Selection of Support Vector Candidates Using Relative Support Distance for Sustainability in Large-Scale Support Vector Machines
Abstract
1. Introduction
2. Preliminaries
2.1. Support Vector Machines
2.2. Decision Tree
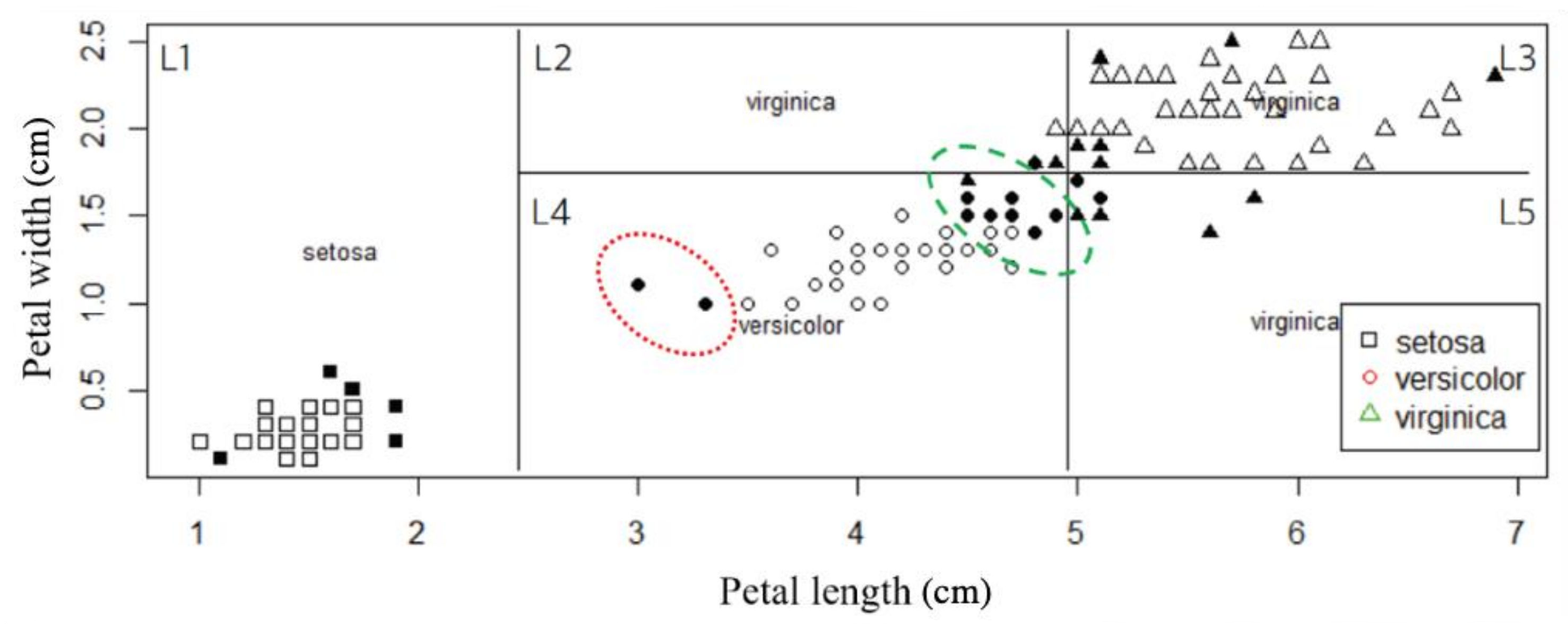
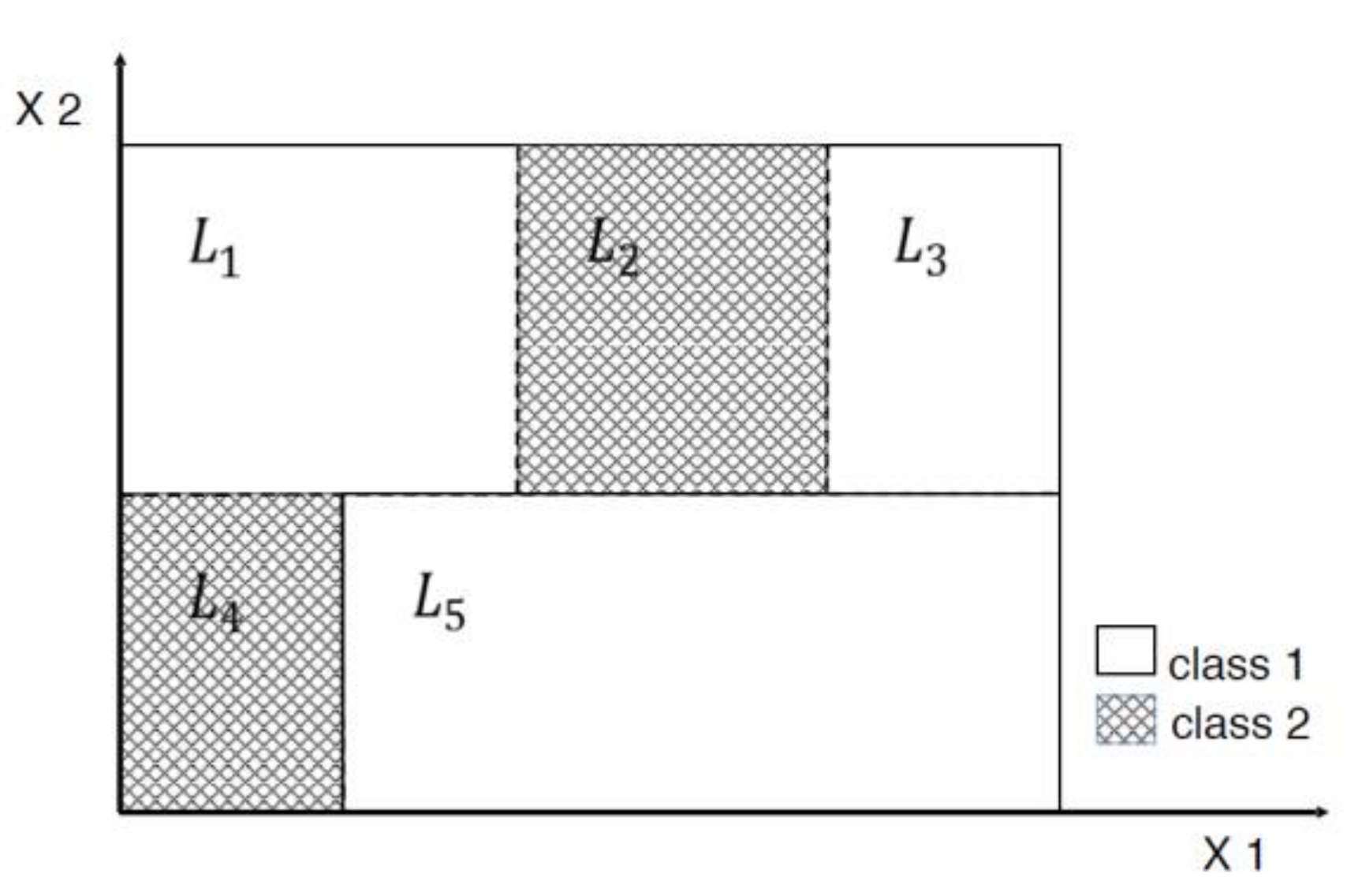
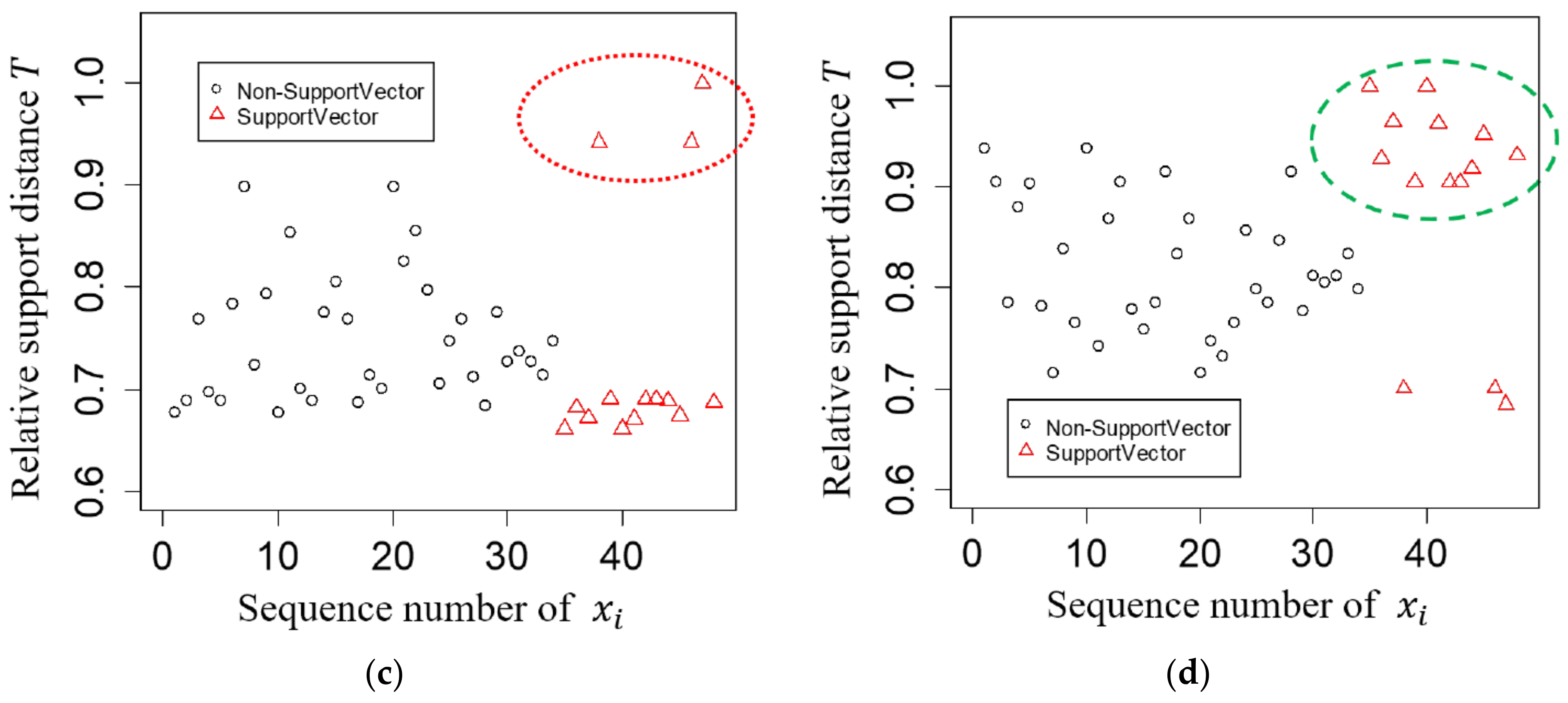
3. Tree-Based Relative Support Distance
- Decompose the input space by decision tree learning.
- Find distinct adjacent regions, which mean adjacent regions whose majority class is different from that of each region.
- Calculate the relative support distances for the data points in the found distinct adjacent regions.
- Select the candidates of support vectors according to the relative support distances.
