3D Dense Separated Convolution Module for Volumetric Medical Image Analysis
Abstract
:1. Introduction
2. Related Work
3. Methods
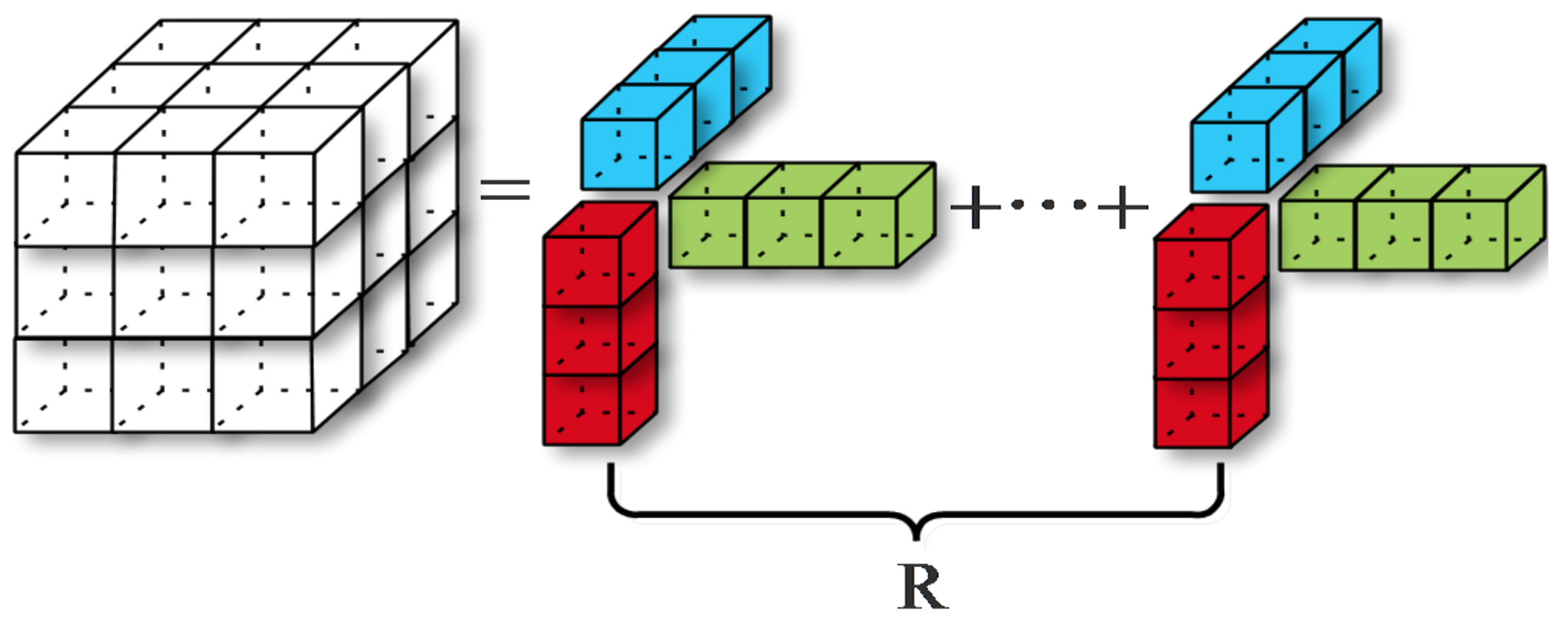
3.1. 3D Separability of Convolutional Kernels
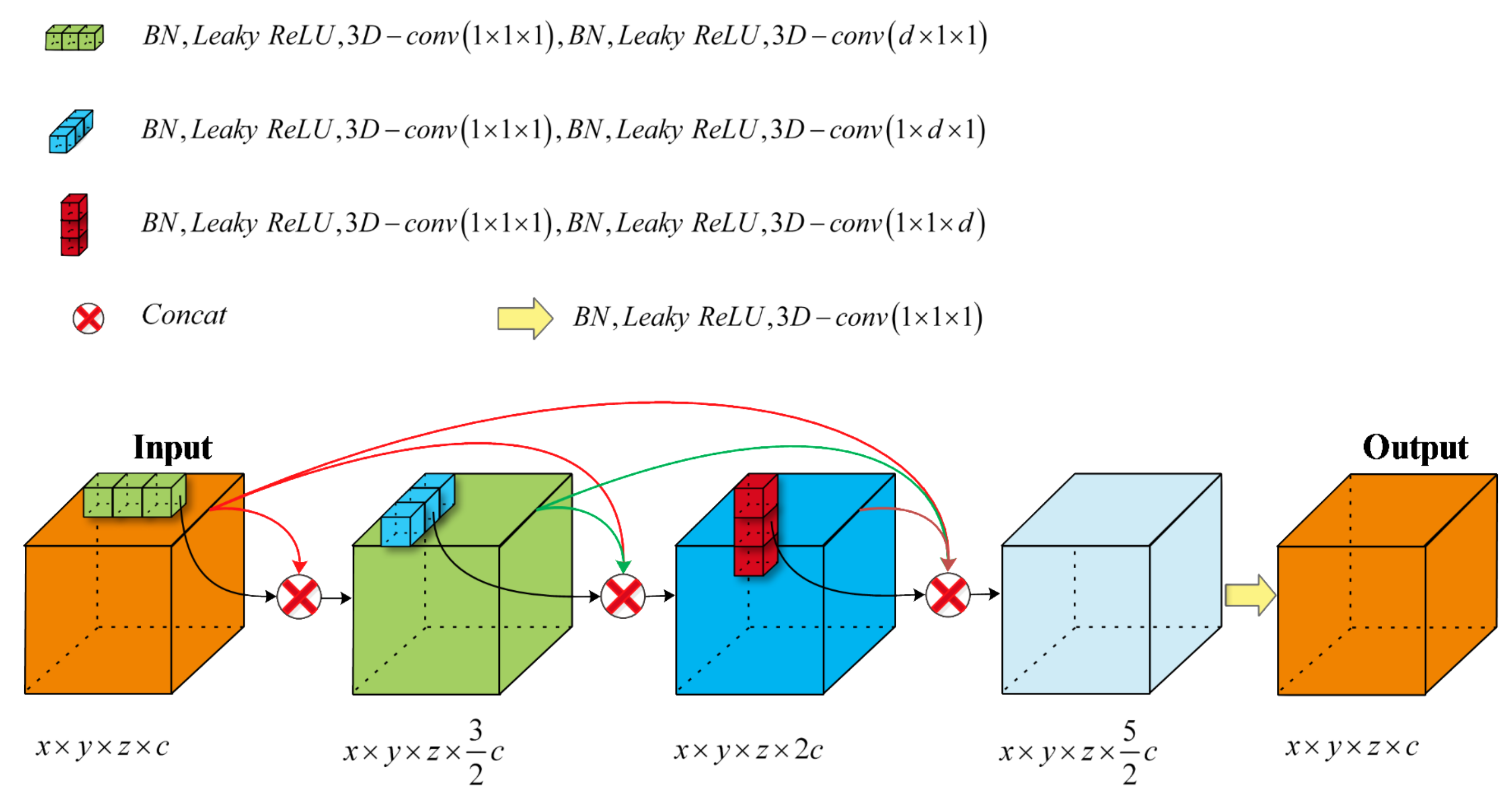
3.2. 3D Dense Separated Convolution Module
3.3. 3D CNN Architecture Based on 3D-DSC
3.4. Training of the 3D CNN Architecture
4. Experiments and Results
4.1. Attention Deficit Hyperactivity Disorder Diagnosis
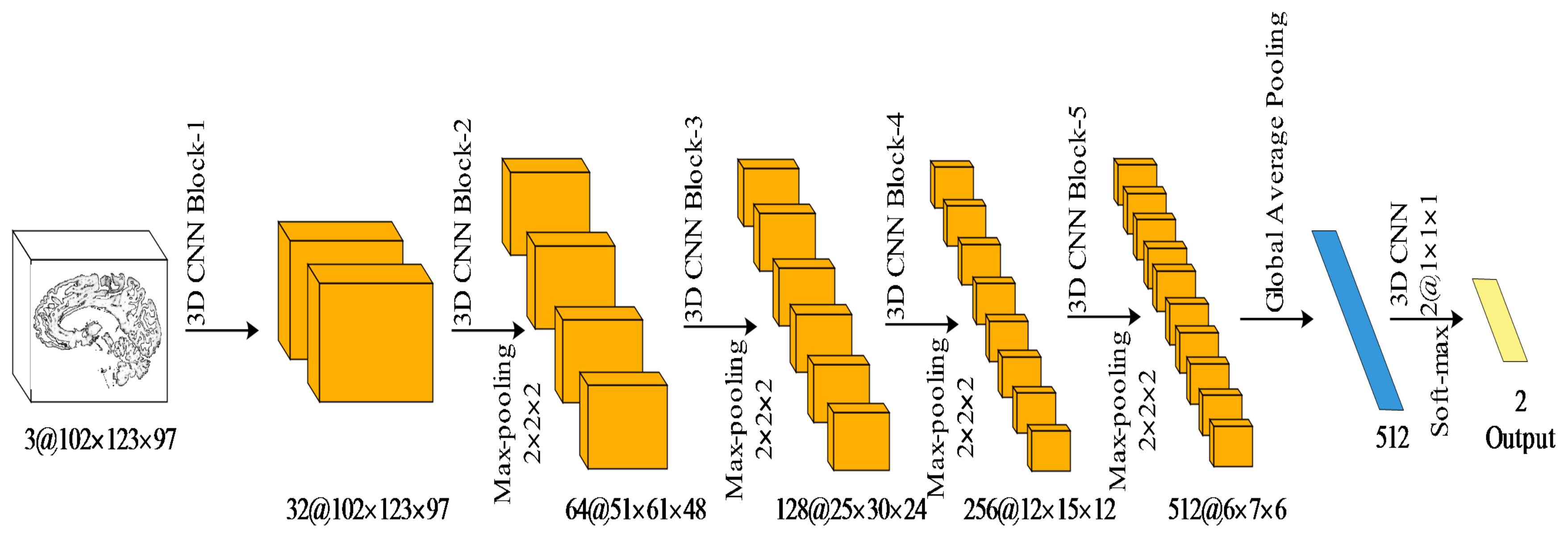
4.1.1. Network Architecture
4.1.2. Accuracy and Analysis of the Network’s Depth
| Method | Classifier | Accuracy |
|---|---|---|
| [32] | MKL | 61.54% |
| [33] | SVM | 69.62% |
| [34] | SVM | 63.57% |
| [8] | SM 3D CNN | 66.04% |
| [8] | MM 3D CNN | 69.15% |
| [35] | 4D CNN | 71.30% |
| 3D-DSC () | SM 3D CNN | 73.68% |
4.1.3. Ablation Studies
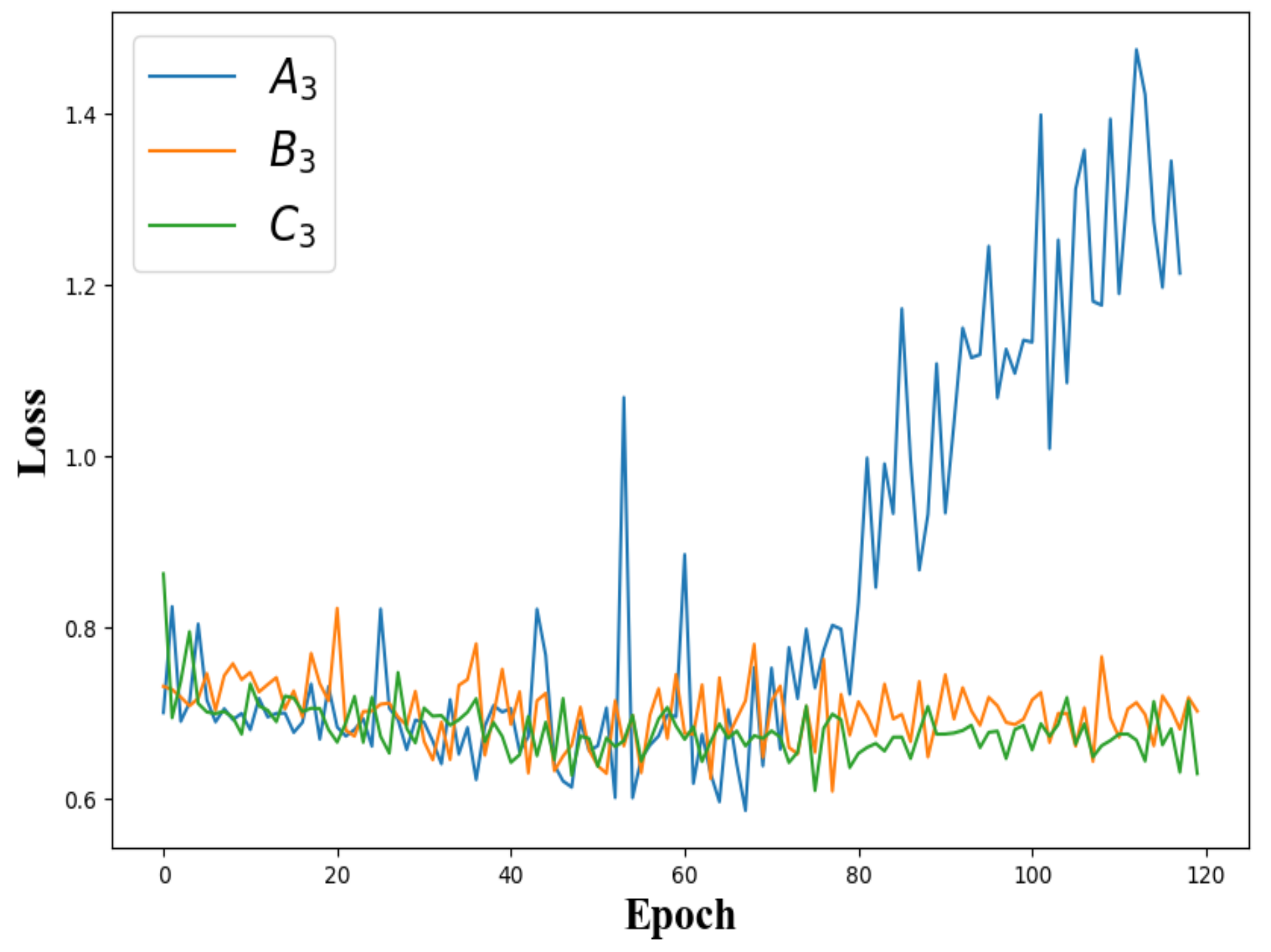
4.1.4. Overfitting
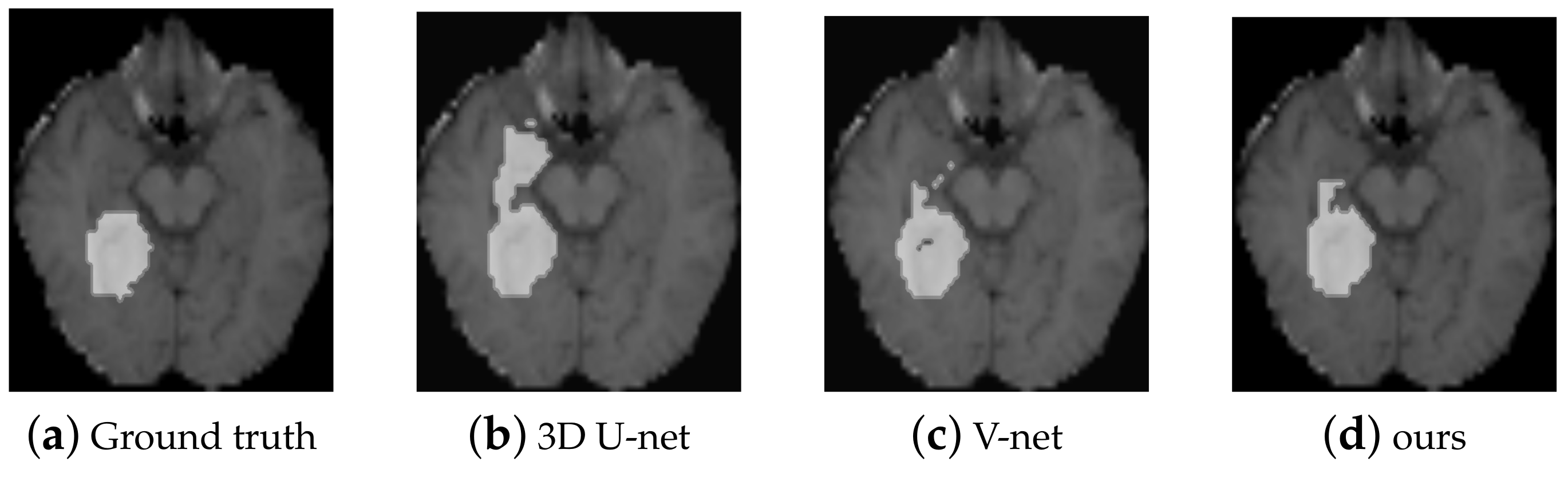
4.2. The Brain Tumour Segmentation on BRATS 2017
5. Conclusions and Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wainberg, M.; Merico, D.; Delong, A.; Frey, B.J. Deep learning in biomedicine. Nat. Biotechnol. 2018, 36, 829. [Google Scholar] [CrossRef] [PubMed]
- Miao, S.; Wang, Z.J.; Liao, R. A CNN regression approach for real-time 2D/3D registration. IEEE Trans. Med. Imaging 2016, 35, 1352–1363. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Dai, W.; Sun, S.Y.; Jonasch, D.; He, C.Y.; Schmid, M.F.; Chiu, W.; Ludtke, S.J. