Urinary 1H-NMR Metabolic Signature in Subjects Undergoing Colonoscopy for Colon Cancer Diagnosis
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Sample Collection
2.2. Urine Samples Preparation
2.3. 1H-NMR Spectroscopic Analysis
2.4. NMR Data Preprocessing and Multivariate Statistical Analysis
2.5. Serum Carcinoembryonic Antigen Level Determination
3. Results
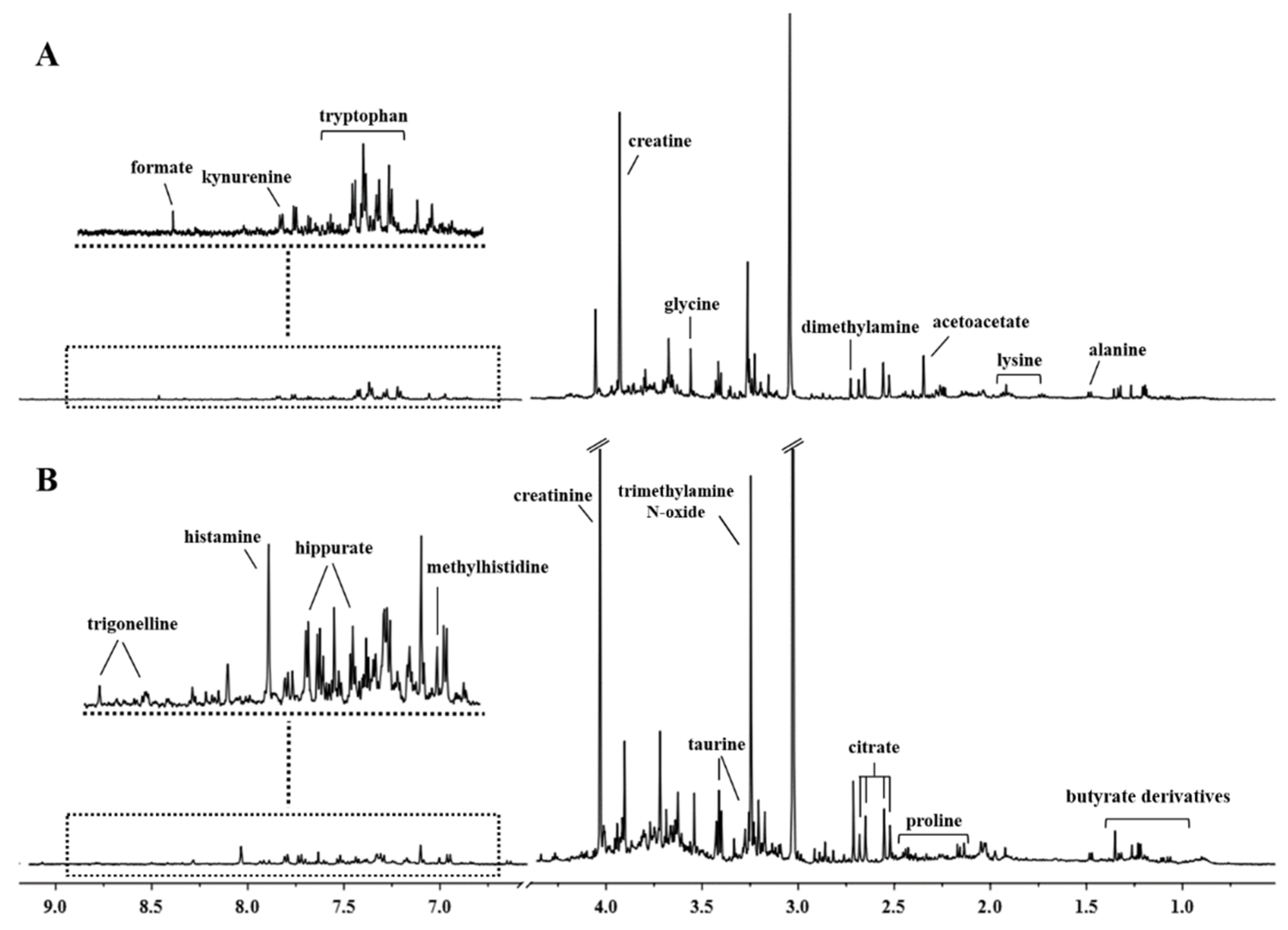
3.1. 1H-NMR Spectra of Urine Samples
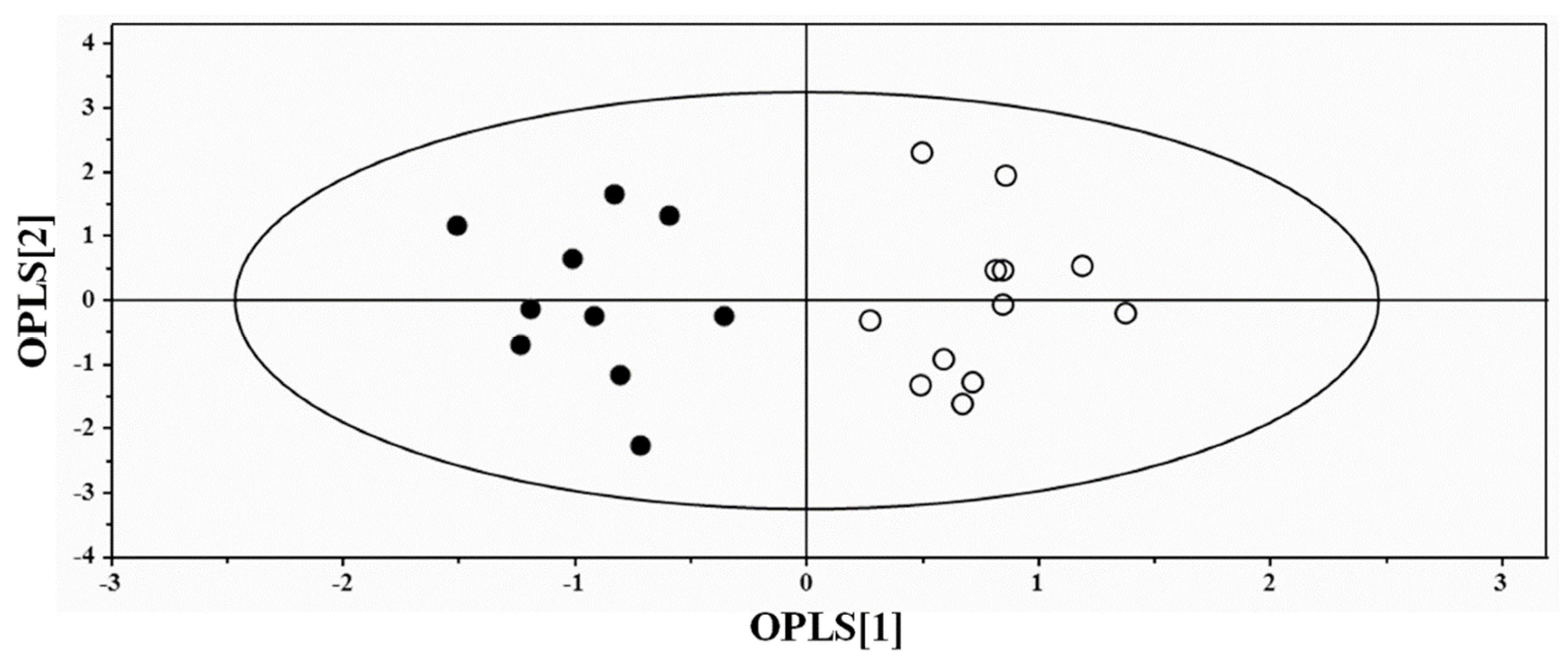
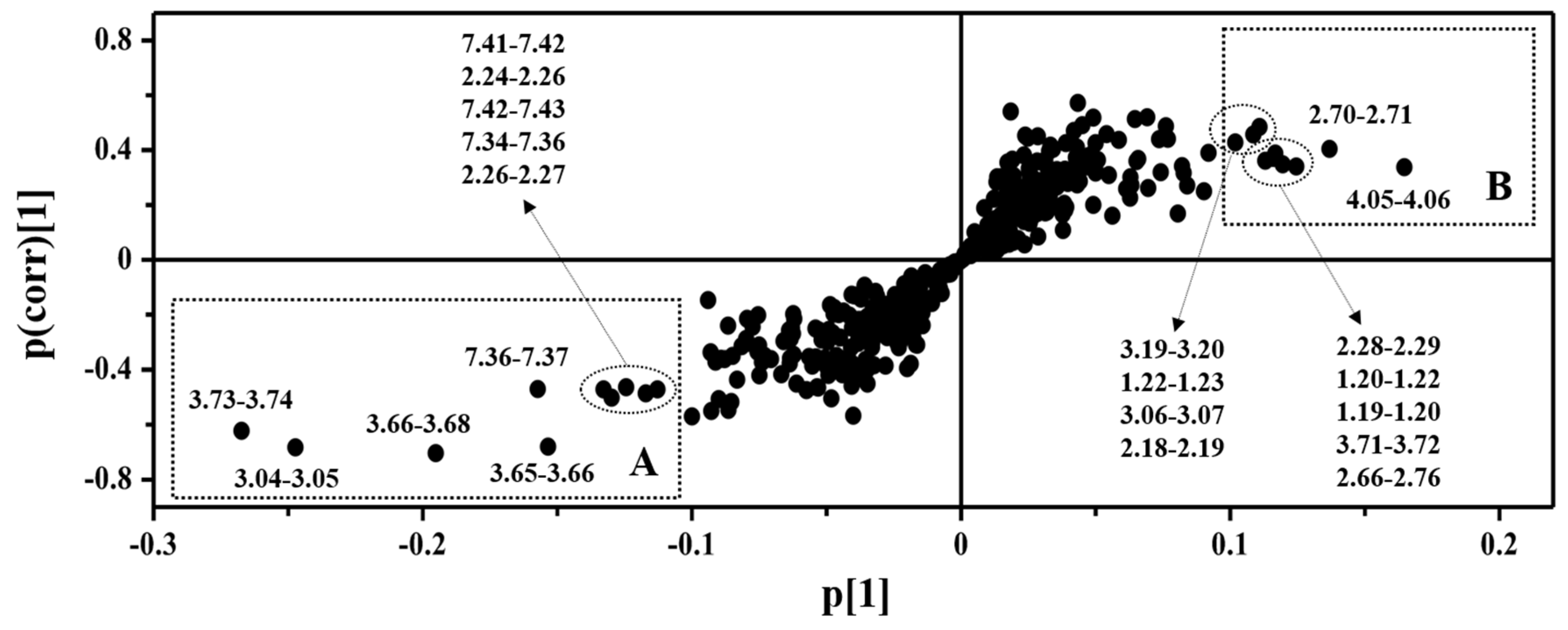
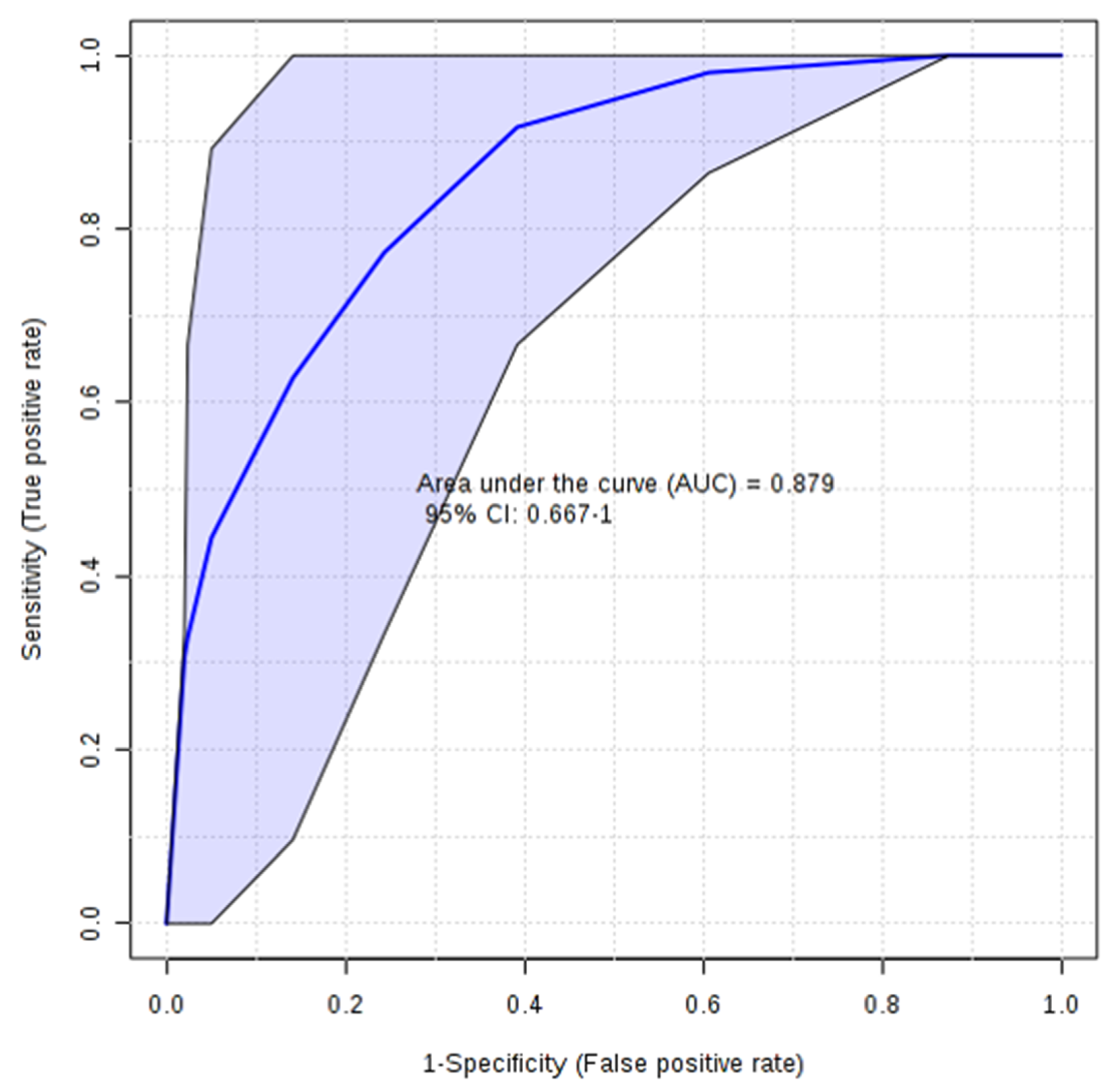
3.2. Multivariate Statistical Analysis of NMR Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Soerjomataram, I.; Lortet-Tieulent, J.; Parkin, D.M.; Ferlay, J.; Mathers, C.; Forman, D.; Bray, F. Global burden of cancer in 2008: A systematic analysis of disability-adjusted life-years in 12 world regions. Lancet 2012, 380, 1840–1850. [Google Scholar] [CrossRef]
- El-Awady, S.; Lithy, R.; Morshed, M.; Khafagy, W.; Abd Monem, H.; Waleed, O.; Badr, S.; Fekry, A.; El Nakeeb, A.; Ghazy, H.; et al. Utility of serum preoperative carcinoembryonic antigen in colorectal cancer patients. Hepato-Gastroenterology 2009, 56, 361–366. [Google Scholar] [PubMed]
