson, L.S.; Dong, S.; Barnes, S.; Allison, D.B. The Effect of Mannan Oligosaccharide Supplementation on Body Weight Gain and Fat Accrual in C57Bl/6J Mice. Obesity 2010, 18, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.S.; Puri, N.; Sharma, P.; Gupta, N. Mannanases: Microbial sources, production, properties and potential biotechnological applications. Appl. Microbiol. Biotechnol. 2012, 93, 1817–1830. [Google Scholar] [CrossRef] [PubMed]
- Araujo, A.; Ward, O. Hemicellulases of Bacillusspecies: Preliminary comparative studies on production and properties of mannanases and galactanases. J. Appl. Bacteriol. 1990, 68, 253–261. [Google Scholar] [CrossRef]
- Bouquelet, S.; Spik, G.; Montreuil, J. Properties of a β-d-mannosidase from Aspergillus niger. Biochim. Biophys. Acta 1978, 522, 521–530. [Google Scholar] [CrossRef]
- Lüthi, E.; Jasmat, N.B.; Grayling, R.A.; Love, D.R.; Bergquist, P.L. Cloning, sequence analysis, and expression in Escherichia coli of a gene coding for a beta-mannanase from the extremely thermophilic bacterium “Caldocellum saccharolyticum”. Appl. Environ. Microbiol. 1991, 57, 694–700. [Google Scholar] [CrossRef]
- Sunna, A.; Gibbs, M.D.; Chin, C.W.J.; Nelson, P.J.; Bergquist, P.L. A Gene Encoding a Novel Multidomain β-1,4-Mannanase from Caldibacillus cellulovorans and Action of the Recombinant Enzyme on Kraft Pulp. Appl. Environ. Microbiol. 2000, 66, 664–670. [Google Scholar] [CrossRef]
- Mai, V.; Wiegel, J. Advances in development of a genetic system for Thermoanaero bacterium spp.: Expression of genes encoding hydrolytic enzymes, development of a second shuttle vector, and integration of genes into the chromosome. Appl. Environ. Microbiol. 2000, 66, 4817–4821. [Google Scholar] [CrossRef] [PubMed]
- Park, G.G.; Kusakabe, I.; Komatsu, Y.; Kobayashi, H.; Yasui, T.; Murakami, K. Purification and some properties of β-mannanase from Penicillium purpurogenum. Agric. Biol. Chem. 1987, 51, 2709–2716. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, Y.; Nie, X.; Yang, W.; Wu, Y. Comparison of expression systems for the extracellular production of mannanase Man23 originated from Bacillus subtilis B23. Microb. Cell Fact. 2013, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.D.; Yu, K.O.; Kim, S.W.; Han, S.O. A celluloytic complex from Clostridium cellulovorans consisting of mannanase B and endoglucanase E has synergistic effects on galactomannan degradation. Appl. Microbiol. Biotechnol. 2011, 90, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.M.; Hong, S.Y.; Lee, S.M.; Kim, Y.H.; Kahng, G.G.; Kim, H.; Yun, H.D. A cel44C-man26A gene of endophytic Paenibacillus polymyxa GS01 has multi-glycosyl hydrolases in two catalytic domains. Appl. Microbiol. Biotechnol. 2006, 73, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Hekmat, O.; Lo Leggio, L.; Rosengren, A.; Kamarauskaite, J.; Kolenova, K.; Stilbrand, H. Rational engineering of mannosyl binding in the distal glycone subsites of cellulomonas fimi endo-β-l,4-mannanase: Mannosyl binding promoted at subsite -2 and demoted at subsite -3. Biochemistry 2010, 49, 4884–4896. [Google Scholar] [CrossRef]
- Celińska, E.; Grajek, W. Biotechnological production of 2,3-butanediol—Current state and prospects. Biotechnol. Adv. 2009, 27, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-M.; Kim, Y.H.; Oh, M.-K. Formate and Nitrate Utilization in Enterobacter aerogenes for Semi-Anaerobic Production of Isobutanol. Biotechnol. J. 2017, 12, 1700121. [Google Scholar] [CrossRef] [PubMed]
- Sunarno, J.N.; Prasertsan, P.; Duangsuwan, W.; Cheirsilp, B.; Sangkharak, K. Improve biotransformation of crude glycerol to ethanol of Enterobacter aerogenes by two-stage redox potential fed-batch process under microaerobic environment. Biomass Bioenergy 2020, 134, 105503. [Google Scholar] [CrossRef]
- Boshagh, F.; Rostami, K.; Moazami, N. Immobilization of Enterobacter aerogenes on carbon fiber and activated carbon to study hydrogen production enhancement. Biochem. Eng. J. 2019, 144, 64–72. [Google Scholar] [CrossRef]