3.1. Distinct Adjacent Regions
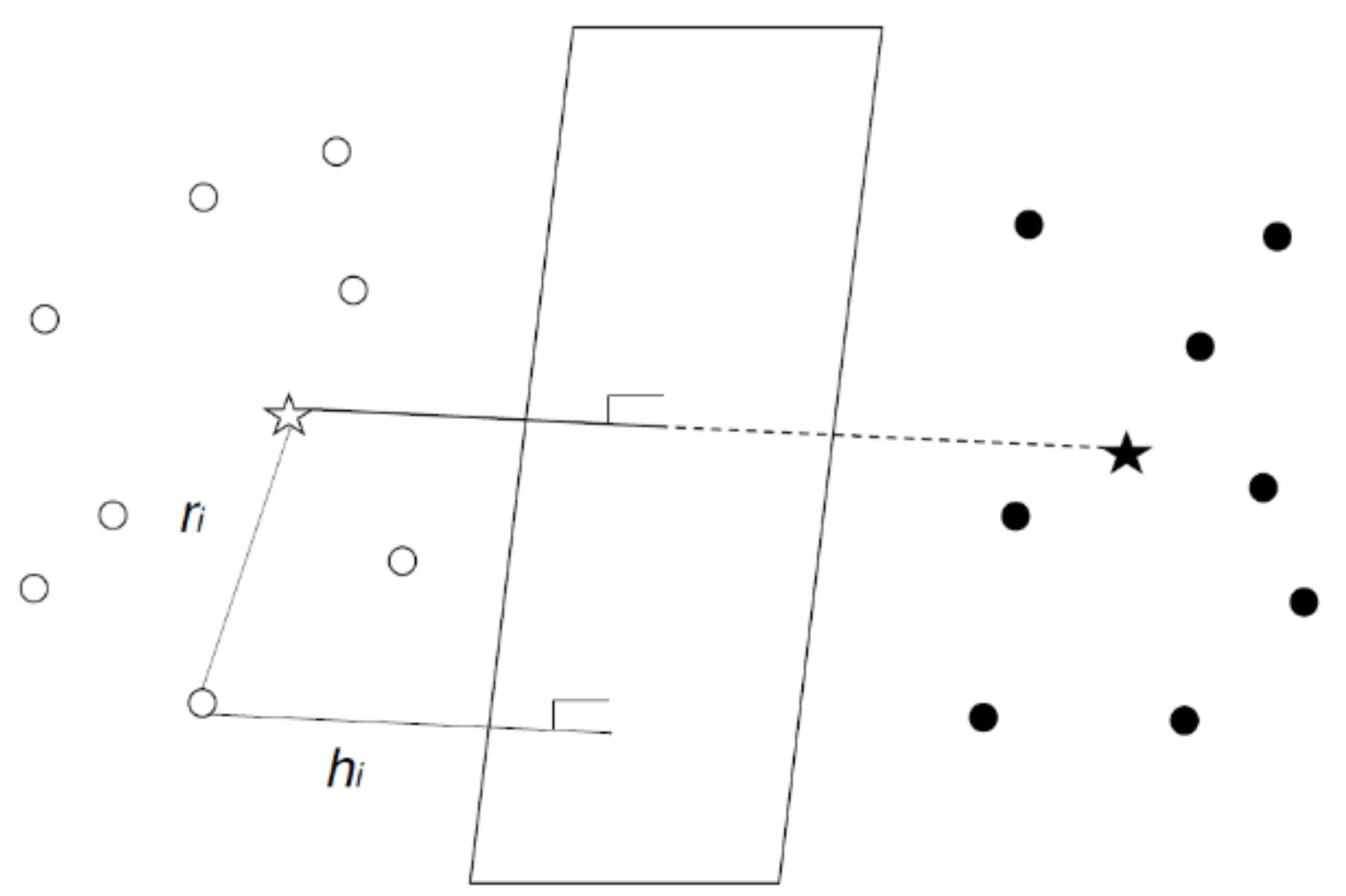
3.2. Relative Support Distance
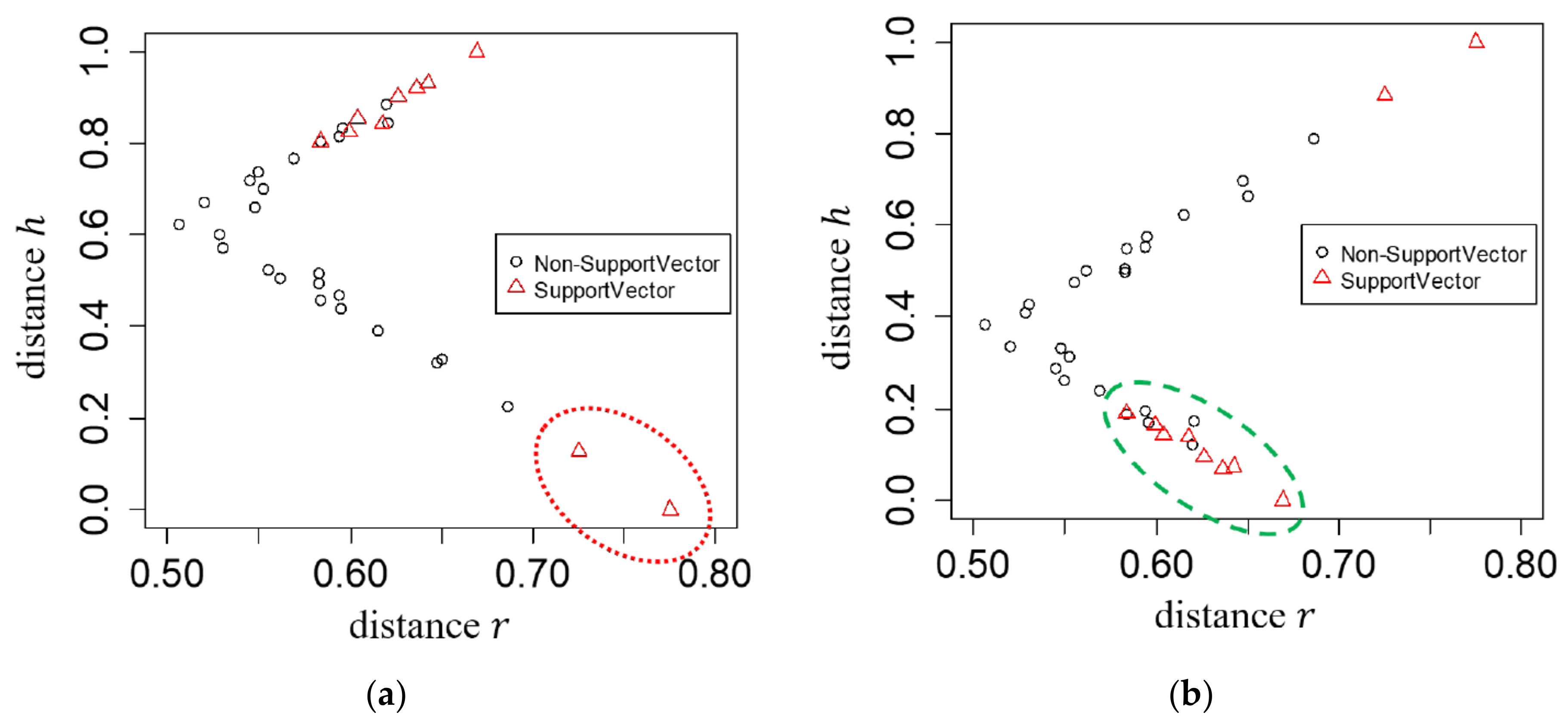
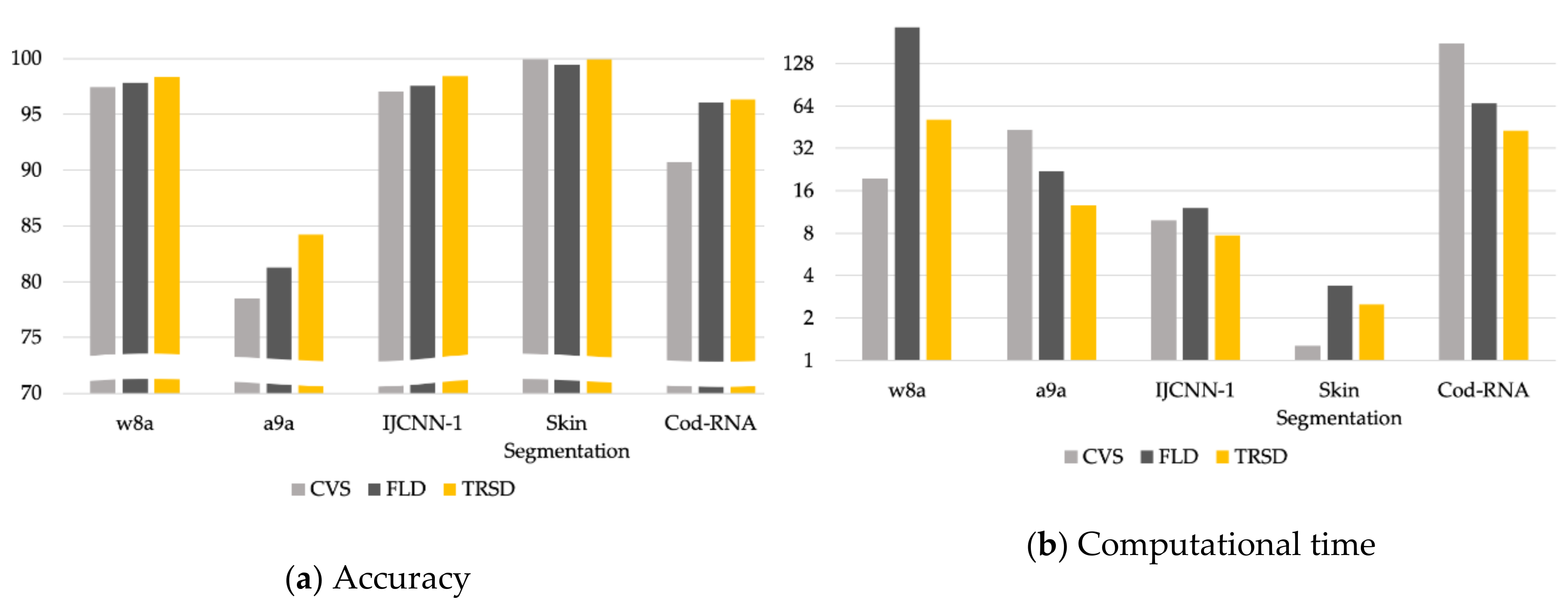
4. Experimental Results
5. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Cai, Y.D.; Liu, X.J.; biao Xu, X.; Zhou, G.P. Support Vector Machines for predicting protein structural class. BMC Bioinform. 2001, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Belongie, S.; Malik, J.; Puzicha, J. Shape Matching and Object Recognition Using Shape Contexts. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 509–522. [Google Scholar] [CrossRef]
- Ahn, H.; Lee, K.; Kim, K.j. Global Optimization of Support Vector Machines Using Genetic Algorithms for Bankruptcy Prediction. In Proceedings of the 13th International Conference on Neural Information Processing—Volume Part III; Springer: Berlin, Germany, 2006; pp. 420–429. [Google Scholar]
- Bayro-Corrochano, E.J.; Arana-Daniel, N. Clifford Support Vector Machines for Classification, Regression, and Recurrence. IEEE Trans. Neural Netw. 2010, 21, 1731–1746. [Google Scholar] [CrossRef] [PubMed]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A