- Hewitson, P.; Glasziou, P.; Watson, E.; Towler, B.; Irwig, L. Cochrane systematic review of colorectal cancer screening using the fecal occult blood test (hemoccult): An update. Am. J. Gastroenterol. 2008, 103, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Ahlquist, D.A.; Hongzhi, Z.; Domanico, M.; Mahoney, D.W.; Yab, T.C.; Taylor, W.R.; Butz, M.L.; Thibodeau, S.N.; Rabeneck, L.; Paszat, L.F.; et al. Next-Generation Stool DNA Test Accurately Detects Colorectal Cancer and Large Adenomas. Gastroenterology 2012, 142, 248–256. [Google Scholar] [CrossRef]
- Grady, W.M.; Markowitz, S.D. The molecular pathogenesis of colorectal cancer and its potential application to colorectal cancer screening. Dig. Dis. Sci. 2015, 60, 762–772. [Google Scholar] [CrossRef]
- Jung, J.; Jung, Y.; Bang, E.J.; Cho, S.; Jang, Y.J.; Kwak, J.M.; Ryu, D.H.; Park, S.; Hwang, G.S. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR-based metabolomic profiling. Ann. Surg. Oncol. 2014, 21, 736–742. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.M.; Darzi, A.W.; Takats, Z.; Lindon, J.C. Metabolic phenotyping in clinical and surgical environments. Nature 2012, 491, 384–392. [Google Scholar] [CrossRef]
- Chan, E.C.Y.; Koh, P.K.; Mal, M.; Cheah, P.Y.; Eu, K.W.; Backshall, A.; Cavill, R.; Nicholson, J.K.; Keun, H.C. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 2009, 8, 352–361. [Google Scholar] [CrossRef]
- Barberini, L.; Restivo, A.; Noto, A.; Deidda, S.; Fattuoni, C.; Fanos, V.; Saba, L.; Zorcolo, L.; Mussap, M. A gas chromatography-mass spectrometry (GC-MS) metabolomics approach in human colorectal cancer (CRC): The emerging role of monosaccharides and amino acids. Ann. Transl. Med. 2019, 7, 727. