Application of Artificial Neural Network for Modeling and Studying In Vitro Genotype-Independent Shoot Regeneration in Wheat
Abstract
1. Introduction
2. Material and Methods
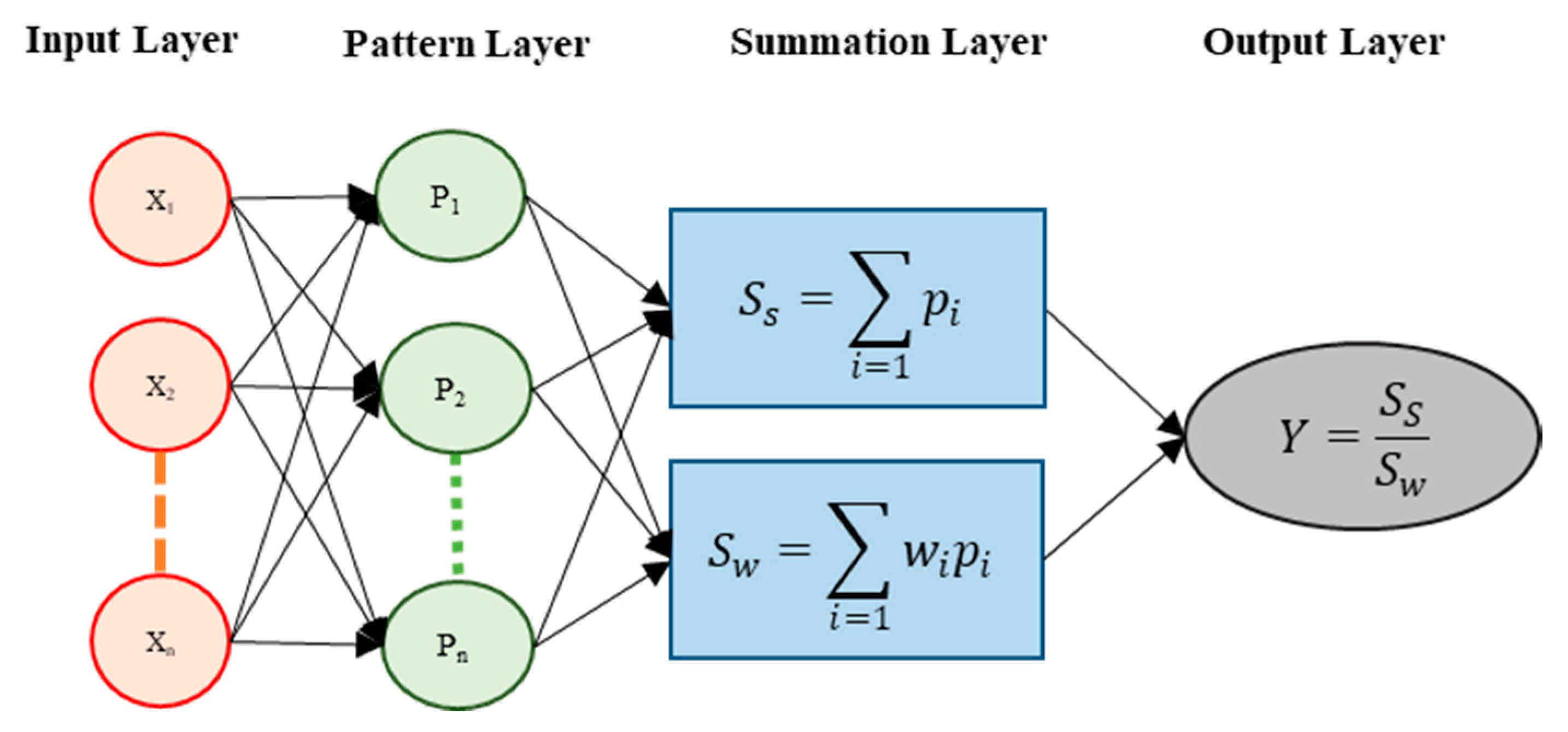
2.1. Data and Model Development
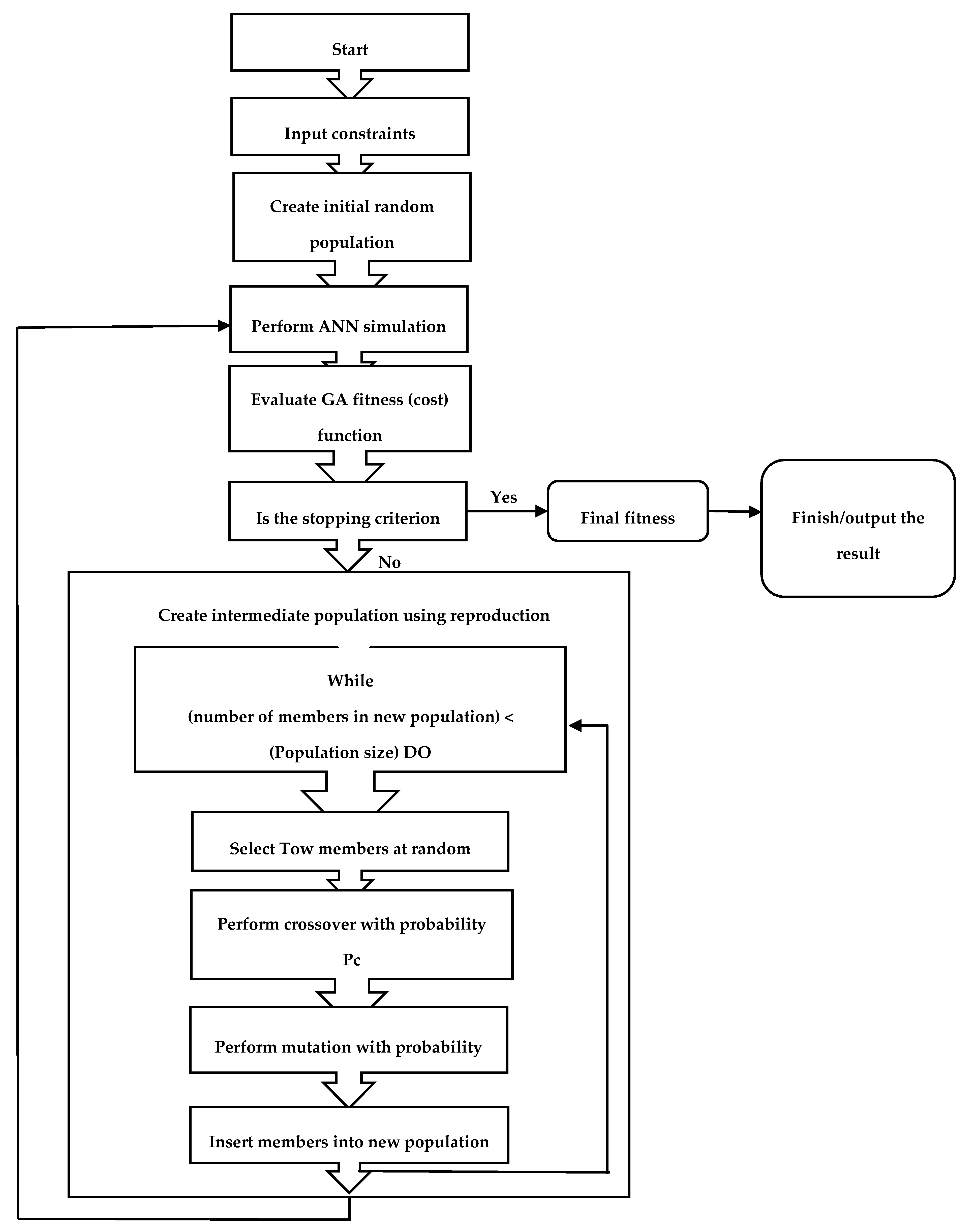
2.2. Process Optimization via Genetic Algorithm (GA)
2.3. Sensitivity Analysis of Shoot Regeneration to Variations in the Input Variables
3. Results
3.1. Artificial Intelligence Accurately Predicted In Vitro Shoot Regeneration
3.2. Determining an Optimized Solution for Regeneration by Using Generalized Regression Neural Network (GRNN)-GA
3.3. The Importance of Input Variables in Shoot Regeneration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.; Malik, K.; Kumar, S.; Jaiwal, P.K. Comparative regeneration in six bread wheat (Triticum aestivum L.) varieties from immature and mature scutella for developing efficient and genotype-independent protocol prerequisite for genetic improvement of wheat. In Vitro Cell. Dev. Biol. Plant 2020. [Google Scholar] [CrossRef]
- Kumari, A.; Baskaran, P.; Plačková, L.; Omámiková, H.; Nisler, J.; Doležal, K.; Van Staden, J. Plant growth regulator interactions in physiological processes for controlling plant regeneration and In Vitro development of Tulbaghia simmleri. J. Plant Physiol. 2018, 223, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Daneshvar, M.H.; Yoosefzadeh-Najafabadi, M.; Alizadeh, M. Effect of plant growth regulators on indirect shoot organogenesis of Ficus religiosa through seedling derived petiole segments. J. Genet. Eng. Biotechnol. 2018, 16, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Daneshvar, M.H.; Yoosefzadeh-Najafabadi, M. Establishment of a protocol for In Vitro seed germination and callus formation of Ficus religiosa L., an important medicinal plant. Jundishapur J. Nat. Pharm. Prod. 2018, 13, e62682. [Google Scholar] [CrossRef]
- Hesami, M.; Daneshvar, M.H. Indirect organogenesis through seedling-derived leaf segments of Ficus religiosa-a multipurpose woody medicinal plant. J. Crop. Sci. Biotechnol. 2018, 21, 129–136. [Google Scholar] [CrossRef]
- Bidabadi, S.S.; Jain, S.M. Cellular, Molecular, and Physiological Aspects of In Vitro Plant Regeneration. Plants 2020, 9, 702. [Google Scholar] [CrossRef]
