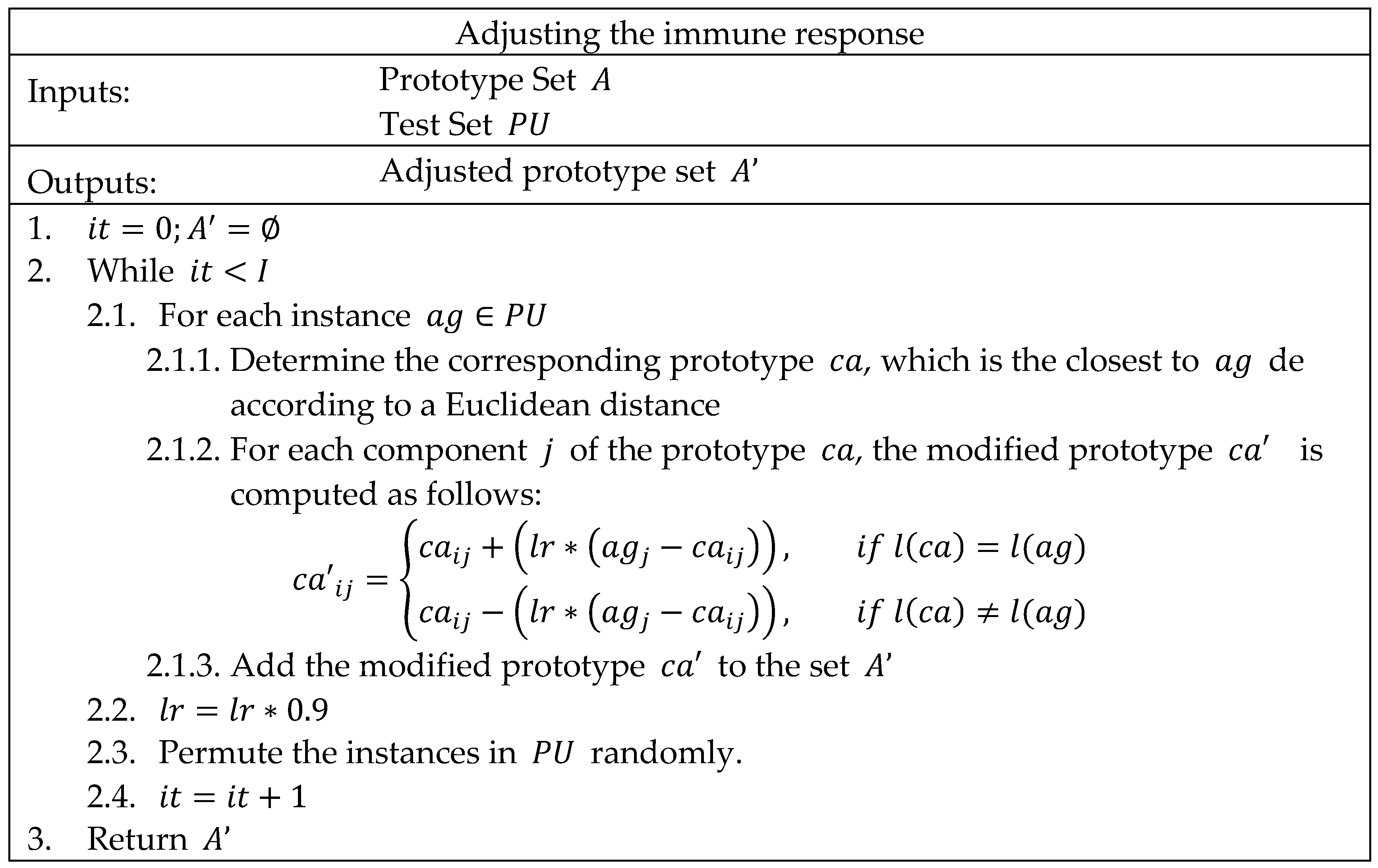
3.1. The Human Immune System
The immune system is a biological system that protects an organism against pathogens, such as biological, chemical, or intern hazards [
33]. The immune system has two main functions: to recognize substances foreign to the body (also called antigens), and to react against them. These substances may be microorganisms that cause infectious diseases, transplanted organs or tissues of another individual, or tumors. The proper functioning of the immune system provides protection against infectious diseases and can protect a person from cancer.
An antigen is any substance that causes the body to create antibodies. It is a substance capable of inducing an immune response. Among the properties of antigens, the following can be highlighted [
34]:
They have to possess the quality of strangers to the human body. That is, the antigens may come from outside (exogenous) or they may be generated in our body (endogenous);
Not all trigger an immune response, because of the amount of inoculum that is introduced. A considerable proportion is needed to trigger a response;
The immune response is under genetic control. Because of this, the immune system decides whether to respond or not, and against whom it will respond and against whom it will not;
The basic structure has an important relevance. This is because T and B lymphocytes are involved in cell-mediated immunity: T lymphocytes regulate the entire immune response, and B lymphocytes are secondary;
Some antigens must be recognized by the T lymphocytes to give a response, which are called antigens of thymus-dependent type; there are others that do not—it is enough for them to reach the B lymphocyte to be recognized as such. These are called thymus-independent antigens.
Antigenic macromolecules have two fundamental elements: the antigenic carrier, which is a macroprotein, and the antigenic determinants (epitopes), which are small molecules attached to them with a particular spatial configuration that can be identified by an antibody; therefore, the epitopes are responsible for the specificity of the antigen for the antibody.
Thus, the same antigenic molecule can induce the production of as many different antibody molecules as different antigenic determinants it possesses. For this reason, antigens are said to be polyvalent. Generally, an antigen has between five and ten antigenic determinants on its surface (although some have 200 or more), which may be different from each other so that they may react with different types of antibodies.
The immune system is divided into two subsystems: the innate immune system and the adaptive immune system. The innate immune system can detect antigens inside the system, while the adaptive immune system is more complex, since it exhibits a response which can be modified to answer back to specific antigens. This response is improved by the repeated presence of the same antigen.