Training Algorithm for Optimal Margin Classifiers. Proceedings of the Fifth Annual Workshop on Computational Learning Theory; Association for Computing Machinery: New York, NY, USA, 1992; pp. 144–152. [Google Scholar]
- Friedman, J.; Hastie, T.; Tibshirani, R. The Elements of Statistical Learning; Springer: Berlin, Germany, 2001; Volume 1. [Google Scholar]
- Qiu, J.; Wu, Q.; Ding, G.; Xu, Y.; Feng, S. A survey of machine learning for big data processing. EURASIP J. Adv. Signal. Process. 2016, 2016, 1–16. [Google Scholar]
- Liu, P.; Choo, K.K.R.; Wang, L.; Huang, F. SVM of Deep Learning? A Comparative Study on Remote Sensing Image Classification. Soft Comput. 2017, 21, 7053–7065. [Google Scholar] [CrossRef]
- Chapelle, O. Training a support vector machine in the primal. Neural Comput. 2007, 19, 1155–1178. [Google Scholar] [CrossRef] [PubMed]
- Platt, J.C. 12 fast training of support vector machines using sequential minimal optimization. Adv. Kernel Methods 1999, 185–208. [Google Scholar]
- Joachims, T. Svmlight: Support Vector Machine. SVM-Light Support. Vector Mach. Univ. Dortm. 1999, 19.. Available online: http://svmlight.joachims.org (accessed on 29 November 2019).