- Niazian, M.; Shariatpanahi, M.E.; Abdipour, M.; Oroojloo, M. Modeling callus induction and regeneration in an anther culture of tomato (Lycopersicon esculentum L.) using image processing and artificial neural network method. Protoplasma 2019, 256, 1317–1332. [Google Scholar] [CrossRef]
- Prasad, V.; Gupta, S.D. Applications and potentials of artificial neural networks in plant tissue culture. In Plan Tissue Culture Engineering; Springer: Berlin, Germany, 2008; pp. 47–67. [Google Scholar]
- Zielinska, S.; Kepczynska, E. Neural modeling of plant tissue cultures: A review. BioTechnologia 2013, 94, 253–268. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Yoosefzadeh-Najafabadi, M.; Rahmati, M. Data-driven modeling in plant tissue culture. J. Appl. Environ. Biol. Sci. 2017, 7, 37–44. [Google Scholar]
- Specht, D.F. A general regression neural network. IEEE Trans. Neural Netw. 1991, 2, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Z.; Guo, S.; Li, C.J.; Sun, J.Q. A hybrid annual power load forecasting model based on generalized regression neural network with fruit fly optimization algorithm. Knowl. Based Syst. 2013, 37, 378–387. [Google Scholar] [CrossRef]
- Xia, C.; Lei, B.; Wang, H.; Li, J. GRNN short-term load forecasting model and virtual instrument design. Energy Procedia 2011, 13, 9150–9158. [Google Scholar] [CrossRef]
- Polat, Ö.; Yıldırım, T. Genetic optimization of GRNN for pattern recognition without feature extraction. Expert. Syst. Appl. 2008, 34, 2444–2448. [Google Scholar] [CrossRef]
- Kulkarni, S.G.; Chaudhary, A.K.; Nandi, S.; Tambe, S.S.; Kulkarni, B.D. Modeling and monitoring of batch processes using principal component analysis (PCA) assisted generalized regression neural networks (GRNN). Biochem. Eng. J. 2004, 18, 193–210. [Google Scholar] [CrossRef]
- Shahlaei, M.; Sabet, R.; Ziari, M.B.; Moeinifard, B.; Fassihi, A.; Karbakhsh, R. QSAR study of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2 using LS-SVM and GRNN based on principal components. Eur. J. Med. Chem. 2010, 45, 4499–4508. [Google Scholar] [CrossRef]
- Leung, M.T.; Chen, A.-S.; Daouk, H. Forecasting exchange rates using general regression neural networks. Comput. Oper. Res. 2000, 27, 1093–1110. [Google Scholar] [CrossRef]
- Guo, Z.H.; Wu, J.; Lu, H.Y.; Wang, J.Z. A case study on a hybrid wind speed forecasting method using BP neural network. Knowl. Based Syst. 2011, 24, 1048–1056. [Google Scholar] [CrossRef]