- Lin, R.; Deng, C.; Zhen, G.; Murphy, J.D. Low concentrations of furfural facilitate biohydrogen production in dark fermentation using Enterobacter aerogenes. Renew. Energy 2020, 150, 23–30. [Google Scholar] [CrossRef]
- Liu, S.; Cui, T.; Song, Y. Expression, homology modeling and enzymatic characterization of a new β-mannanase belonging to glycoside hydrolase family 1 from Enterobacter aerogenes B19. Microb. Cell Fact. 2020, 19, 1–19. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Teather, R.M.; Wood, P.J. Use of congo red-polysaccharide interactions in enumeration and characterization of from the bovine rument. Appl. Environ. Microbiol. 1982, 43, 777–780. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Lineweaver, H.; Burk, D. The Determination of Enzyme Dissociation Constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Joo, J.; Lee, S.J.; Yoo, H.Y.; Kim, Y.; Jang, M.; Lee, J.; Han, S.O.; Kim, S.W.; Park, C. Improved fermentation of lignocellulosic hydrolysates to 2,3-butanediol through investigation of effects of inhibitory compounds by Enterobacter aerogenes. Chem. Eng. J. 2016, 306, 916–924. [Google Scholar] [CrossRef]
- Singh, S.; Singh, G.; Khatri, M.; Kaur, A.; Arya, S.K. Thermo and alkali stable β-mannanase: Characterization and application for removal of food (mannans based) stain. Int. J. Biol. Macromol. 2019, 134, 536–546. [Google Scholar] [CrossRef]
- Pongsapipatana, N.; Damrongteerapap, P.; Chantorn, S.; Sintuprapa, W.; Keawsompong, S.; Nitisinprasert, S. Molecular cloning of kman coding for mannanase from Klebsiella oxytoca KUB-CW2-3 and its hybrid mannanase characters. Enzym. Microb. Technol. 2016, 89, 39–51. [Google Scholar] [CrossRef]
- You, J.; Liu, J.-F.; Yang, S.-Z.; Mu, B.-Z. Low-temperature-active and salt-tolerant β-mannanase from a newly isolated Enterobacter sp. strain N18. J. Biosci. Bioeng. 2016, 121, 140–146. [Google Scholar] [CrossRef]
- Yang, M.; Wang, C.; Du, X.; Lin, J.; Cai, J. Characterization of endo-β-mannanase from Enterobacter ludwigii MY271 and application in pulp industry. Bioprocess Biosyst. Eng. 2017, 40, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, S.; Singh, S.; Singh, G.; Khatri, M.; Arya, S.K. Biochemical characterization and thermodynamic study of β-mannanase from Enterobacter asburiae. Biocatal. Agric. Biotechnol. 2019, 20, 101211. [Google Scholar] [CrossRef]
- Wang, J.; Shao, Z.; Hong, Y.; Li, C.; Fu, X.; Liu, Z. A novel β-mannanase from Pantoea agglomerans A021: Gene cloning, expression, purification and characterization. World J. Microbiol. Biotechnol. 2010, 26, 1777–1784. [Google Scholar] [CrossRef]
- Hilge, M.; Perrakis, A.; Abrahams, J.P.; Winterhalter, K.; Pionteka, K.; Gloor, S.M. Structure elucidation of β-mannanase: From the electron-density map to the DNA sequence. Acta Crystallogr. D 2001, 57, 37–43. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Yan, X.-X.; An, X.-M.; Gui, L.-L.; Liang, D. From Structure to Function: Insights into the Catalytic Substrate Specificity and Thermostability Displayed by Bacillus subtilis Mannanase BCman. J. Mol. Biol. 2008, 379, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Tailford, L.E.; Ducros, V.M.-A.; Flint, J.E.; Roberts, S.M.; Morland, C.; Zechel, D.L.; Smith, N.; Bjo̷rnvad, M.E.; Borchert, T.V.; Wilson, K.S.; et al. Understanding How Diverse β-Mannanases Recognize Heterogeneous Substrates. Biochemistry 2009, 48, 7009–7018. [Google Scholar] [CrossRef]
- Cartmell, A.; Topakas, E.; Ducros, V.M.A.; Suits, M.D.L.; Davies, G.J.; Gilbert, H.J. The Cellvibrio japonicus Mannanase CjMan26C displays a unique exo-mode of action that is conferred by subtle changes to the distal region of the active site. J. Biol. Chem. 2008, 283, 34403–34413. [Google Scholar] [CrossRef]
