A Smartphone-Based Cell Segmentation to Support Nasal Cytology
Abstract
1. Introduction
2. Rhino-Cytology
2.1. A. The Cytodiagnostic Technique
- Sampling: Consists of collecting a sample of nasal mucosa containing the superficial cells. It is carried out using a disposable plastic curette, called nasal scraping, or a simple nasal swab is preferred for smaller patients;
- Processing: The material collected is placed on a slide and dried in the open air. Then, the slide is stained using the May Grunwald–Giemsa method, which provides the cells with the classic purple staining and highlights all the cytotypes present in the nasal mucosa. Usually, the complete staining procedure takes about 20–30 s with rapid staining techniques;
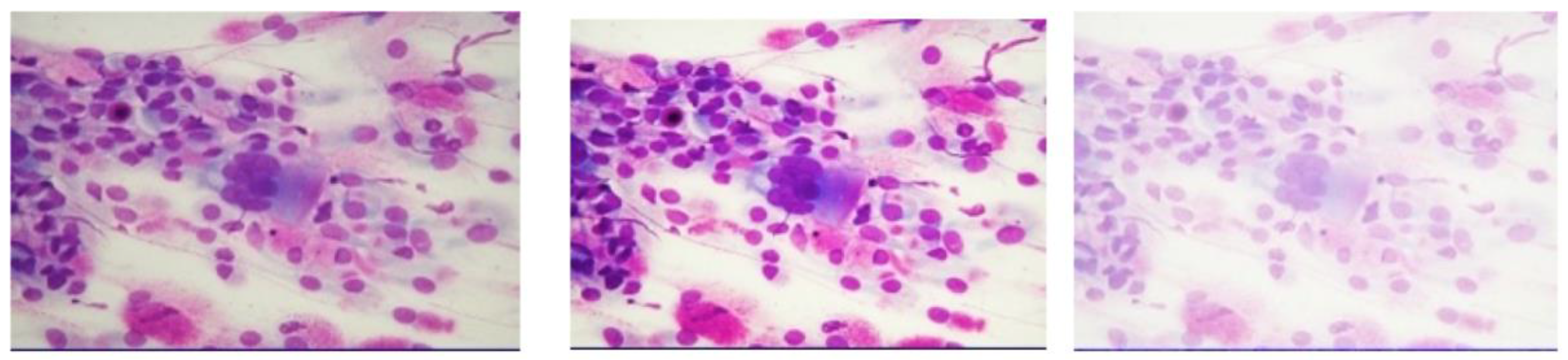
- Microscopic observation: An optical microscope is used, mainly connected to a special camera to view the cells on a monitor. The diagnostic protocol involves viewing and analyzing 50 digital images for each slide, called fields, usually at 1000X magnification.