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods 2017, 14, 983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Išgum, I. Dilated convolutional neural networks for cardiovascular MR segmentation in congenital heart disease. In Reconstruction, Segmentation, and Analysis of Medical Images; Springer: Cham, Switzerland, 2016; pp. 95–102. [Google Scholar]
- Zheng, H.; Zhang, Y.; Yang, L.; Liang, P.; Zhao, Z.; Wang, C.; Chen, D.Z. A new ensemble learning framework for 3D biomedical image segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 5909–5916. [Google Scholar]
- Chen, J.; Yang, L.; Zhang, Y.; Alber, M.; Chen, D.Z. Combining fully convolutional and recurrent neural networks for 3D biomedical image segmentation. In Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 3044–3052. [Google Scholar]
- Khosravan, N.; Bagci, U. S4ND: Single-shot single-scale lung nodule detection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 794–802. [Google Scholar]
- Zou, L.; Zheng, J.; Miao, C.; Mckeown, M.J.; Wang, Z.J. 3D CNN based automatic diagnosis of attention deficit hyperactivity disorder using functional and structural MRI. IEEE Access 2017, 5, 23626–23636. [Google Scholar] [CrossRef]
- Lee, K.; Zlateski, A.; Vishwanathan, A.; Seung, H.S. Recursive training of 2D-3D convolutional networks for neuronal boundary detection. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montréal, QC, Canada, 7–12 December 2015; pp. 3573–3581. [Google Scholar]