- Vishwanathan, S.; Murty, M.N. SSVM: A simple SVM algorithm. 2002 International Joint Conference on Neural Networks. IJCNN’02, Honolulu, HI, USA, 12–17 May 2002. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. (TIST) 2011, 2, 27. [Google Scholar] [CrossRef]
- Lee, Y.J.; Mangasarian, O.L. RSVM: Reduced Support Vector Machines. SDM. In Proceedings of the 2001 SIAM International Conference on Data Mining, Chicago, IL, USA, 5–7 April 2001; pp. 325–361. [Google Scholar]
- Collobert, R.; Bengio, S.; Bengio, Y. A parallel mixture of SVMs for very large scale problems. Neural Comput. 2002, 14, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, F.; Kou, J. Candidate vectors selection for training support vector machines. In Natural Computation, 2007. ICNC 2007. In Proceedings of the Third International Conference on Natural Computation, Haikou, China, 24–27 August 2007; pp. 538–542. [Google Scholar]
- Nishida, K.; Kurita, T. RANSAC-SVM for large-scale datasets. In Proceedings of the 19th International Conference on Pattern Recognition, Tampa, FL, USA, 8–11 December 2008; pp. 1–4. [Google Scholar]
- Kawulok, M.; Nalepa, J. Support Vector Machines Training Data Selection Using a Genetic Algorithm. In Proceedings of the 2012 Joint IAPR International Conference on Structural, Syntactic, and Statistical Pattern Recognition; Springer: Berlin, Germany, 2012; pp. 557–565. [Google Scholar]
- Nalepa, J.; Kawulok, M. A Memetic Algorithm to Select Training Data for Support Vector Machines. In Proceedings of the 2014 Annual Conference on Genetic and Evolutionary Computation, Association for Computing Machinery, New York, NY, USA, 12–16 July 2014; pp. 573–580. [Google Scholar]
- Nalepa, J.; Kawulok, M. Adaptive Memetic Algorithm Enhanced with Data Geometry Analysis to Select Training Data for SVMs. Neurocomputing 2016, 185, 113–132. [Google Scholar] [CrossRef]
- Martin, J.K.; Hirschberg, D. On the complexity of learning decision trees. International Symposium on Artificial Intelligence and Mathematics. Citeseer 1996, 112–115. [Google Scholar]
- Chang, F.; Guo, C.Y.; Lin, X.R.; Lu, C.J. Tree decomposition for large-scale SVM problems. J. Mach. Learn. Res. 2010, 11, 2935–2972. [Google Scholar]
- Chau, A.L.; Li, X.; Yu, W. Support vector machine classification for large datasets using decision tree and fisher linear discriminant. Future Gener. Comput. Syst. 2014, 36, 57–65. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia, F.; Chau, A.L.; Rodriguez-Mazahua, L.; Castilla, J.S.R. Data selection based on decision tree for SVM classification on large data sets. Appl. Soft Comput. 2015, 37, 787–798. [Google Scholar] [CrossRef]
- Radial Basis Function Kernel, Wikipedia, Wikipedia Foundation. Available online: https://en.wikipedia.org/wiki/Radial_basis_function_kernel (accessed on 29 November 2019).
- Kass, G.V. An Exploratory Technique for Investigating Large Quantities of Categorical Data. J. R. Stat. Soc. 1980, 29, 119–127. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Wadsworth and Brooks: Monterey, CA, USA, 1984. [Google Scholar]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 1993. [Google Scholar]
- Loh, W.Y.; Shih, Y.S. Split Selection Methods for Classification Trees. Stat. Sin. 1997, 7, 815–840. [Google Scholar]
- Martin, J.K.; Hirschberg, D.S. The Time Complexity of Decision Tree Induction; University of California: Irvine, CA, USA, 1995. [Google Scholar]
- Li, X.B.; Sweigart, J.; Teng, J.; Donohue, J.; Thombs, L. A dynamic programming based pruning method for decision trees. Inf. J. Comput. 2001, 13, 332–344. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Dua, D.; Graff, C. UCI Machine Learning Repository; University of California, School of Information and Computer Science: Irvine, CA, USA, 2019. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM: A Library for Support Vector Machines. ACM Transactions on Intelligent Systems and Technology, 2:27:1—27:27. 2011. Available online: https://www.csie.ntu.edu.tw/cjlin/libsvmtools/datasets/binary.html (accessed on 29 November 2019).
- Ho, T.K.; Kleinberg, E.M. Checkerboard Data Set. 1996. Available online: https://research.cs.wisc.edu/math-prog/mpml.html (accessed on 29 November 2019).