- Songsiriritthigul, C.; Buranabanyat, B.; Haltrich, D.; Yamabhai, M. Efficient recombinant expression and secretion of a thermostable GH26 mannan endo-1,4-β-mannosidase from Bacillus licheniformis in Escherichia coli. Microb. Cell Fact. 2010, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Hatada, Y.; Takeda, N.; Hirasawa, K.; Ohta, Y.; Usami, R.; Yoshida, Y.; Grant, W.D.; Ito, S.; Horikoshi, K. Sequence of the gene for a high-alkaline mannanase from an alkaliphilic Bacillus sp. strain JAMB-750, its expression in Bacillus subtilis and characterization of the recombinant enzyme. Extremophiles 2005, 9, 497–500. [Google Scholar] [CrossRef]
- Kurokawa, J.; Hemjinda, E.; Arai, T.; Karita, S.; Kimura, T.; Sakka, K.; Ohmiya, K. Sequence of the Clostridium thermocellum mannanase gene man26B and characterization of the translated product. Biosci. Biotechnol. Biochem. 2001, 65, 548–554. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Kapoor, M. Production, properties, and applications of endo-β-mannanases. Biotechnol. Adv. 2017, 35, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Verma, A.K.; Tingirikari, J.R.; Shukla, R.; Goyal, A. Recovery and Purification of Oligosaccharides from Copra Meal by Recombinant Endo-β-mannanase and Deciphering Molecular Mechanism Involved and Its Role as Potent Therapeutic Agent. Mol. Biotechnol. 2015, 57, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Cann, I.K.O.; Kocherginskaya, S.; King, M.R.; White, B.A.; Mackie, R.I. Molecular Cloning, Sequencing, and Expression of a Novel Multidomain Mannanase Gene from Thermoanaerobacterium polysaccharolyticum. J. Bacteriol. 1999, 181, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Eom, G.T.; Oh, J.Y.; Park, J.H.; Jegal, J.; Song, J.K. Secretory production of enzymatically active endo-β-1,4-mannanasefrom Bacillus subtilis by ABC exporter in Escherichia coli. Process Biochem. 2016, 51, 999–1005. [Google Scholar] [CrossRef]
- Yoon, K.-H.; Chung, S.; Lim, B.-L. Characterization of the Bacillus subtilis WL-3 mannanase from a recombinant Escherichia coli. J. Microbiol. 2008, 46, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Seesom, W.; Thongket, P.; Yamamoto, T.; Takenaka, S.; Sakamoto, T.; Sukhumsirichart, W. Purification, characterization, and overexpression of an endo-1,4-β-mannanase from thermotolerant Bacillus sp. SWU60. World J. Microbiol. Biotechnol. 2017, 33, 53. [Google Scholar] [CrossRef]
- Luo, Z.; Miao, J.; Li, G.; Du, Y.; Yu, X. A Recombinant Highly Thermostable β-Mannanase (ReTMan26) from Thermophilic Bacillus subtilis (TBS2) Expressed in Pichia pastoris and Its pH and Temperature Stability. Appl. Biochem. Biotechnol. 2017, 182, 1259–1275. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Kapoor, M. Recombinant GH-26 endo-mannanase from Bacillus sp. CFR1601: Biochemical characterization and application in preparation of partially hydrolysed guar gum. Lebensm. Wiss Technol. 2015, 64, 809–816. [Google Scholar] [CrossRef]
- Chung, S.; Lim, B.L. Characterizationg, gene cloning, and heterologous expression of β-mannanase from a thermophilic Bacillus subtilis. J. Microbiol. 2011, 49, 86–93. [Google Scholar]
- Zhang, J.; He, Z.; Hu, K. Purification and characterization of β-mannanase from Bacillus licheniformis for industrial use. Biotechnol. Lett. 2000, 22, 1375–1378. [Google Scholar] [CrossRef]
- Lin, S.-S.; Dou, W.-F.; Xu, H.-Y.; Li, H.-Z.; Xu, Z.-H.; Ma, Y.-H. Optimization of medium composition for the production of alkaline β-mannanase by alkaliphilic Bacillus sp. N16-5 using response surface methodology. Appl. Microbiol. Biotechnol. 2007, 75, 1015–1022. [Google Scholar] [CrossRef]
- Kanjanavas, P.; Khawsak, P.; Pakpitcharoen, A.; Areekit, S.; Sriyaphai, T.; Pothivejkul, K.; Santiwatanakul, S.; Matsui, K.; Kajiwara, T.; Chansiri, K. Over-expression and characterization of the alkalophilic, organic solventtolerant, and thermotolerant endo-1,4-mannanase from Bacillus licheniformis isolate THCM 3.1. Sci. Asia 2009, 35, 17–23. [Google Scholar] [CrossRef]
- Takeda, N.; Hirasawa, K.; Uchimura, K. Purification and enzymatic properties of a highly alkaline mannanase from alkaliphilic Bacillus sp. strain JAMB-750. Int. J. Biol. Macromol. 2004, 4, 67–74. [Google Scholar]