- Lai, M. Deep learning for medical image segmentation. arXiv 2015, arXiv:1505.02000. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Choromanska, A.; Henaff, M.; Mathieu, M.; Arous, G.B.; LeCun, Y. The loss surfaces of multilayer networks. J. Mach. Learn. Res. 2015, 38, 192–204. [Google Scholar]
- Rigamonti, R.; Sironi, A.; Lepetit, V.; Fua, P. Learning separable filters. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Portland, OR, USA, 23–28 June 2013; pp. 2754–2761. [Google Scholar]
- Denton, E.; Zaremba, W.; Bruna, J.; LeCun, Y.; Fergus, R. Exploiting linear structure within convolutional networks for efficient evaluation. In Proceedings of the 28th Annual Conference on Neural Information Processing Systems, Neural Information Processing Systems Foundation, Montréal, QC, Canada, 8–13 December 2014; pp. 1269–1277. [Google Scholar]
- Jaderberg, M.; Vedaldi, A.; Zisserman, A. Speeding up convolutional neural networks with low rank expansions. arXiv 2014, arXiv:1405.3866. [Google Scholar]
- Lebedev, V.; Ganin, Y.; Rakhuba, M.; Oseledets, I.; Lempitsky, V. Speeding-up convolutional neural networks using fine-tuned cp-decomposition. arXiv 2014, arXiv:1412.6553. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Jin, J.; Dundar, A.; Culurciello, E. Flattened convolutional neural networks for feedforward acceleration. arXiv 2014, arXiv:1412.5474. [Google Scholar]