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, G.; Chen, T.; Qiu, Y.; Zou, X.; Zheng, M.; Tan, B.; Feng, B.; Don, T.; He, P.; et al. Distinct urinary metabolic profile of human colorectal cancer. J. Proteome Res. 2012, 11, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lin, Y.; Liang, J.; Huang, Y.; Ma, C.; Liu, X.; Yang, J. NMR-based metabolomic techniques identify potential urinary biomarkers for early colorectal cancer detection. Oncotarget 2017, 8, 105819–105831. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.D.; Shinkins, B.; Pathiraja, I.; Roberts, N.W.; James, T.J.; Mallett, S.; Perera, R.; Primrose, J.N.; Mant, D. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst. Rev. 2015, 12, CD011134. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Pintus, R.; Pruna, D.; Dessì, A.; Atzori, L.; Fanos, V. Pediatric Acute-onset Neuropsychiatric Syndrome and Mycoplasma PneumoniaeInfection: A Case Report Analysis with a Metabolomics Approach. Curr. Pediatr. Rev. 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.A.A.; Magalhaes, A.; Ferreira, M.M.C. Optimized bucketing for NMR spectra: Three case studies. Chemometr. Intell. Lab. 2013, 122, 93–102. [Google Scholar] [CrossRef]
- Wu, Y.; Li, L. Sample normalization methods in quantitative metabolomics. J. Chromatogr. A 2016, 1430, 80–95. [Google Scholar] [CrossRef]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted Profiling: Quantitative Analysis of 1HNMR Metabolomics Data. Anal. Chem. 2006, 2, 4430–4442. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucl. Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Schicho, R.; Shaykhutdinov, R.; Ngo, J.; Nazyrova, A.; Schneider, C.; Panaccione, R.; Kaplan, G.G.; Vogel, H.J.; Storr, M. Quantitative metabolomic profiling of serum, plasma, and urine by 1H-NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J. Proteome Res. 2012, 11, 3344–3357. [Google Scholar] [CrossRef]
- Bathe, O.F.; Farshidfar, F. From Genotype to Functional Phenotype: Unraveling the Metabolomic Features of Colorectal Cancer. Genes 2014, 5, 536–560. [Google Scholar] [CrossRef]
- Lin, J.K.; Lin, C.C.; Yang, S.H.; Wang, H.S.; Jiang, J.K.; Lan, Y.T.; Lin, T.C.; Li, A.F.Y.; Chen, W.S.; Chang, S.C. Early postoperative CEA level is a better prognostic indicator than is preoperative CEA level in predicting prognosis of patients with curable colorectal cancer. Int. J. Colorectal Dis. 2011, 26, 1135–1141. [Google Scholar] [CrossRef]
- Wang, H.; Tso, V.; Wong, C.; Sadowski, D.; Fedorak, R.N. Development and Validation of a Highly Sensitive Urine-Based Test to Identify Patients with Colonic Adenomatous Polyps. Clin. Trans. Gastroenterol. 2014, 5, e54. [Google Scholar] [CrossRef] [PubMed]