The innate immune system has an immediate response, but this is not specific and there are no memory cells involved. In contrast, the adaptive immune system response takes more time to be activated but is specific to the antigen because memory cells are involved.
Adaptive immunity or acquired immunity is the ability of the immune system to adapt, over time, to the recognition of specific pathogens with greater efficiency [
35]. Immunological memory is created from the primary response to a specific pathogen and allows the system to develop a better response to eventual future encounters.
Antibodies are chemicals that help destroy pathogens and neutralize their toxins. An antibody is a protein produced by the body in response to the presence of an antigen, and it is able to combine effectively with it. An antibody is essentially the complement of an antigen.
The specific adjustment of the antibody to the antigen depends not only on the size and shape of the antigenic determinant site, but also on the site corresponding to the antibody, more similar to the analogy of a lock and key. An antibody, as well as an antigen, also has a valence. While most of the antigens are polyvalent, the antibodies are bivalent or polyvalent. Most human antibodies are bivalent.
Various cell types carry out immune responses by way of the soluble molecules they secrete. Although lymphocytes are essential in all immune responses, other cell types also play a role [
35]. Lymphocytes are a special group of white blood cells: they are the cells that intervene in the defense mechanisms and in the immune reactions of the organism.
There are two main categories of lymphocytes: B and T [
35].
B lymphocytes, which represent between 10% and 20% of the total population, circulate in the blood and are transformed into antibody-producing plasma cells in the event of infection. They are responsible for humoral immunity. T lymphocytes are divided into two groups that perform different functions:
T lymphocyte killers (killer cells or suppressor cells) are activated by abnormal cells (tumor or virus-infected); they attach to these cells and release toxic substances called lymphokines to destroy them;
T helper cells (collaborators) stimulate the activity of T-killer cells and intervene in other varied aspects of the immune reaction.
Macrophages (from Greek “big eaters”) are cells of the immune system that are located within tissues. These phagocytic cells process and present the antigens to the immune system. They come from precursors of bone marrow that pass into the blood (monocytes) and migrate to sites of inflammation or immune reactions. They differ greatly in size and shape depending on their location. They are mobile, adhere to surfaces, emit pseudopodia, and are capable of phagocytosis-pinocytosis or have the capacity to store foreign bodies.
When macrophages phagocyte a microbe, they process and secrete the antigens on their surface, which are recognized by helper T lymphocytes, which produce lymphokines that activate B lymphocytes. This is why macrophages are part of the antigen-presenting cells. Activated B lymphocytes produce and release antibodies specific for the antigens presented by the macrophage. These antibodies adhere to the antigens of the microbes or cells invaded by viruses, and thus attract with greater avidity the macrophages to phagocyte them [
34].
Subsequently, the regulation phase controls the cells generated in the immune response in order to avoid damaging the system itself and preventing autoimmune responses. Finally, in the resolution phase, the harmful agent is removed and the cells generated to destroy the antigen die, only storing the memory cells.
The immune system has been the inspiration for numerous researchers in order to develop learning classifier systems [
36,
37,
38,
39]. Similarly, the immune system inspired the model proposed in this paper, the Artificial Immune System for Associative Classification, which is described in the next section.
3.2. AISAC: Artificial Immune System for Associative Classification
The main goal of classifier systems is to assign or predict a class label for an unseen pattern according to its attributes after training it with similar patterns [
40].
Many classifier systems are based on two steps: model construction and model operation [
41]. Model construction is a representation of the training set, which is used to generate a structure to classify the patterns presented. Model operation classifies data whose class labels are not known using the previously constructed model. In this paper, the classification problem is addressed by proposing a new model based on the immune system. Although numerous computational models have been developed based on the human immune system [
42], this research presents a new model that incorporates additional elements of the immune response. The model is called Artificial Immune System for Associative Classification (AISAC). This model is proposed within the supervised classification paradigm, and therefore constitutes a new supervised classifier.
Among the characteristics of the proposed model, it can be highlighted that it is an eager classifier, since it generates internal data structures to classify the new instances. Thus, the training set is replaced by other structures. The proposed artificial immune system model includes two types of functions: the acquired (adaptive) immune response, and the innate immune response. The acquired immune response consists of five phases:
Detection of antigenic macromolecules;
Activation of B lymphocytes;
Immune response regulation;
Development of adaptive immunity;
Resolution of the threat.
The innate immune response has only one phase:
In general, the acquired immunological response begins with the detection of antigenic macromolecules by macrophages. Each macrophage will phagocytose a number of antigenic determinants of the antigenic molecule in which it specializes. Subsequently, each macrophage will present the antigenic determinants that it phagocytosed to the T lymphocyte helpers.
In Phase 2, these lymphocytes will generate an immune response, activating a certain number of B lymphocytes. Activated B lymphocytes will produce and release specific antibodies to the antigens presented by the macrophage. Then, the immune response will be monitored (Phase 3). If the immune response is satisfactory, the generated antibodies are conserved. Otherwise, a readjustment of the generated antibodies is performed so that they are able to combine with the antigenic determinants presented.
To guarantee the development of adaptive or acquired immunity (Phase 4), each of the antibodies will undergo a reconstitution phase so that it is capable of improving its immune response. Finally, in Phase 5, the antigenic macromolecules are completely removed and the antigens are stored in the immune memory.
In the case of the innate immune response a set of antibodies is already in memory; when an antigen is present, it is automatically detected and eliminated.