- Gonda, F.; Wei, D.; Parag, T.; Pfister, H. Parallel Separable 3D Convolution for Video and Volumetric Data Understanding. arXiv 2018, arXiv:1809.04096. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Wang, M.; Liu, B.; Foroosh, H. Factorized convolutional neural networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 545–553. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Zhang, J.; Xie, Y.; Zhang, P.; Chen, H.; Xia, Y.; Shen, C. Light-weight hybrid convolutional network for liver tumour segmentation. In Proceedings of the 28th International Joint Conference on Artificial Intelligence, Macao, China, 10–16 August 2019; pp. 4271–4277. [Google Scholar]
- Zou, L.; Chen, X.; Wang, Z.J. Underdetermined joint blind source separation for two datasets based on tensor decomposition. IEEE Signal Process. Lett. 2016, 23, 673–677. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Milham, M.P.; Fair, D.; Mennes, M.; Mostofsky, S.H. The ADHD-200 consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Front. Syst. Neurosci. 2012, 6, 62. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathycramer, J.; Farahani, K.; Kirby, J.S.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Miao, B.; Zhang, L.; Guan, J.; Meng, Q.; Zhang, Y. Classification of ADHD Individuals and Neurotypicals Using Reliable RELIEF: A Resting-State Study. IEEE Access 2019, 7, 62163–62171. [Google Scholar] [CrossRef]
- The Magnetic Resonance Imaging Research Center, IPCAS. The R-fmri Maps Project. 2018. Available online: http://mrirc.psych.ac.cn/RfMRIMaps (accessed on 1 October 2018).