- Jamshidi, S.; Yadollahi, A.; Arab, M.M.; Soltani, M.; Eftekhari, M.; Sabzalipoor, H.; Sheikhi, A.; Shiri, J. Combining gene expression programming and genetic algorithm as a powerful hybrid modeling approach for pear rootstocks tissue culture media formulation. Plant Meth. 2019, 15, 136. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M. Modeling and optimizing In Vitro sterilization of chrysanthemum via multilayer perceptron-non-dominated sorting genetic algorithm-II (MLP-NSGAII). Front. Plant Sci. 2019, 10, 282. [Google Scholar] [CrossRef] [PubMed]
- Gago, J.; Landín, M.; Gallego, P.P. A neurofuzzy logic approach for modeling plant processes: A practical case of In Vitro direct rooting and acclimatization of Vitis vinifera L. Plant Sci. 2010, 179, 241–249. [Google Scholar] [CrossRef]
- Gago, J.; Landín, M.; Gallego, P.P. Artificial neural networks modeling the In Vitro rhizogenesis and acclimatization of Vitis vinifera L. J. Plant Physiol. 2010, 167, 1226–1231. [Google Scholar] [CrossRef] [PubMed]
- Gago, J.; Martinez-Nunez, L.; Landin, M.; Flexas, J.; Gallego, P.P. Modeling the effects of light and sucrose on In Vitro propagated plants: A multiscale system analysis using artificial intelligence technology. PLoS ONE 2014, 9, e85989. [Google Scholar] [CrossRef] [PubMed]
- García-Pérez, P.; Lozano-Milo, E.; Landín, M.; Gallego, P.P. Machine Learning Technology Reveals the Concealed Interactions of Phytohormones on Medicinal Plant In Vitro Organogenesis. Biomolecules 2020, 10, 746. [Google Scholar] [CrossRef]
- García-Pérez, P.; Lozano-Milo, E.; Landín, M.; Gallego, P.P. Combining Medicinal Plant In Vitro Culture with Machine Learning Technologies for Maximizing the Production of Phenolic Compounds. Antioxidants 2020, 9, 210. [Google Scholar] [CrossRef]
- Przetakiewicz, A.; Orczyk, W.; Nadolska-Orczyk, A. The effect of auxin on plant regeneration of wheat, barley and triticale. Plant Cell. Tiss. Org. Cult. 2003, 73, 245–256. [Google Scholar] [CrossRef]
- Rahman, M.; Shamsuddin, A.; Asad, U. In Vitro regeneration from mature embryos in spring wheat. Int. J. Sustain. Crop Prod. 2008, 3, 76–80. [Google Scholar]
- Ahmad, A.; Zhong, H.; Wang, W.; Sticklen, M.B. Shoot apical meristem: In Vitro regeneration and morphogenesis in wheat (Triticum aestivum L.). In Vitro Cell. Dev. Biol. Plant 2002, 38, 163–167. [Google Scholar] [CrossRef]