3.3. AISAC Graphic Example
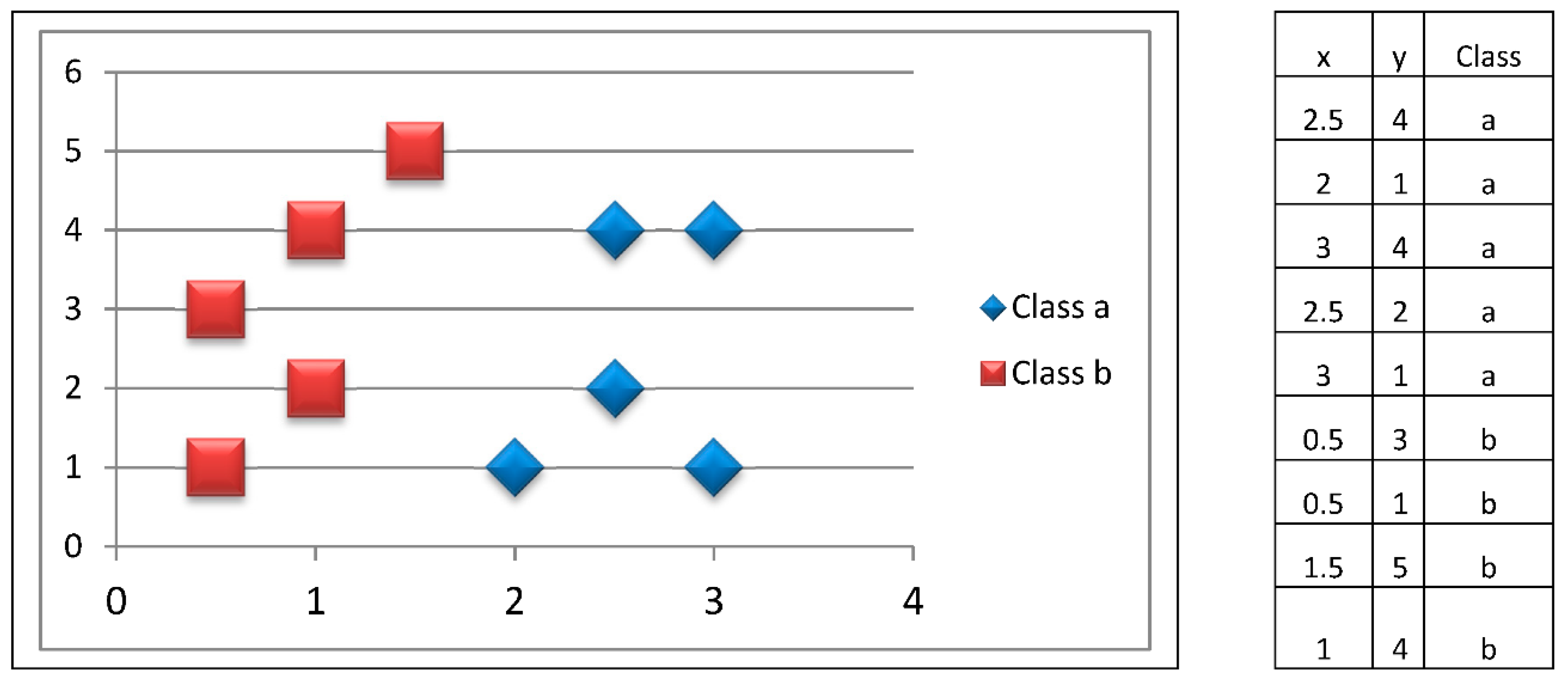
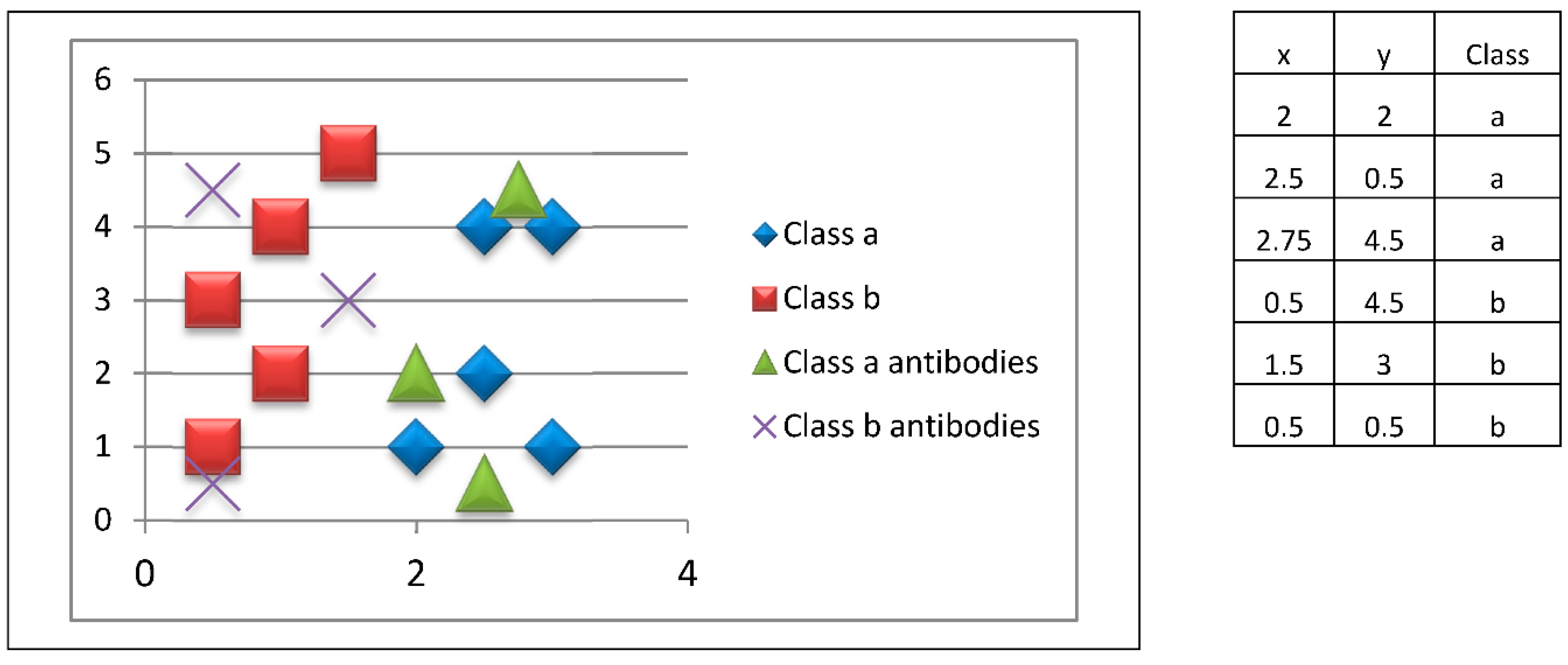
The following is an example of adaptive and innate immune responses in the proposed model. Suppose we have a training set that has 10 two-dimensional patterns, evenly distributed among two classes, as shown in
Figure 4. Let us also consider that our immune system has six macrophages.