- Prokhorov, D. IJCNN 2001 Neural Network Competition.Slide Presentation in IJCNN’01, Ford Research Laboratory. 2001. Available online: https://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html (accessed on 29 November 2019).
| Region | Distinct Adjacent Regions |
|---|---|
| Dataset | Size | Dim | ||
|---|---|---|---|---|
| Iris-Setosa | 150 | 4 | 50 | 100 |
| Iris-Versicolor | 150 | 4 | 50 | 100 |
| Iris-Virginia | 150 | 4 | 50 | 100 |
| Breast Cancer | 683 | 10 | 444 | 239 |
| Four-class | 862 | 2 | 555 | 307 |
| Checkerboard | 1000 | 2 | 514 | 486 |
| German Credit | 1000 | 24 | 700 | 300 |
| Waveform-0 | 5000 | 21 | 1657 | 3343 |
| Waveform-1 | 5000 | 21 | 1657 | 3343 |
| Waveform-2 | 5000 | 21 | 1657 | 3343 |
| Banana | 5300 | 2 | 2924 | 2376 |
| Mushroom | 8124 | 112 | 3916 | 4208 |
| Phishing | 11,055 | 68 | 4898 | 6157 |
| w8a | 45,546 | 300 | 44,226 | 1320 |
| a9a | 48,842 | 123 | 37,155 | 11,687 |
| IJCNN-1 | 141,691 | 22 | 128,126 | 13,565 |
| Skin Segmentation | 245,057 | 3 | 50,859 | 194,198 |
| Cod-RNA | 488,565 | 8 | 325,710 | 162,855 |
| Method | Dataset | Dataset | Penalty Factor C | RBF Kernel r | ||
|---|---|---|---|---|---|---|
| Iris-Setosa | 10 | 0.001 | Waveform-2 | 0.1 | ||
| 100 | 0.001 | 10 | 0.01 | |||
| 100 | 0.001 | 100 | 0.001 | |||
| 100 | 0.001 | 1 | ||||
| Iris-Versicolor | 10 | 0.1 | Banana | 1 | 1 | |
| 100 | 0.1 | 100 | 1 | |||
| 0.1 | 0.01 | 10 | ||||
| 10 | 1 | 1 | 1 | |||
| Iris-Virginia | 100 | 0.01 | Mushroom | 100 | 0.001 | |
| 100 | 0.01 | 10 | 0.001 | |||
| 100 | 0.001 | 0.1 | 0.01 | |||
| 100 | 0.1 | 100 | 0.001 | |||
| Breast Cancer | 1 | 0.01 | Phishing | 10 | ||
| 10 | 0.001 | 1 | 0.01 | |||
| 100 | 0.001 | 100 | 0.001 | |||
| 1 | 0.1 | 100 | 0.001 | |||
| Four-class | 10 | 1 | w8a | 10 | 0.001 | |
| 100 | 1 | 10 | 0.01 | |||
| 100 | 1 | 1 | 0.001 | |||
| 10 | 1 | 10 | 0.001 | |||
| Checkerboard | 100 | 1 | a9a | 10 | 0.001 | |
| 100 | 1 | 10 | 0.01 | |||
| 100 | 1 | 100 | 0.001 | |||
| 100 | 1 | 10 | 0.001 | |||
| German Credit | 100 | 0.001 | IJCNN-1 | 10 | 0.1 | |
| 1 | 100 | |||||
| 0.1 | 0.001 | 100 | 0.01 | |||
| 1 | 0.01 | 10 | ||||
| Waveform-0 | 1 | 0.01 | Skin Segmentation | 100 | 1 | |
| 10 | 0.01 | 100 | 1 | |||
| 1 | 0.01 | 100 | 0.1 | |||
| 1 | 0.01 | 100 | 1 | |||
| Waveform-1 | 1 | Cod-RNA | 10 | 1 | ||
| 10 | 100 | 0.001 | ||||
| 1 | 100 | 0.01 | ||||
| 1 | 10 |
| Dataset | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| acc | Time | acc | Time | Acc | Time | Acc | Time | |||||
| Iris-Setosa | 100 | 0 | 0.01 | 100 | 0 | 0.01 | 100 | 0 | 0.01 | 100 | 0 | 0.01 |
| Iris-Versicolor | 95.43 | 0.98 | 0.01 | 78.54 | 12.71 | 0.01 | 52.29 | 17.53 | 0.02 | 87.71 | 2.22 | 0.02 |
| Iris-Virginia | 96.57 | 2.24 | 0.01 | 87.99 | 6.55 | 0.01 | 91.71 | 4.07 | 0.02 | 91.5 | 4.72 | 0.01 |
| Breast Cancer | 96.99 | 0.89 | 0.01 | 95.36 | 0.5 | 0.01 | 96.3 | 0.95 | 0.08 | 96.24 | 1.04 | 0.04 |
| Four-class | 100 | 0 | 0.01 | 93.19 | 0.08 | 0.01 | 93.28 | 3.58 | 0.07 | 93.24 | 2.4 | 0.05 |
| Checkerboard | 94.3 | 0.63 | 0.03 | 84.61 | 1.47 | 0.01 | - | - | - | 79.34 | 3.13 | 0.08 |
| German Credit | 75.7 | 0.7 | 0.09 | 69.87 | 0.75 | 0.01 | 70 | 0.04 | 0.42 | 70.55 | 0.97 | 0.21 |
| Waveform-0 | 89.85 | 0.63 | 1.03 | 85.5 | 0.73 | 0.21 | 87.51 | 0.65 | 0.58 | 89.01 | 0.51 | 0.34 |
| Waveform-1 | 91.26 | 0.3 | 0.68 | 83.7 | 0.87 | 0.2 | 89.46 | 0.38 | 0.66 | 90.06 | 0.5 | 0.35 |
| Waveform-2 | 91.76 | 0.48 | 0.94 | 84.52 | 1.23 | 0.21 | 89.77 | 0.97 | 0.74 | 90.31 | 0.48 | 0.38 |