- Ba, L.J.; Caruana, R. Do deep nets really need to be deep? In Proceedings of the 27th International Conference on Neural Information Processing Systems-Volume 2, Montreal, QC, Canada, 8–13 December 2014; pp. 2654–2662. [Google Scholar]
- Dai, D.; Wang, J.; Hua, J.; He, H. Classification of ADHD children through multimodal magnetic resonance imaging. Front. Syst. Neurosci. 2012, 6, 63. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Tang, Y.; Chen, Y.; Zhou, L.; Wang, C. ADHD classification by feature space separation with sparse representation. In Proceedings of the 2018 IEEE 23rd International Conference on Digital Signal Processing (DSP), Shanghai, China, 19–21 November 2018; pp. 1–5. [Google Scholar]
- Guo, X.; An, X.; Kuang, D.; Zhao, Y.; He, L. ADHD-200 classification based on social network method. In Proceedings of the International Conference on Intelligent Computing, Taiyuan, China, 3–6 August 2014; pp. 233–240. [Google Scholar]
- Mao, Z.; Su, Y.; Xu, G.; Wang, X.; Huang, Y.; Yue, W.; Xiong, N. Spatio-temporal deep learning method for ADHD fMRI classification. Inf. Sci. 2019, 499, 1–11. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. Brain tumour segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge. In Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 14 September 2017; pp. 287–297. [Google Scholar]
- Liu, H.; Shen, X.; Shang, F.; Ge, F.; Wang, F. CU-Net: Cascaded U-Net with Loss Weighted Sampling for Brain Tumor Segmentation. In Multimodal Brain Image Analysis and Mathematical Foundations of Computational Anatomy; Springer: Cham, Switzerland, 2019; pp. 102–111. [Google Scholar]