The adaptive immune response is developed as follows.
Phase 1: Detection of antigenic macromolecules
The adaptive immune response begins by determining the number of macrophages necessary to phagocyte the antigenic determinants of each antigenic macromolecule, such as:
Thus, the macrophages are divided in such a way that they can phagocyte equitably to the antigenic determinants. Later, these antigenic determinants are presented to the T-Helper lymphocytes. In the example of
Figure 4, we have two antigenic macromolecules, which correspond to two classes. Thus:
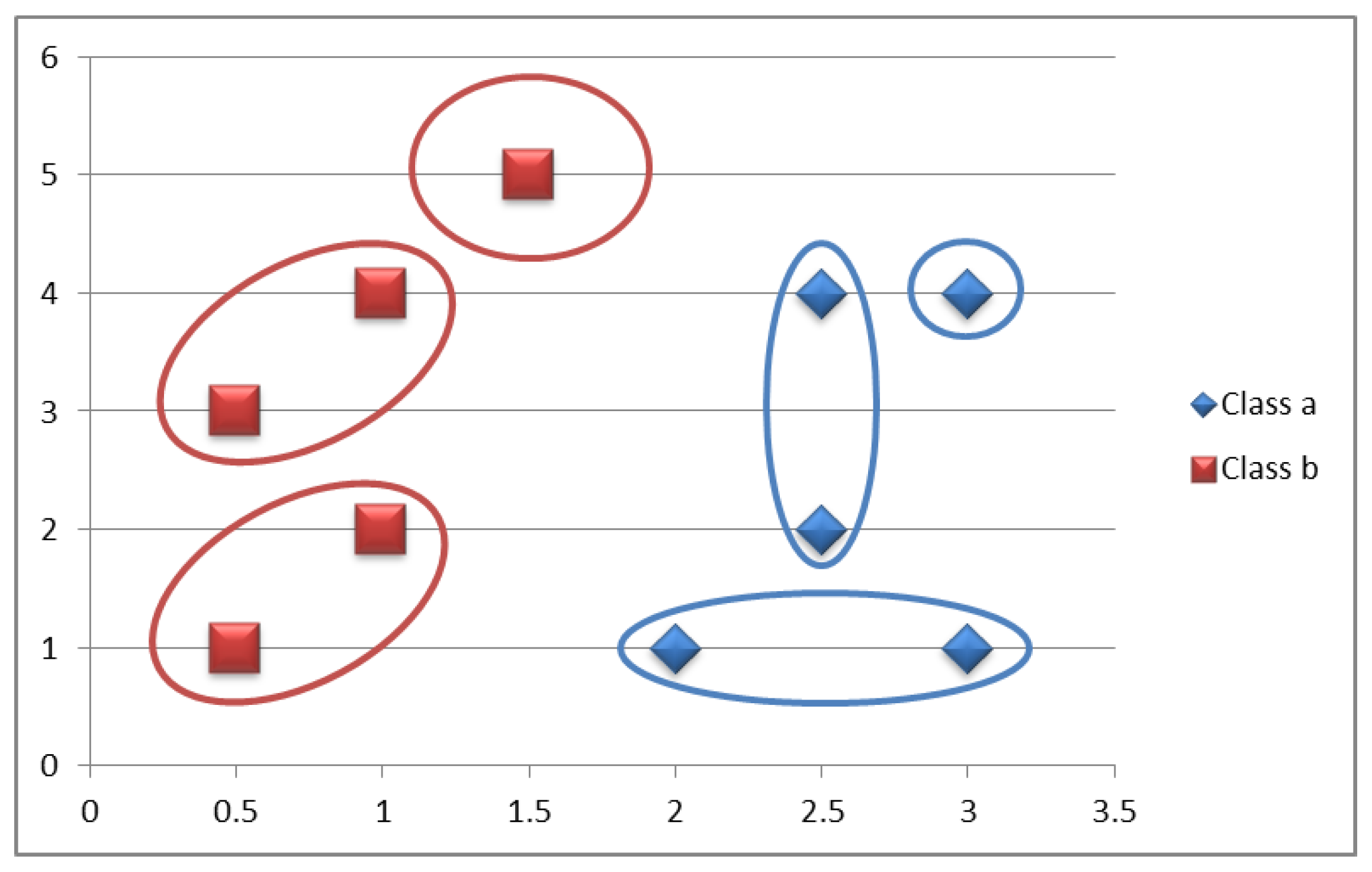
Accordingly, three bags (macrophages) will be assigned to phagocyte the antigenic determinants of each antigenic macromolecule, that is, to group the class data, through random sampling without replacement. In the example, two instances (antigenic determinants) of class a (antigenic macromolecule a) will be assigned to bag (macrophage) 1, two will be assigned to bag 2, and the remainder to bag 3. This process will be repeated for class b (antigenic macromolecule b) (
Figure 5).
The initial training patterns are shown in
Figure 4. We have a balanced distribution of five patterns belonging to class a, and five patterns of class b. These patterns are kept in consistent figures; however, it is important to keep in mind what the ten initial patterns are.
In
Figure 5, the training patterns are grouped into equally distributed bags for each class, that is, all classes will have the same number of bags. As a consequence of the above, each class will be represented by the same number of prototypes.