- Xhulaj, D.B. Effect of plant growth regulators on In Vitro plant regeneration of wheat (Triticum aestivum L.) from embryo explants. J. Anim. Plant Sci. 2019, 29, 1616–1621. [Google Scholar]
- Mokhtari, A.; Alizadeh, H.; Samadi, B.Y.; Omidi, M.; Otroshy, M.; Moeini, Z. Effect of plant growth regulators on direct shoot regeneration of wheat immature embryonic explants. J. Agric. Eng. Biotech. 2013, 1, 74–80. [Google Scholar] [CrossRef]
- Hafeez, I.; Sadia, B.; Sadaqat, N.; Kainth, R.A.; Iqbal, M.Z.; Khan, I.A. Establishment of efficient In Vitro culture protocol for wheat land races of Pakistan. Afr. J. Biotechnol. 2012, 11, 2782–2790. [Google Scholar] [CrossRef]
- Kumar, R.; Mamrutha, H.M.; Kaur, A.; Venkatesh, K.; Grewal, A.; Kumar, R.; Tiwari, V. Development of an efficient and reproducible regeneration system in wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plants 2017, 23, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Ivashchuk, O.A.; Fedorova, V.; Shcherbinina, N.V.; Maslova, E.V.; Shamraeva, E. Microclonal propagation of plant process modeling and optimization of its parameters based on neural network. Drug Invent. Today 2018, 10, 3170–3175. [Google Scholar]
- Mansouri, A.; Fadavi, A.; Mortazavian, S.M.M. An artificial intelligence approach for modeling volume and fresh weight of callus—A case study of cumin (Cuminum cyminum L.). J. Theor. Biol. 2016, 397, 199–205. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat-Noori, S.A.; Abdipour, M.; Tohidfar, M.; Mortazavian, S.M.M. Image processing and artificial neural network-based models to measure and predict physical properties of embryogenic callus and number of somatic embryos in ajowan (Trachyspermum ammi (L.) Sprague). In Vitro Cell. Dev. Biol. Plant 2018, 54, 54–68. [Google Scholar] [CrossRef]
- Munasinghe, S.P.; Somaratne, S.; Weerakoon, S.R.; Ranasinghe, C. Prediction of chemical composition for callus production in Gyrinops walla Gaetner through machine learning. Inf. Process. Agric. 2020, 7, 1–12. [Google Scholar] [CrossRef]
- Albiol, J.; Campmajó, C.; Casas, C.; Poch, M. Biomass estimation in plant cell cultures: A neural network approach. Biotechnol. Prog. 1995, 11, 88–92. [Google Scholar] [CrossRef]