- Jobard, E.; Pontoizeau, C.; Blaise, B.J.; Bachelot, T.; Elena-Herrmann, B.; Trédan, O. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 2014, 343, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Argiles, J.; Azco Bieto, J. The metabolic environment of cancer. Mol. Cell. Biochem. 1988, 81, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Eisner, R.; Stretch, C.; Eastman, T.; Xia, J.; Hau, D.; Damaraju, S.; Greiner, R.; Wishart, D.S.; Baracos, V.E. Learning to predict cancer-associated skeletal muscle wasting from 1H-NMR profiles of urinary metabolites. Metabolomics 2011, 7, 25–34. [Google Scholar] [CrossRef]
- Wyss, M.; Kaddurah-Daouk, R. Creatine and creatinine metabolism. Physiol. Rev. 2000, 80, 1107–1213. [Google Scholar] [CrossRef]
- Bryan, G.T. The role of urinary tryptophan metabolites in the etiology of bladder cancer. Am. J. Clin. Nutr. 1971, 24, 841–847. [Google Scholar] [CrossRef]
- Denz, H.; Orth, B.; Weiss, G.; Herrmann, R.; Huber, P.; Wachter, H.; Fuchs, D. Weight loss in patients with hematological neoplasias is associated with immune system stimulation. Clin. Investig. 1993, 71, 37–41. [Google Scholar] [CrossRef]
- Carlin, J.M.; Ozaki, Y.; Byrne, G.I.; Brown, R.R.; Borden, E.C. Interferons and indoleamine 2,3-dioxygenase: Role in antimicrobial and antitumor effects. Experientia 1989, 45, 535–541. [Google Scholar] [CrossRef]
- Murr, C.; Bergant, A.; Widschwendter, M.; Heim, K.; Schrocksnadel, H.; Fuchs, D. Neopterin is an independent prognostic variable in females with breast cancer. Clin. Chem. 1999, 45, 1998–2004. [Google Scholar] [CrossRef]