| Banana | 90.6 | 0.69 | 0.59 | 64.13 | 2.69 | 0.03 | 83.46 | 1.77 | 0.09 | 88.8 | 0.4 | 0.06 |
| Mushroom | 100 | 0 | 1.47 | 90.67 | 0.62 | 2.08 | 84.99 | 7.23 | 1.11 | 99.83 | 0.12 | 0.79 |
| Phishing | 96.69 | 0.26 | 4.59 | 90 | 0.49 | 2.28 | 93.28 | 0.69 | 1.46 | 93.96 | 0.26 | 0.79 |
| w8a | 99.11 | 0.04 | 114.85 | 97.4 | 0.05 | 19.64 | 97.83 | 0.3 | 231.89 | 98.38 | 0.09 | 51.18 |
| a9a | 84.7 | 0.14 | 373.24 | 78.48 | 0.29 | 43.19 | 81.27 | 0.72 | 21.91 | 84.24 | 0.16 | 12.59 |
| IJCNN-1 | 99.27 | 0.7 | 224.29 | 97.02 | 0.12 | 9.83 | 97.55 | 0.1 | 12 | 98.39 | 0.05 | 7.71 |
| Skin Segmentation | 99.94 | 0 | 20.23 | 99.93 | 0.01 | 1.27 | 99.45 | 0.11 | 3.39 | 99.89 | 0.01 | 2.51 |
| Cod-RNA | 96.98 | 0.03 | 4425.55 | 90.68 | 0.08 | 178.3 | 96.05 | 0.08 | 66.93 | 96.32 | 0.03 | 42.85 |
| Iris-Setosa | 100 | 0 | 0.01 | 100 | 0 | 0.01 | 100 | 0 | 0.01 | 100 | 0 | 0.01 |
| Iris-Versicolor | 95.43 | 0.98 | 0.01 | 89.44 | 3.92 | 0.01 | 51.72 | 17.02 | 0.02 | 93.43 | 1.48 | 0.02 |
| Iris-Virginia | 96.57 | 2.24 | 0.01 | 93.43 | 2.2 | 0.01 | 94.26 | 2.17 | 0.02 | 90.31 | 3.03 | 0.02 |
| Breast Cancer | 96.99 | 0.89 | 0.01 | 95.55 | 1.09 | 0.01 | 96.49 | 0.91 | 0.07 | 96.3 | 0.95 | 0.06 |
| Four-class | 100 | 0 | 0.01 | 96.42 | 1.78 | 0.01 | 95.38 | 1.63 | 0.07 | 98.81 | 0.8 | 0.05 |
| Checkerboard | 94.3 | 0.63 | 0.03 | 93.66 | 1.09 | 0.01 | - | - | - | 83.66 | 2.48 | 0.08 |
| German Credit | 75.7 | 0.7 | 0.09 | 70.04 | 0.01 | 0.02 | 70 | 0.04 | 0.35 | 70.56 | 0.62 | 0.25 |
| Waveform-0 | 89.85 | 0.63 | 1.03 | 88.35 | 0.37 | 0.27 | 88.55 | 0.73 | 0.76 | 89.28 | 0.73 | 0.54 |
| Waveform-1 | 91.26 | 0.3 | 0.68 | 86.45 | 0.56 | 0.26 | 89.89 | 0.37 | 0.78 | 90.52 | 0.49 | 0.5 |
| Waveform-2 | 91.76 | 0.48 | 0.94 | 88.21 | 0.5 | 0.27 | 90.39 | 0.54 | 0.99 | 90.62 | 0.58 | 0.51 |
| Banana | 90.6 | 0.69 | 0.59 | 79.86 | 2.02 | 0.1 | 85.14 | 1.5 | 0.1 | 89.69 | 0.36 | 0.07 |
| Mushroom | 100 | 0 | 1.47 | 97.09 | 1.12 | 2.22 | 97.37 | 0.88 | 1.33 | 99.91 | 0.1 | 0.85 |
| Phishing | 96.69 | 0.26 | 4.59 | 93.91 | 0.2 | 2.58 | 94.42 | 0.38 | 1.63 | 94.62 | 0.3 | 0.99 |
| w8a | 99.11 | 0.04 | 114.85 | 97.5 | 0.07 | 27.22 | 98.12 | 0.12 | 233.78 | 98.63 | 0.05 | 63.04 |
| a9a | 84.7 | 0.14 | 373.24 | 78.86 | 0.26 | 74.22 | 82.19 | 0.42 | 39.37 | 84.61 | 0.18 | 24.27 |
| IJCNN-1 | 99.27 | 0.7 | 224.29 | 98.41 | 0.05 | 23.56 | 98.07 | 0.09 | 18.7 | 98.72 | 0.07 | 14.45 |
| Skin Segmentation | 99.94 | 0 | 20.23 | 99.94 | 0.01 | 2.36 | 99.73 | 0.06 | 4.82 | 99.9 | 0.01 | 3.64 |
| Cod-RNA | 96.98 | 0.03 | 4425.55 | 95.44 | 0.03 | 514.9 | 96.19 | 0.03 | 293.29 | 96.43 | 0.03 | 152.98 |
| Iris-Setosa | 100 | 0 | 0.01 | 100 | 0 | 0.01 | 100 | 0 | 0.01 | 100 | 0 | 0.01 |
| Iris-Versicolor | 95.43 | 0.98 | 0.01 | 92.3 | 2.44 | 0.01 | 59.87 | 14.47 | 0.01 | 94.01 | 2.57 | 0.02 |
| Iris-Virginia | 96.57 | 2.24 | 0.01 | 93.98 | 2.03 | 0.01 | 95.51 | 2.53 | 0.01 | 94.31 | 1.77 | 0.02 |
| Breast Cancer | 96.99 | 0.89 | 0.01 | 96.42 | 0.69 | 0.01 | 96.36 | 0.55 | 0.08 | 96.17 | 0.79 | 0.05 |
| Four-class | 100 | 0 | 0.01 | 96.62 | 1.52 | 0.01 | 95.62 | 1.6 | 0.07 | 99.5 | 0.66 | 0.05 |
| Checkerboard | 94.3 | 0.63 | 0.03 | 93.96 | 1 | 0.01 | - | - | - | 87.18 | 2.3 | 0.1 |
| German Credit | 75.7 | 0.7 | 0.09 | 69.95 | 0.22 | 0.03 | 70 | 0.04 | 0.35 | 70.81 | 0.87 | 0.29 |
| Waveform-0 | 89.85 | 0.63 | 1.03 | 89.1 | 0.38 | 0.35 | 88.93 | 0.71 | 0.71 | 89.47 | 0.34 | 0.41 |