| Method | Structure | Domain |
|---|---|---|
| [17] | 2D to 1D | general image |
| [18] | 2D to 1D, depthwise separation | general image |
| [20,21,22] | depthwise separation | general image |
| [13,14,15,16] | fine tune the trained model (LRA) | general image |
| [19,23] | 3D to 2D + 1D | medical image |
| 3D-DSC | 3D to 1D | medical image |
| Input (3D multi-channel MRI) | ||
| 3D-conv3-32 | 3D-conv3-32 | 3D-conv3-32 |
| 3D-conv3-32 | 3D-conv3-32 | 3D-conv3-32 |
| Max pooling | ||
| 3D-conv3-64 | 3D-conv3-64 | 3D-conv3-64 |
| 3D-conv3-64 | 3D-conv3-64 | 3D-DSC-64 |
| Max pooling | ||
| 3D-conv3-128 | 3D-conv3-128 | 3D-conv3-128 |
| 3D-conv3-128 | 3D-DSC-128 | 3D-DSC-128 |
| Max pooling | ||
| 3D-conv3-256 | 3D-conv3-256 | 3D-conv3-256 |
| 3D-conv3-256 | 3D-DSC-256 | 3D-DSC-256 |
| Max pooling | ||
| 3D-conv3-512 | 3D-conv3-512 | 3D-conv3-512 |
| 3D-conv3-512 | 3D-DSC-512 | 3D-DSC-512 |
| Global Average Pooling | ||
| 3D-conv1-2 | ||
| Softmax | ||
| Network | Depth | Params | Accuracy | OD |
|---|---|---|---|---|
| 8 | 4.8 M | 73.17% | 0.2785 | |
| 11 | 14.1 M | 74.89% | 0.2806 | |
| 14 | 23.4 M | 74.53% | 0.2807 | |
| 17 | 32.7 M | 71.94% | 0.2961 | |
| 20 | 42.0 M | 70.78% | 0.3368 | |
| 23 | 51.3 M | 69.91% | 0.3390 | |
| 26 | 60.6 M | 69.22% | 0.3567 | |
| 11 | 7.2 M | 73.45% | 0.2415 | |
| 14 | 9.6 M | 74.58% | 0.2498 | |
| 17 | 12.1 M | 75.22% | 0.2526 | |
| 20 | 14.4 M | 75.57% | 0.2610 | |