- Platten, M.; Wick, W.; Van den Eynde, B.J. Tryptophan catabolism in cancer: Beyond IDO and tryptophan depletion. Cancer Res. 2012, 72, 5435–5440. [Google Scholar] [CrossRef]
- Medina, V.A.; Rivera, E.S. Histamine receptors and cancer pharmacology. Br. J. Pharmacol. 2010, 161, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Cricco, G.P.; Mohamad, N.A.; Sambuco, L.A.; Genre, F.; Croci, M.; Gutiérrez, A.S.; Medina, V.A.; Bergoc, R.M.; Rivera, E.S.; Martín, G.A. Histamine regulates pancreatic carcinoma cell growth through H3 and H4 receptors. Inflamm. Res. 2008, 57, 23–24. [Google Scholar] [CrossRef] [PubMed]
- Medina, V.; Croci, M.; Crescenti, E.; Mohamad, N.; Sanchez-Jiménez, F.; Massari, N.; Nuñez, M.; Cricco, G.; Martin, G.; Bergoc, R.; et al. The role of histamine in human mammary carcinogenesis: H3 and H4 receptors as potential therapeutic targets for breast cancer treatment. Cancer Biol. Ther. 2008, 7, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Cai, G.; Su, M.; Chen, T.; Liu, Y.; Xu, Y.; Ni, Y.; Zhao, A.; Cai, S.; Xu, L.X. Urinary metabonomic study on colorectal cancer. J. Proteome Res. 2010, 9, 1627–1634. [Google Scholar] [CrossRef]
- Deng, L.; Ismond, K.; Liu, Z.; Constable, J.; Wang, H.; Alatise, O.I.; Weiser, M.R.; Kingham, T.P.; Chang, D. Urinary Metabolomics to Identify a Unique Biomarker Panel for Detecting Colorectal Cancer: A Multicentre Study. Cancer Epidemiol. Prev. Biomark. 2019, 28, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