- Shiotani, S.; Fukuda, T.; Arai, F.; Takeuchi, N.; Sasaki, K.; Kinosita, T. Cell recognition by image processing: Recognition of dead or living plant cells by neural network. JSME Int. J. 1994, 37, 202–208. [Google Scholar] [CrossRef][Green Version]
- Molto, E.; Harrell, R.C. Neural network classification of sweet potato embryos. In Optics in Agriculture and Forestry; SPIE: Bellingham, WA, USA, 1993; pp. 239–249. [Google Scholar]
- Zhang, C.; Timmis, R.; Hu, W.-S. A neural network based pattern recognition system for somatic embryos of Douglas fir. Plant Cell. Tiss. Org. Cult. 1999, 56, 25–35. [Google Scholar] [CrossRef]
- Arab, M.M.; Yadollahi, A.; Shojaeiyan, A.; Ahmadi, H. Artificial neural network genetic algorithm as powerful tool to predict and optimize In Vitro proliferation mineral medium for G× N15 rootstock. Front. Plant Sci. 2016, 7, e1526. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, S.; Yadollahi, A.; Ahmadi, H.; Arab, M.M.; Eftekhari, M. Predicting In Vitro culture medium macro-nutrients composition for pear rootstocks using regression analysis and neural network models. Front. Plant Sci. 2016, 7, 274. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.D.; Pattanayak, A. Intelligent image analysis (IIA) using artificial neural network (ANN) for non-invasive estimation of chlorophyll content in micropropagated plants of potato. In Vitro Cell. Dev. Biol. Plant 2017, 53, 520–526. [Google Scholar] [CrossRef]
- Barone, J.O. Use of multiple regression analysis and artificial neural networks to model the effect of nitrogen in the organogenesis of Pinus taeda L. Plant Cell. Tiss. Organ. Cult. 2019, 137, 455–464. [Google Scholar] [CrossRef]
- Mehrotra, S.; Prakash, O.; Khan, F.; Kukreja, A. Efficiency of neural network-based combinatorial model predicting optimal culture conditions for maximum biomass yields in hairy root cultures. Plant Cell Rep. 2013, 32, 309–317. [Google Scholar] [CrossRef]
- Osama, K.; Somvanshi, P.; Pandey, A.K.; Mishra, B.N. Modelling of nutrient mist reactor for hairy root growth using artificial neural network. Eur. J. Sci. Res. 2013, 97, 516–526. [Google Scholar]
- Arab, M.M.; Yadollahi, A.; Eftekhari, M.; Ahmadi, H.; Akbari, M.; Khorami, S.S. Modeling and Optimizing a New Culture Medium for In Vitro Rooting of G× N15 Prunus Rootstock using Artificial Neural Network-Genetic Algorithm. Sci. Rep. 2018, 8, e9977. [Google Scholar] [CrossRef]