- Titapoka, S.; Keawsompong, S.; Haltrich, D.; Nitisinprasert, S. Selection and characterization of mannanase-producing bacteria useful for the formation of prebiotic manno-oligosaccharides from copra meal. World J. Microbiol. Biotechnol. 2008, 24, 1425–1433. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Chen, X. Cloning and expression of a mannanase gene from Erwinia carotovora CXJZ95-198. Ann. Microbiol. 2007, 57, 623–628. [Google Scholar] [CrossRef]
- Duffaud, G.D.; McCutchen, C.M.; Leduc, P.; Parker, K.N.; Kelly, R.M. Purification and characterization of extremely thermostable β-mannanase, β-mannosidase, and β-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl. Environ. Microbiol. 1997, 63, 169–177. [Google Scholar] [CrossRef]
- Kim, D.Y.; Ham, S.-J.; Lee, H.J.; Kim, Y.-J.; Shin, D.-H.; Rhee, Y.H.; Son, K.-H.; Park, H.-Y. A highly active endo-β-1,4-mannanase produced by Cellulosimicrobium sp. strain HY-13, a hemicellulolytic bacterium in the gut of Eisenia fetida. Enzym. Microb. Technol. 2011, 48, 365–370. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, R.; Gao, Y.; Li, J.; Tang, X.; Mu, Y.; Wang, F.; Li, C.; Dong, Y.; Huang, Z. Novel low-temperature-active, salt-tolerant and proteases-resistant endo-1,4-β-mannanase from a new Sphingomonas strain. J. Biosci. Bioeng. 2012, 113, 568–574. [Google Scholar] [CrossRef]
- Jiang, Z.; Wei, Y.; Li, D.; Li, L.; Chai, P.; Kusakabe, I. High-level production, purification and characterization of a thermostable beta-mannanase from the newly isolated Bacillus subtilis WY34. Carbohydr. Polym. 2006, 66, 88–96. [Google Scholar] [CrossRef]
- Yin, L.-J.; Tai, H.-M.; Jiang, S.-T. Characterization of Mannanase from a Novel Mannanase-Producing Bacterium. J. Agric. Food Chem. 2012, 60, 6425–6431. [Google Scholar] [CrossRef]
- Katrolia, P.; Yan, Q.; Zhang, P.; Zhou, P.; Yang, S.; Jiang, Z. Gene Cloning and Enzymatic Characterization of an Alkali-Tolerant Endo-1,4-β-mannanase from Rhizomucor miehei. J. Agric. Food Chem. 2013, 61, 394–401. [Google Scholar] [CrossRef]
- Zang, H.; Xie, S.; Wu, H.; Wang, W.; Shao, X.; Wu, L.; Rajer, F.U.; Gao, X. A novel thermostable GH5_7 beta-mannanase from Bacillus pumilus GBSW19 and its application in manno-oligosaccharides (MOS) production. Enzym. Microb. Tech. 2015, 78, 1–9. [Google Scholar] [CrossRef]
- Piwpankaew, Y.; Sakulsirirat, S.; Nitisinprasert, S.; Nguyen, T.-H.; Nguyen, T.-H.; Keawsompong, S. Cloning, secretory expression and characterization of recombinant β-mannanase from Bacillus circulans NT 6.7. SpringerPlus 2014, 3, 430. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Talbot, G.; Sygusch, J. Purification and characterization of thermostable beta-mannanase and alpha-galactosidase from Bacillus stearothermophilus. Appl. Environ. Microbiol. 1990, 56, 3505–3510. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, P.; Meng, K.; Wang, Y.; Luo, H.; Wu, N.; Fan, Y.; Yao, B. Gene cloning, expression, and characterization of a novel beta-mannanase from Bacillus circulans CGMCC 1416. J. Microbiol. Biotechnol. 2008, 18, 160–166. [Google Scholar] [PubMed]
- Li, H.L.; Liu, Z.Y.; Wang, C.L.; Huang, S.C.; Zhao, M. Secretory expression and characterization of a novel thermo-stable, salt-tolerant endo-1,4-β-mannanase of Bacillus subtilis WD23 by Pichia pastoris. Eur. Food Res. Technol. 2015, 240, 671–677. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Sharma, P.; Puri, N.; Gupta, N. Purification and characterization of an alkali-thermostable β-mannanase from Bacillus nealsonii PN-11 and its application in mannooligosaccharides preparation having prebiotic potential. Eur. Food Res. Technol. 2014, 238, 927–936. [Google Scholar] [CrossRef]
- Liu, H.-X.; Gong, J.-S.; Li, H.; Lu, Z.-M.; Li, H.; Qian, J.-Y.; Xu, Z.-H.; Shi, J.-S. Biochemical characterization and cloning of an endo-1,4-β-mannanase from Bacillus subtilis YH12 with unusually broad substrate profile. Process. Biochem. 2015, 50, 712–721. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).