| CRC Patients | Controls | |
|---|---|---|
| Number | 14 | 10 |
| Age (median, range) | 67.4, 40–88 | 56.3, 47–67 |
| Male/female ratio | 11/3 | 7/3 |
| CEA (>2.5 ng/mL) | 5 | 0 |
| Cancer site | ||
| Colon | 6 | - |
| Rectum | 7 | - |
| Caecum | 1 | - |
| FOBT | ||
| Positive | 9 | N/A |
| Negative | 5 | N/A |
| Dukes’ Stage | ||
| A | 3 | - |
| B | 7 | - |
| C | 3 | - |
| D | 1 | - |
| Metabolites | Mean (SD) of Group (mM) a | p-Value b | FC c (log2 CRC/C) | |
|---|---|---|---|---|
| C | CRC | |||
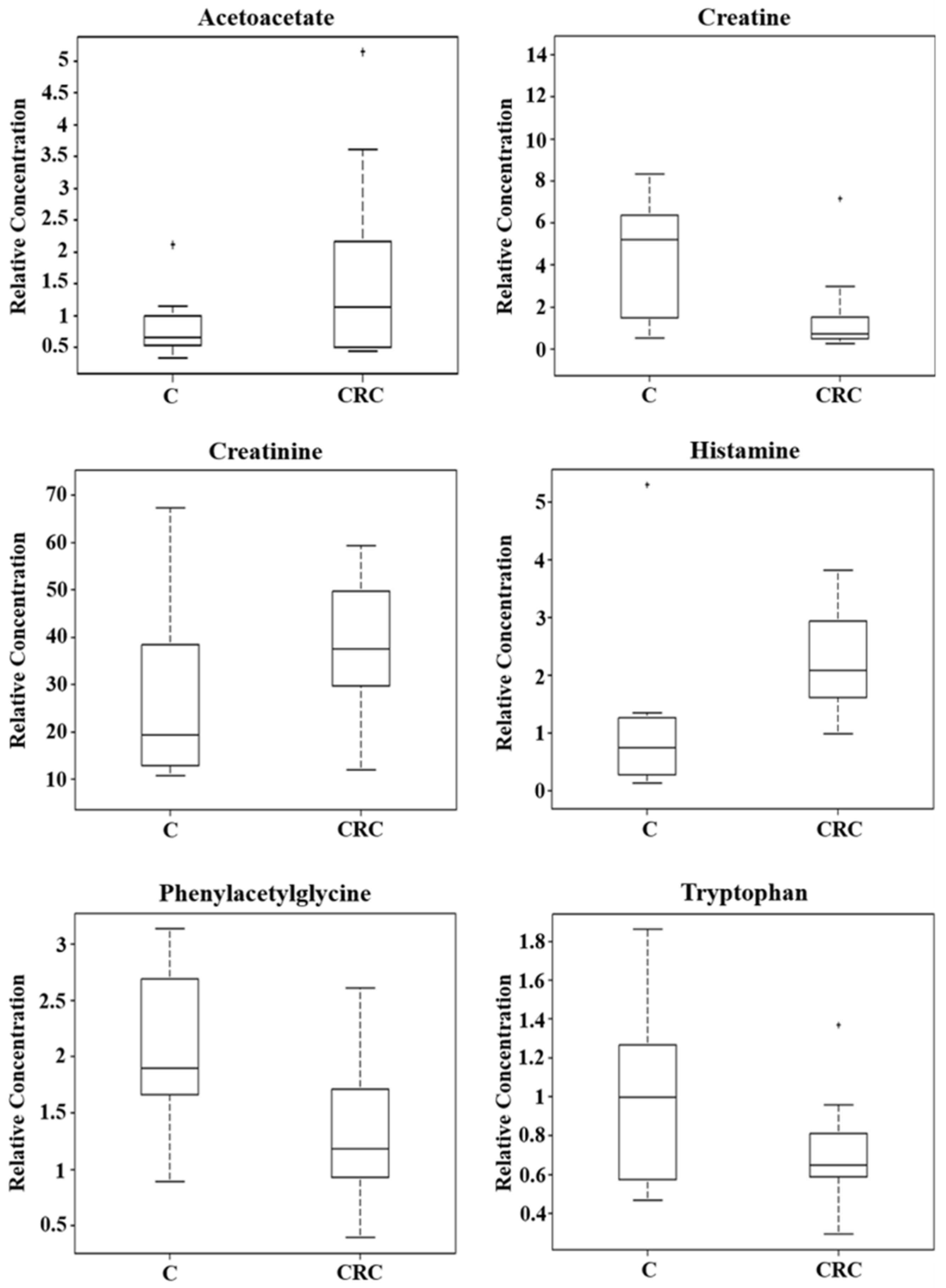
| Phenylacetylglycine | 2.07 ± 0.65 | 1.34 ± 0.72 | 0.01 | −0.623 |
| Histamine | 1.14 ± 0.49 | 2.26 ± 0.85 | 0.02 | 0.988 |
| Creatine | 6.64 ± 3.34 | 1.49 ± 0.98 | 0.03 | −2.150 |
| Tryptophan | 1.01 ± 0.46 | 0.71 ± 0.27 | 0.03 | −0.517 |
| Acetoacetate | 0.85 ± 0.28 | 1.64 ± 1.37 | 0.05 | 0.970 |
| Creatinine | 26.12 ± 17.96 | 37.83 ± 14.59 | 0.05 | 0.535 |
| 3-hydroxybutyrate | 1.00 ± 0.54 | 1.64 ± 1.45 | 0.10 | 0.706 |
| Tyrosine | 0.82 ± 0.41 | 0.62 ± 0.37 | 0.12 | −0.409 |
| Proline | 2.93 ± 1.12 | 3.38 ± 0.59 | 0.13 | 0.202 |
| 3-Aminoisobutyrate | 1.09 ± 0.54 | 2.08 ± 2.87 | 0.15 | 0.933 |
| Sn-glycero-3-phosphocholine | 1.62 ± 0.67 | 1.88 ± 0.71 | 0.19 | −0.215 |
| Fucose | 1.28 ± 0.54 | 1.16 ± 2.87 | 0.25 | 0.145 |