Phase 2: Activation of B lymphocytes
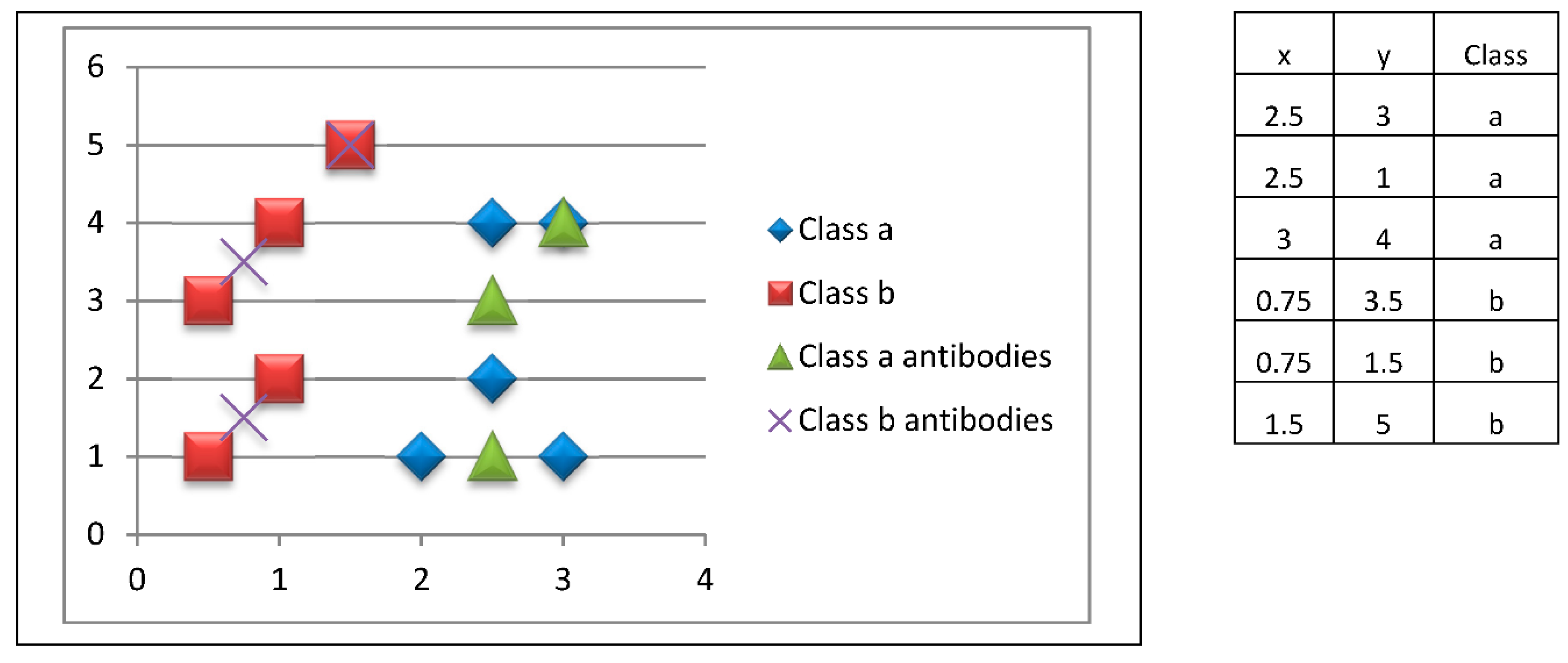
Subsequently, each bag (macrophage) activates the corresponding merging procedure (B lymphocyte), which will release a prototype (antibody)
corresponding to the instances (antigenic determinants) presented by the macrophage, which is determined by the mean of the instances (
Figure 6).
At this stage, a unique prototype is generated with the mean value of the patterns of each bag. In this example, three prototypes are created for each class. As we can notice, there are two prototypes similar to an original training pattern; this is because in their respective bag there was only one pattern to average their values.
Phase 3: Control of the immune response
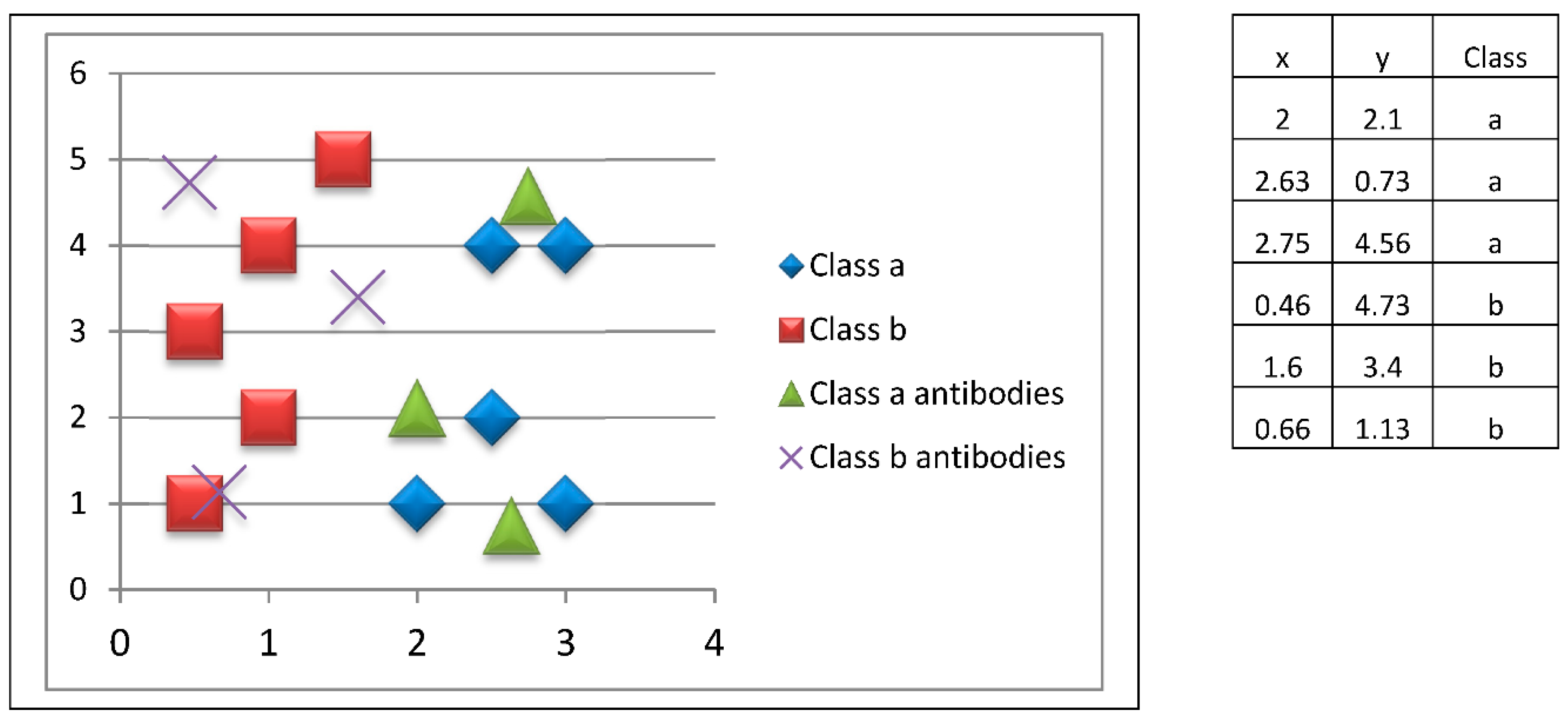
Estimation of the current prototypes’ (antibodies) ability is performed by calculating the weighted performance in order to reduce bias a little due to the possible imbalance in the data set. To do this, the instances (antigenic determinants) of the validation set are presented to the prototypes. Each prototype responds to its nearest instance (antigenic determinant).