- Araghinejad, S.; Fayaz, N.; Hosseini-Moghari, S.-M. Development of a Hybrid Data Driven Model for Hydrological Estimation. Water Resour. Manag. 2018, 32, 3737–3750. [Google Scholar] [CrossRef]
- Fayaz, N.; Condon, L.E.; Chandler, D.G. Evaluating the Sensitivity of Projected Reservoir Reliability to the Choice of Climate Projection: A Case Study of Bull Run Watershed, Portland, Oregon. Water Resour. Manag. 2020, 34, 1991–2009. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M. Modeling and Optimizing Medium Composition for Shoot Regeneration of Chrysanthemum via Radial Basis Function-Non-dominated Sorting Genetic Algorithm-II (RBF-NSGAII). Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Moravej, M.; Amani, P.; Hosseini-Moghari, S.-M. Groundwater level simulation and forecasting using interior search algorithm-least square support vector regression (ISA-LSSVR). Groundw. Sustain. Dev. 2020, 11, 100447. [Google Scholar] [CrossRef]
- Moravej, M. Discussion of “Modified Firefly Algorithm for Solving Multireservoir Operation in Continuous and Discrete Domains” by Irene Garousi-Nejad, Omid Bozorg-Haddad, and Hugo, A. Loáiciga. J. Water Resour. Plan. Manag. 2017, 143, 07017004. [Google Scholar] [CrossRef]
- Dezfooli, D.; Abdollahi, B.; Hosseini-Moghari, S.-M.; Ebrahimi, K. A comparison between high-resolution satellite precipitation estimates and gauge measured data: Case study of Gorganrood basin, Iran. J. Water Supply: Res. Technol. Aqua 2018, 67, 236–251. [Google Scholar] [CrossRef]
- Soleimani, S.; Haddad, O.B.; Moravej, M. Modeling water quality parameters using data-driven methods. J. Water Soil 2016, 30, Pe743–Pe757. [Google Scholar]
- Salehi, M.; Farhadi, S.; Moieni, A.; Safaie, N.; Ahmadi, H. Mathematical Modeling of Growth and Paclitaxel Biosynthesis in Corylus avellana Cell Culture Responding to Fungal Elicitors using Multilayer Perceptron-Genetic Algorithm. Front. Plant Sci. 2020, 11, 1148. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M.; Yoosefzadeh-Najafabadi, M. Application of adaptive neuro-fuzzy inference system-non-dominated sorting genetic Algorithm-II (ANFIS-NSGAII) for modeling and optimizing somatic embryogenesis of Chrysanthemum. Front. Plant Sci. 2019, 10, 869. [Google Scholar] [CrossRef]