| Methylhistidine | 1.09 ± 0.67 | 1.23 ± 0.37 | 0.26 | −0.180 |
| Citrate | 8.57 ± 4.12 | 7.53 ± 6.94 | 0.34 | 0.187 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piras, C.; Pibiri, M.; Leoni, V.P.; Cabras, F.; Restivo, A.; Griffin, J.L.; Fanos, V.; Mussap, M.; Zorcolo, L.; Atzori, L. Urinary 1H-NMR Metabolic Signature in Subjects Undergoing Colonoscopy for Colon Cancer Diagnosis. Appl. Sci. 2020, 10, 5401. https://doi.org/10.3390/app10165401
Piras C, Pibiri M, Leoni VP, Cabras F, Restivo A, Griffin JL, Fanos V, Mussap M, Zorcolo L, Atzori L. Urinary 1H-NMR Metabolic Signature in Subjects Undergoing Colonoscopy for Colon Cancer Diagnosis. Applied Sciences. 2020; 10(16):5401. https://doi.org/10.3390/app10165401
Chicago/Turabian StylePiras, Cristina, Monica Pibiri, Vera Piera Leoni, Francesco Cabras, Angelo Restivo, Julian Leether Griffin, Vassilios Fanos, Michele Mussap, Luigi Zorcolo, and Luigi Atzori. 2020. "Urinary 1H-NMR Metabolic Signature in Subjects Undergoing Colonoscopy for Colon Cancer Diagnosis" Applied Sciences 10, no. 16: 5401. https://doi.org/10.3390/app10165401
APA StylePiras, C., Pibiri, M., Leoni, V. P., Cabras, F., Restivo, A., Griffin, J. L., Fanos, V., Mussap, M., Zorcolo, L., & Atzori, L. (2020). Urinary 1H-NMR Metabolic Signature in Subjects Undergoing Colonoscopy for Colon Cancer Diagnosis. Applied Sciences, 10(16), 5401. https://doi.org/10.3390/app10165401