The immune response is then adjusted so that the prototypes (antibodies) are able to correctly classify the instances (to combine with the antigenic determinants presented). To do this, the prototypes “approach” the instances of the corresponding class, and “move away” from the instances of other classes. Thus, the prototypes move in the search space, so that they obtain a better performance compared to being in their previous positions. If the new prototypes (antibodies) have a better immune response than the previous antibodies, they are replaced. This process is shown in
Figure 7.
Phase 4: Development of adaptive immunity
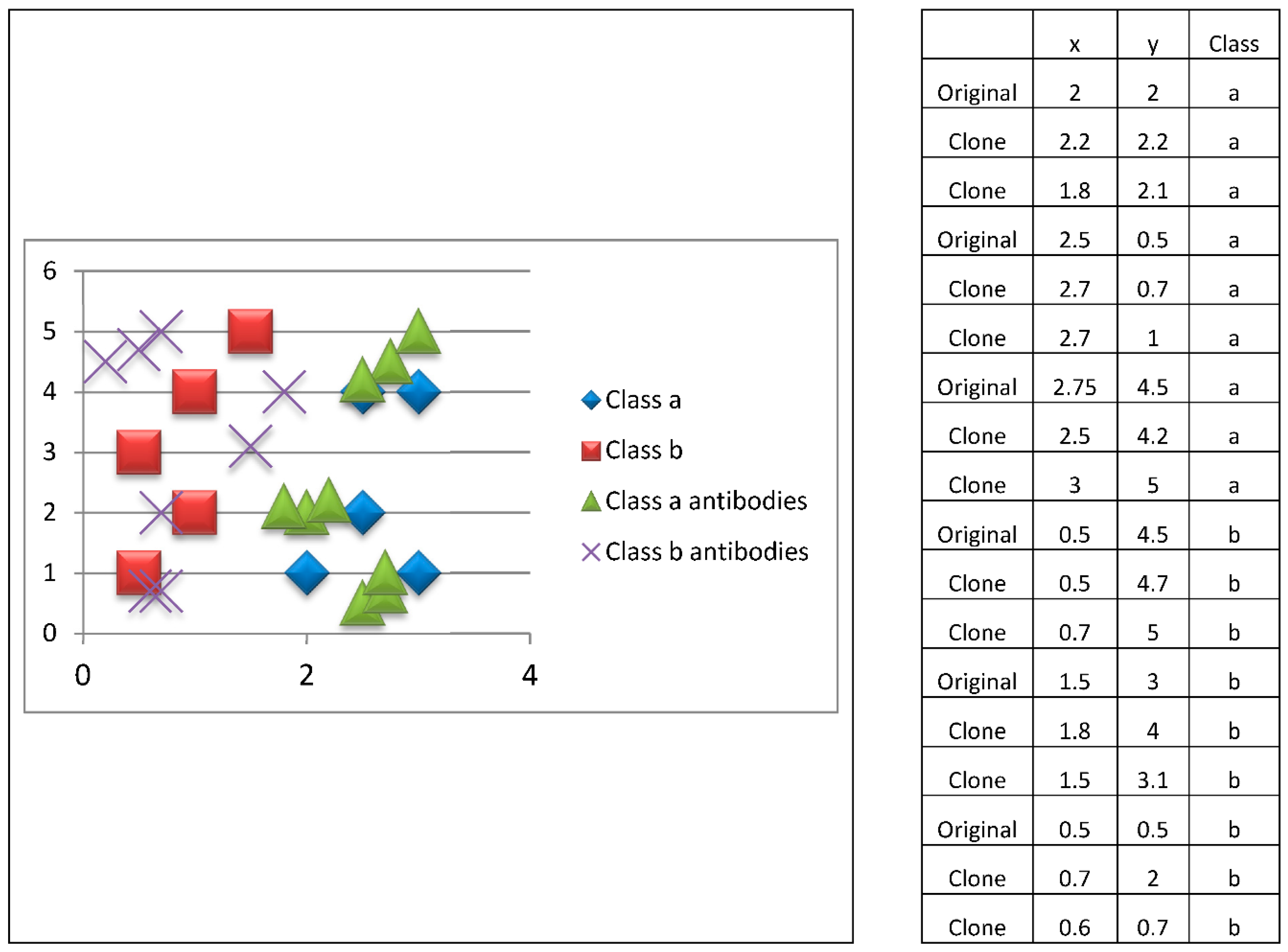
To develop the adaptive response, the prototypes (antibodies) are cloned. This allows the algorithm to explore the search space. To achieve this, the position of the prototype is slightly modified. This process is shown in
Figure 8, where two clones are generated for each prototype (antibody).