| 23 | 16.9 M | 75.79% | 0.2689 | |
| 26 | 19.3 M | 75.74% | 0.2809 | |
| 26 | 19.2 M | 76.70% | 0.2580 |
| Network | DC | Activation | Orders | Accuracy | OD |
|---|---|---|---|---|---|
| no | no | H, V, L | 74.26% | 0.2733 | |
| no | yes | H, V, L | 74.83% | 0.2638 | |
| yes | yes | H, V, L | 75.22% | 0.2526 | |
| yes | yes | V, L, H | 75.28% | 0.2507 | |
| yes | yes | L, H, V | 75.19% | 0.2514 |
| Method | Depth | Params | Dice Score |
|---|---|---|---|
| 3D U-net | 19 | 23.5M | 0.8554 |
| 3D U-net (Dropout) | 19 | 23.5 M | 0.8592 |
| 3D U-net (s2-conv) | 23 | 25.9 M | 0.8593 |
| [39] | 23 | 25.9 M | 0.8655 |
| V-net [38] | 19 | 23.5 M | 0.8685 |
| 3D U-net (3D-DSC) | 28 | 17.8 M | 0.8932 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, L.; Wu, C.; Zou, L. 3D Dense Separated Convolution Module for Volumetric Medical Image Analysis. Appl. Sci. 2020, 10, 485. https://doi.org/10.3390/app10020485
Qu L, Wu C, Zou L. 3D Dense Separated Convolution Module for Volumetric Medical Image Analysis. Applied Sciences. 2020; 10(2):485. https://doi.org/10.3390/app10020485
Chicago/Turabian StyleQu, Lei, Changfeng Wu, and Liang Zou. 2020. "3D Dense Separated Convolution Module for Volumetric Medical Image Analysis" Applied Sciences 10, no. 2: 485. https://doi.org/10.3390/app10020485
APA StyleQu, L., Wu, C., & Zou, L. (2020). 3D Dense Separated Convolution Module for Volumetric Medical Image Analysis. Applied Sciences, 10(2), 485. https://doi.org/10.3390/app10020485