| Waveform-1 | 91.26 | 0.3 | 0.68 | 87.49 | 0.33 | 0.34 | 90.18 | 0.46 | 0.74 | 90.8 | 0.34 | 0.38 |
| Waveform-2 | 91.76 | 0.48 | 0.94 | 89.63 | 0.43 | 0.33 | 90.71 | 0.66 | 0.84 | 91.09 | 0.63 | 0.41 |
| Banana | 90.6 | 0.69 | 0.59 | 81.47 | 0.74 | 0.17 | 86.63 | 1.83 | 0.12 | 90.09 | 0.49 | 0.1 |
| Mushroom | 100 | 0 | 1.47 | 97.79 | 0.71 | 2.17 | 97.87 | 0.28 | 1.71 | 99.95 | 0.07 | 0.97 |
| Phishing | 96.69 | 0.26 | 4.59 | 94.51 | 0.16 | 3 | 94.75 | 0.3 | 1.99 | 94.93 | 0.14 | 1.29 |
| w8a | 99.11 | 0.04 | 114.85 | 97.71 | 0.08 | 49.63 | 98.36 | 0.07 | 237.01 | 98.83 | 0.06 | 55.09 |
| a9a | 84.7 | 0.14 | 373.24 | 80.35 | 0.2 | 127.4 | 83.18 | 0.28 | 77.18 | 84.68 | 0.13 | 43.02 |
| IJCNN-1 | 99.27 | 0.7 | 224.29 | 98.68 | 0.06 | 37.79 | 98.3 | 0.08 | 34.75 | 98.86 | 0.06 | 25.43 |
| Skin Segmentation | 99.94 | 0 | 20.23 | 99.94 | 0.01 | 3.84 | 99.75 | 0.08 | 7.01 | 99.91 | 0 | 3.78 |
| Cod-RNA | 96.98 | 0.03 | 4425.55 | 95.92 | 0.04 | 899.1 | 96.27 | 0.01 | 598.23 | 96.48 | 0.02 | 256.71 |
| 0.1 | 10 | 6 | 2 | 9 | 9 | 0 |
| 0.2 | 11 | 4 | 3 | 9 | 8 | 1 |
| 0.3 | 11 | 4 | 3 | 10 | 8 | 0 |
| Total | 32 | 14 | 8 | 28 | 25 | 1 |
| Dataset | SVM | CVS | FLD | TRSD | ||||
|---|---|---|---|---|---|---|---|---|
| SC | TS | SC | TS | SC | TS | SC | TS | |
| Iris-Setosa | 0 | 0.01 | 0 | 0.01 | 0 | 0.01 | 0 | 0.01 |
| Iris-Versicolor | 0 | 0.01 | 0 | 0.01 | 0 | 0.01 | 0 | 0.01 |
| Iris-Virginia | 0 | 0.01 | 0 | 0.01 | 0 | 0.01 | 0 | 0.01 |
| Breast Cancer | 0 | 0.01 | 0 | 0.01 | 0.07 | 0.01 | 0.04 | 0.01 |
| Four-class | 0 | 0.01 | 0 | 0.01 | 0.06 | 0.01 | 0.04 | 0.01 |
| Checkerboard | 0 | 0.03 | 0 | 0.01 | - | - | 0.09 | 0.01 |
| German Credit | 0 | 0.09 | 0.01 | 0.02 | 0.33 | 0.02 | 0.27 | 0.02 |
| Waveform-0 | 0 | 1.03 | 0.18 | 0.17 | 0.58 | 0.13 | 0.3 | 0.11 |
| Waveform-1 | 0 | 0.68 | 0.17 | 0.17 | 0.65 | 0.09 | 0.3 | 0.08 |
| Waveform-2 | 0 | 0.94 | 0.17 | 0.16 | 0.73 | 0.11 | 0.34 | 0.07 |
| Banana | 0 | 0.59 | 0 | 0.17 | 0.08 | 0.04 | 0.05 | 0.05 |
| Mushroom | 0 | 1.47 | 1.9 | 0.27 | 0.99 | 0.72 | 0.71 | 0.26 |
| Phishing | 0 | 4.59 | 2.08 | 0.92 | 1.3 | 0.69 | 0.72 | 0.57 |
| w8a | 0 | 114.85 | 17.48 | 32.15 | 228.83 | 8.18 | 46.35 | 8.74 |
| a9a | 0 | 373.24 | 44.5 | 82.88 | 16.52 | 60.66 | 8.84 | 34.18 |
| IJCNN-1 | 0 | 224.29 | 6.06 | 31.73 | 10.08 | 24.67 | 5.77 | 19.66 |
| Skin Segmentation | 0 | 20.23 | 0.3 | 3.54 | 3.22 | 3.79 | 2.3 | 1.48 |
| Cod-RNA | 0 | 4425.55 | 0.7 | 898.4 | 15.46 | 582.77 | 11.24 | 245.47 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, M.; Lee, K. Selection of Support Vector Candidates Using Relative Support Distance for Sustainability in Large-Scale Support Vector Machines. Appl. Sci. 2020, 10, 6979. https://doi.org/10.3390/app10196979
Ryu M, Lee K. Selection of Support Vector Candidates Using Relative Support Distance for Sustainability in Large-Scale Support Vector Machines. Applied Sciences. 2020; 10(19):6979. https://doi.org/10.3390/app10196979
Chicago/Turabian StyleRyu, Minho, and Kichun Lee. 2020. "Selection of Support Vector Candidates Using Relative Support Distance for Sustainability in Large-Scale Support Vector Machines" Applied Sciences 10, no. 19: 6979. https://doi.org/10.3390/app10196979
APA StyleRyu, M., & Lee, K. (2020). Selection of Support Vector Candidates Using Relative Support Distance for Sustainability in Large-Scale Support Vector Machines. Applied Sciences, 10(19), 6979. https://doi.org/10.3390/app10196979