Subsequently, the antibody survival phase is performed, where a mean of the clones generated from each prototype (antibody) is obtained so that we return to the six antibodies (three from class a and three from class b). This survival process is shown in
Figure 9.
If the new clones of the prototypes (antibodies) exhibit a better immune response than the previous antibodies, they are replaced. This process is iterative and elitist because it only retains the best antibodies generated. Then, these prototypes are used in the immune response. Phases 3 and 4 are repeated for a predefined number of iterations.
Phase 5: Threat resolution
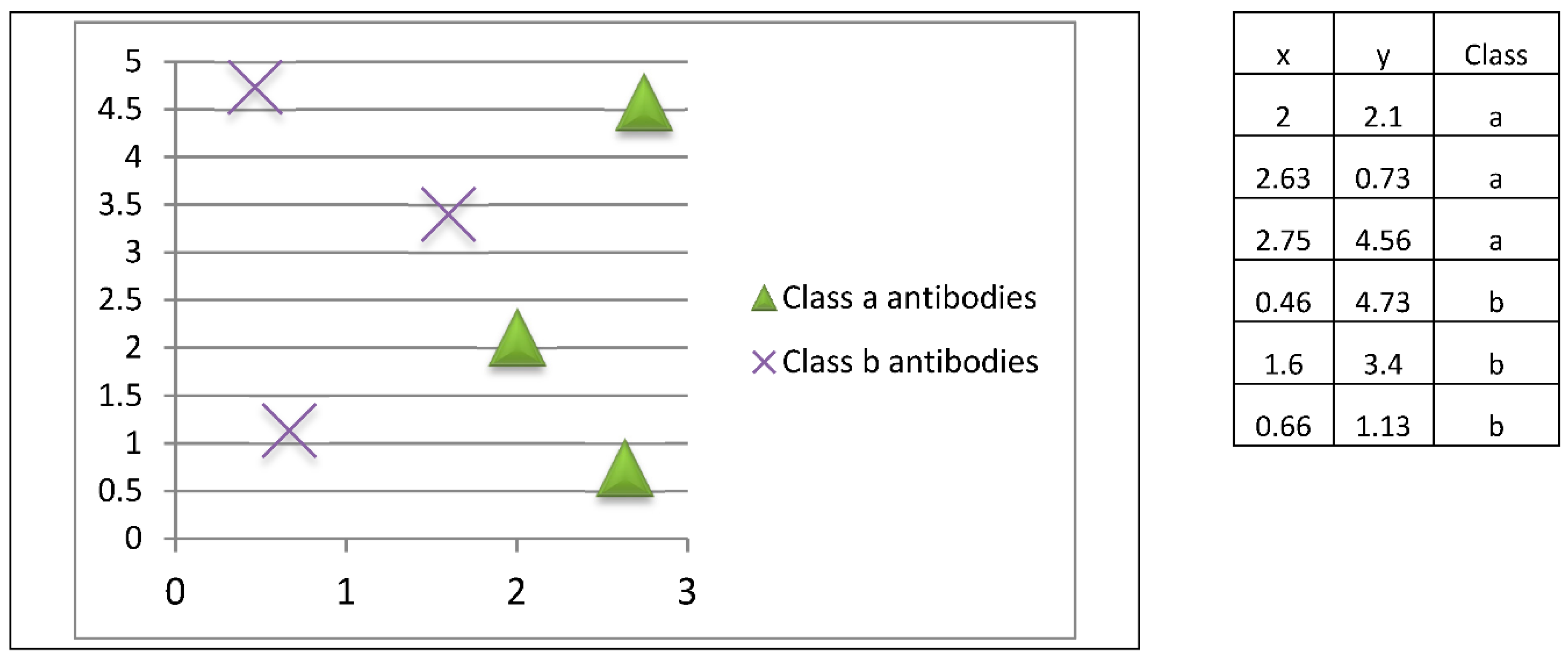
At this stage, the final prototypes (antibodies) are stored in the immune memory (
Figure 10).
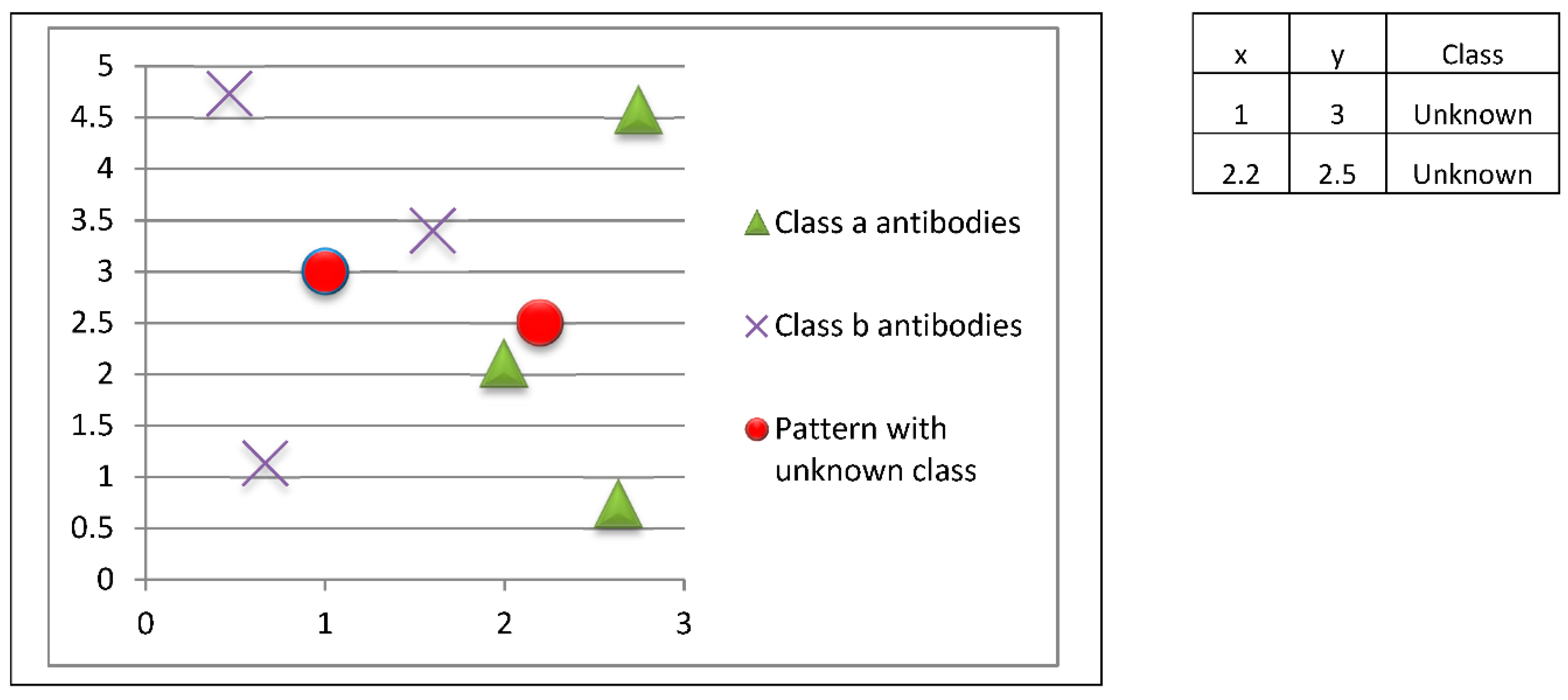
Upon completion of the model construction phase (adaptive immune response), it is possible to perform the classification or model operation (innate immune response). In this case, let us suppose that we have two new patterns whose classes are unknown, as shown in
Figure 11. These patterns correspond to unknown classes (antigenic determinants), and we want to respond to this threat using the prototypes (antibodies) previously stored in the immune memory.