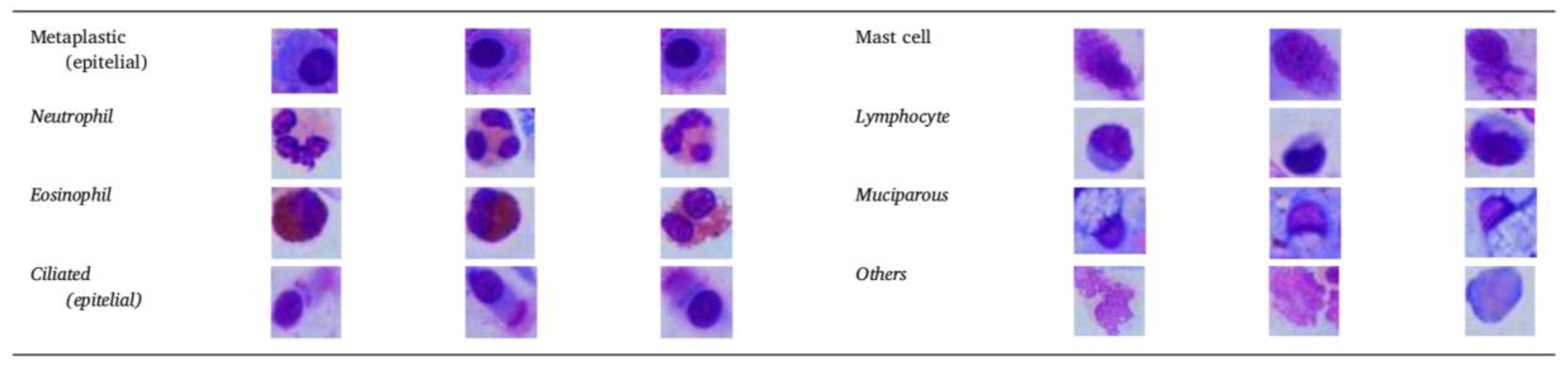
2.2. B. Types of Cells Involved
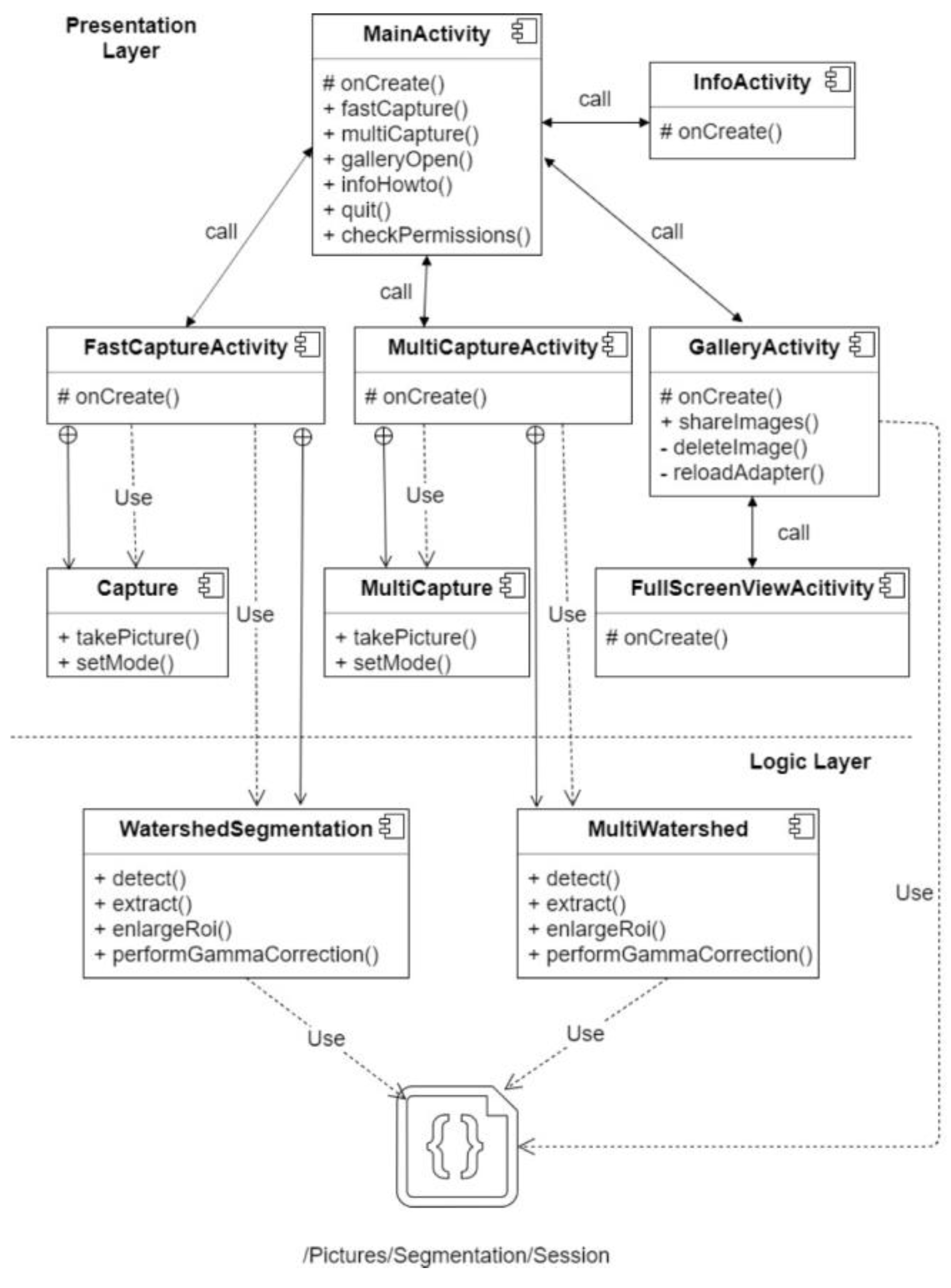
3. Image Acquisition and Processing
3.1. Image Enhancement
3.2. Image Segmentation
3.2.1. Mean Shift
3.2.2. Otsu Segmentation
3.2.3. Watershed
3.2.4. Canny Edge Detector
3.3. Morphological Image Processing