- Zhao, Y. The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant. Biol. 2008, 11, 16–22. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Yoosefzadeh-Najafabadi, M. Optimizing sterilization conditions and growth regulator effects on In Vitro shoot regeneration through direct organogenesis in Chenopodium quinoa. BioTechnologia 2018, 99, 49–57. [Google Scholar] [CrossRef]
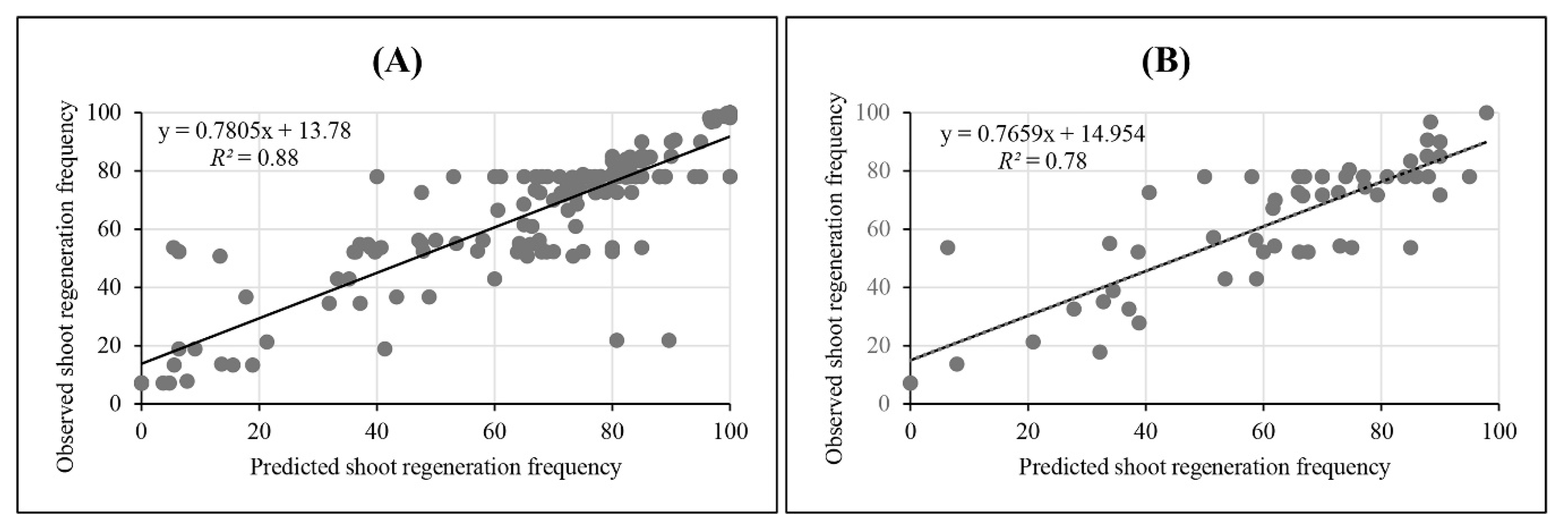
| Performance Measure | Training | Validation |
|---|---|---|
| R2 | 0.88 | 0.78 |
| RMSE | 14.12 | 14.76 |
| MBE | −0.33 | −0.89 |
| Type of Explant | BAP (mg/L) | Kin (mg/L) | 2,4-D (mg/L) | IAA (mg/L) | IBA (mg/L) | NAA (mg/L) | Zeatin (mg/L) | CuSO4 (mg/L) | Shoot Regeneration Frequency (%) |
|---|---|---|---|---|---|---|---|---|---|
| Immature embryo | 0.15 | 0.73 | 0.17 | 0.37 | 0.04 | 0.01 | 4.51 | 13.08 | 97.63 |
| Item | Genotype | Explant | BAP | Kin | 2,4-D | IAA | IBA | NAA | Zeatin | CuSO4 |
|---|---|---|---|---|---|---|---|---|---|---|
| Variable sensitivity ratio (VSR) | 1.35 | 1.83 | 1.22 | 1.08 | 1.97 | 1.17 | 0.93 | 0.54 | 1.29 | 1.02 |
| Rank | 3 | 2 | 5 | 7 | 1 | 6 | 9 | 10 | 4 | 8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hesami, M.; Condori-Apfata, J.A.; Valderrama Valencia, M.; Mohammadi, M. Application of Artificial Neural Network for Modeling and Studying In Vitro Genotype-Independent Shoot Regeneration in Wheat. Appl. Sci. 2020, 10, 5370. https://doi.org/10.3390/app10155370
Hesami M, Condori-Apfata JA, Valderrama Valencia M, Mohammadi M. Application of Artificial Neural Network for Modeling and Studying In Vitro Genotype-Independent Shoot Regeneration in Wheat. Applied Sciences. 2020; 10(15):5370. https://doi.org/10.3390/app10155370
Chicago/Turabian StyleHesami, Mohsen, Jorge A. Condori-Apfata, Maria Valderrama Valencia, and Mohsen Mohammadi. 2020. "Application of Artificial Neural Network for Modeling and Studying In Vitro Genotype-Independent Shoot Regeneration in Wheat" Applied Sciences 10, no. 15: 5370. https://doi.org/10.3390/app10155370
APA StyleHesami, M., Condori-Apfata, J. A., Valderrama Valencia, M., & Mohammadi, M. (2020). Application of Artificial Neural Network for Modeling and Studying In Vitro Genotype-Independent Shoot Regeneration in Wheat. Applied Sciences, 10(15), 5370. https://doi.org/10.3390/app10155370