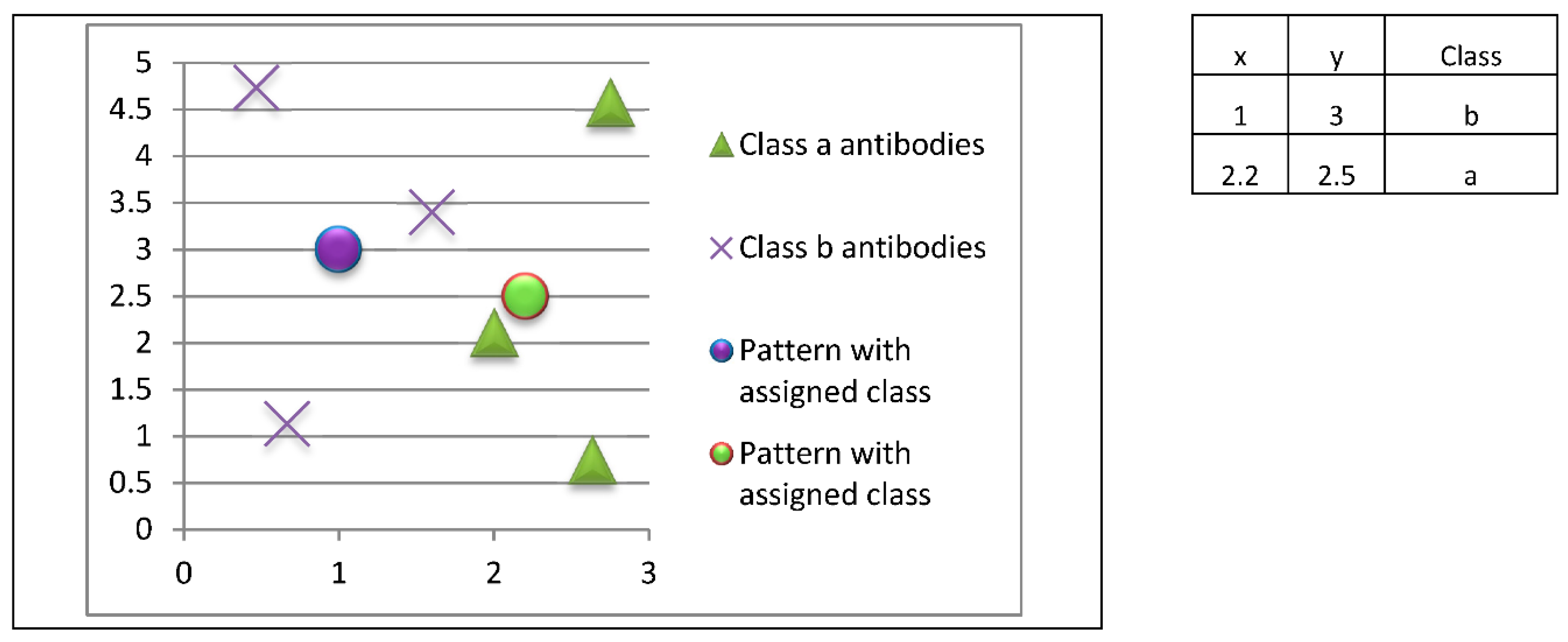
Antibodies stored in memory will be used to classify new patterns whose class is unknown. In this example, three class a patterns and three class b patterns are stored, so each class is represented by the same number of prototypes.
The innate immune response will look for the most closely related prototypes (antibodies) to each unknown instance (antigenic determinant), so the classification would be as shown in
Figure 12.
In this example, the pattern at coordinates (1.0, 3.0) would be classified as class b, while the pattern at coordinates (2.2, 2.5) would be classified as class a.
The proposed AISAC model is a contribution to the state-of-art of artificial immune systems. It provides a new modeling of the biological behavior of the immune system, and has a low computational cost. In addition, AISAC is simple and able to fit the training data using few antibodies. We consider that this model enhances the frontier of classification systems. In the next section, we test the performance of the AISAC model in a very important scenario: cancer detection.