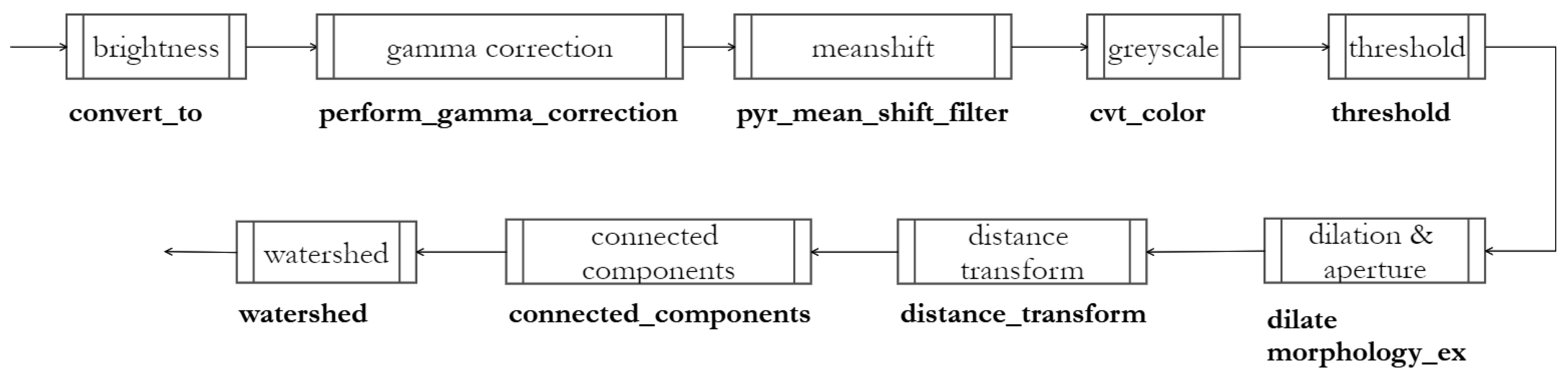
4. Methods
4.1. Increase in Brightness and Contrast
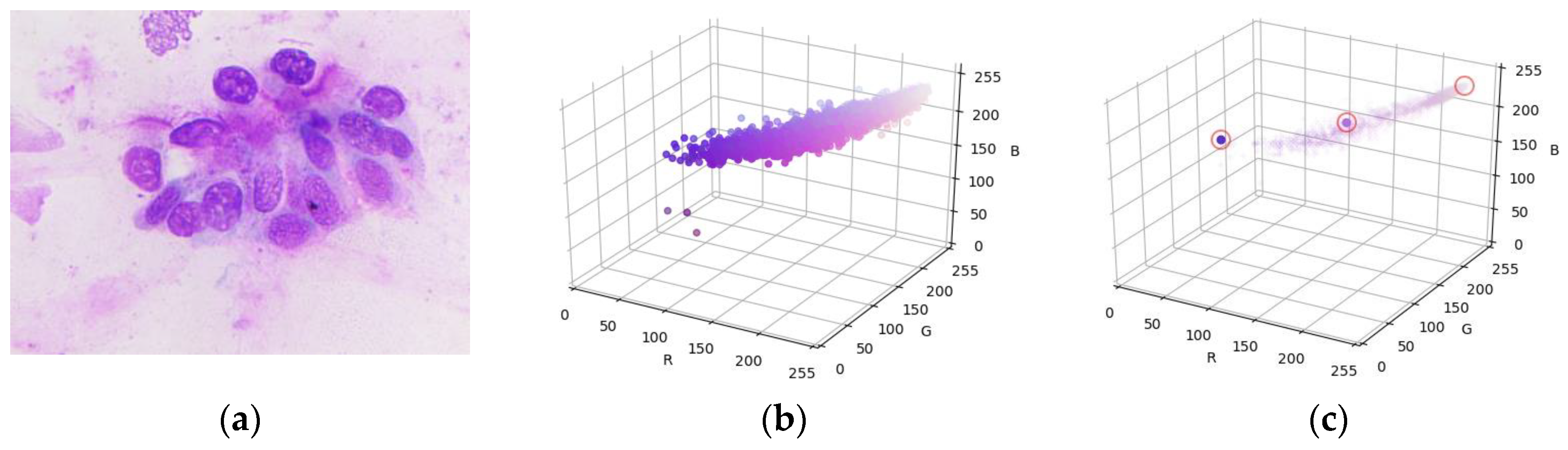
4.2. Gamma and Mean Shift Correction
4.3. Otsu Binarization
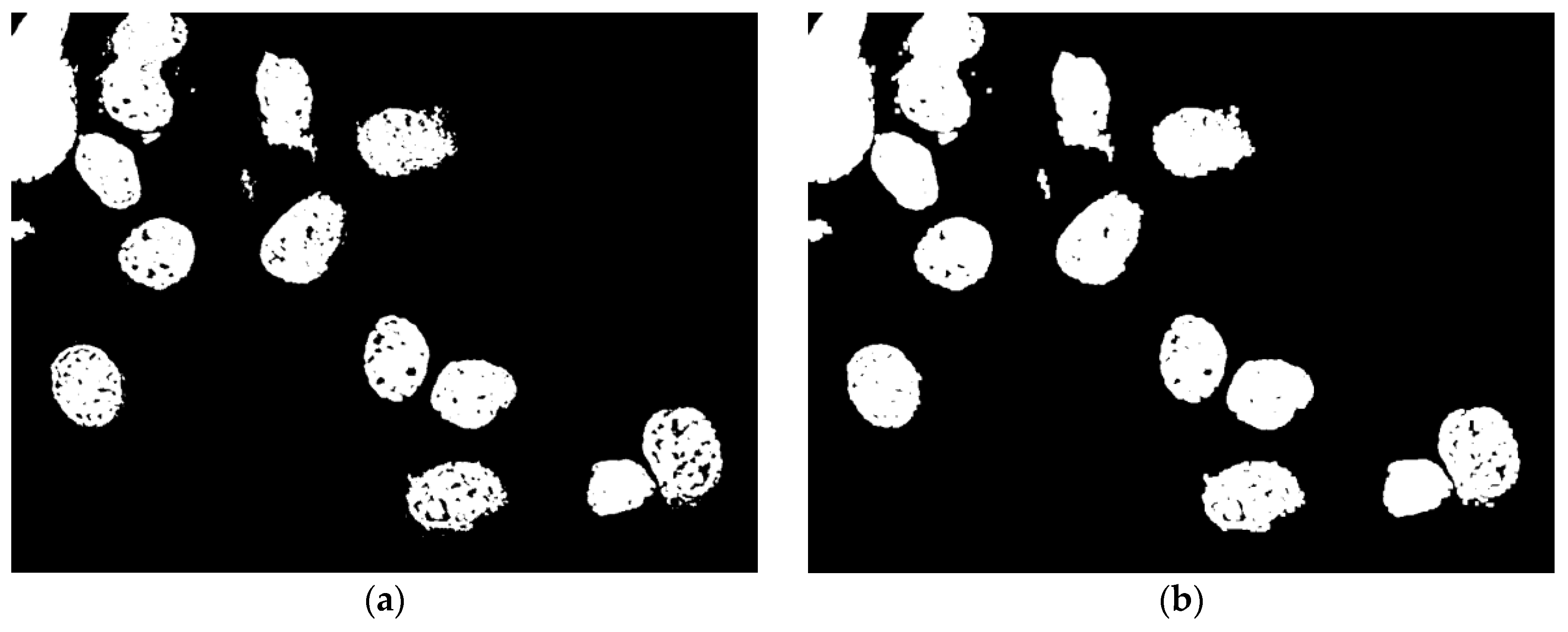
4.4. Identification of Markers, Watershed
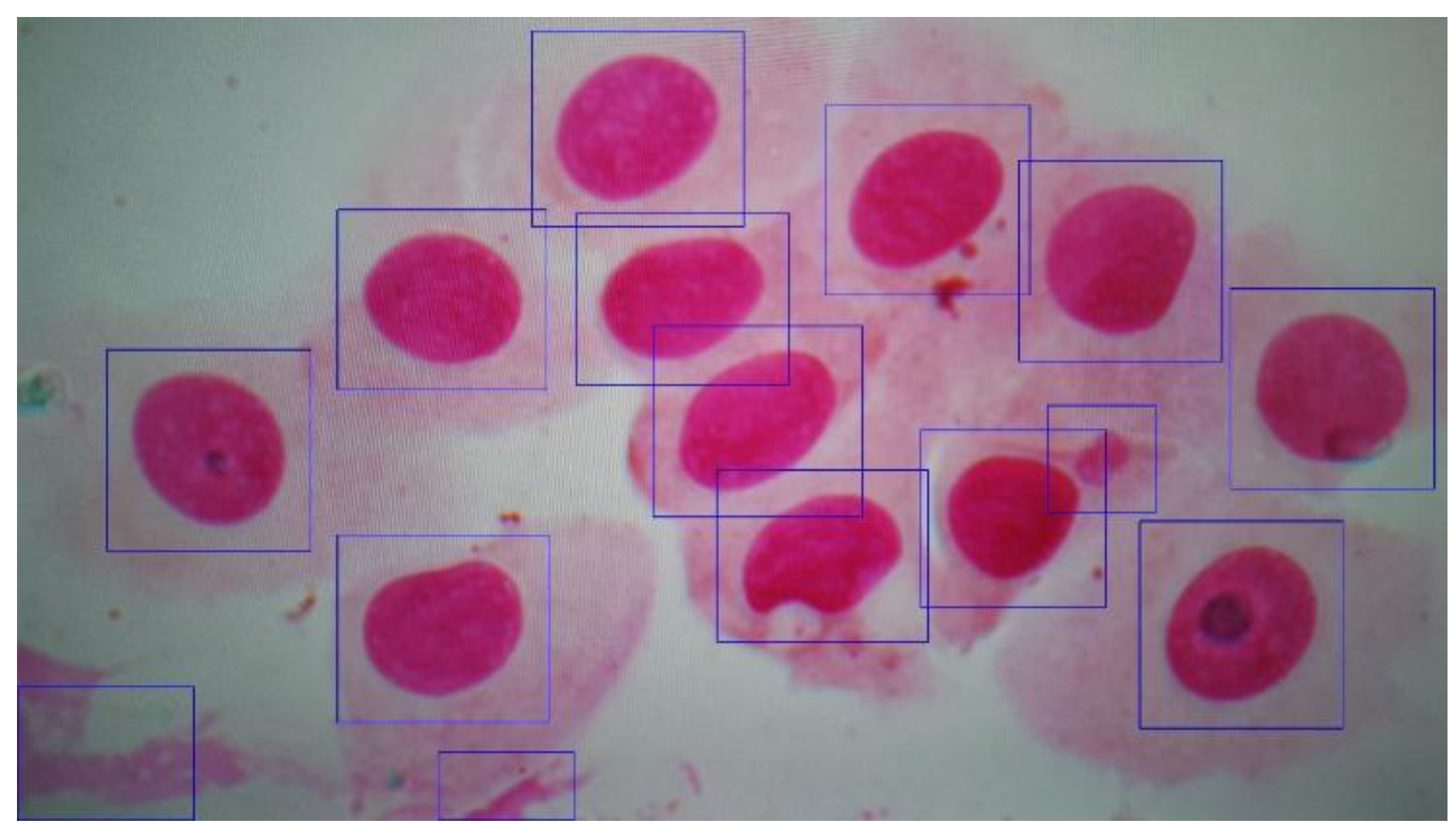
4.5. Cropping
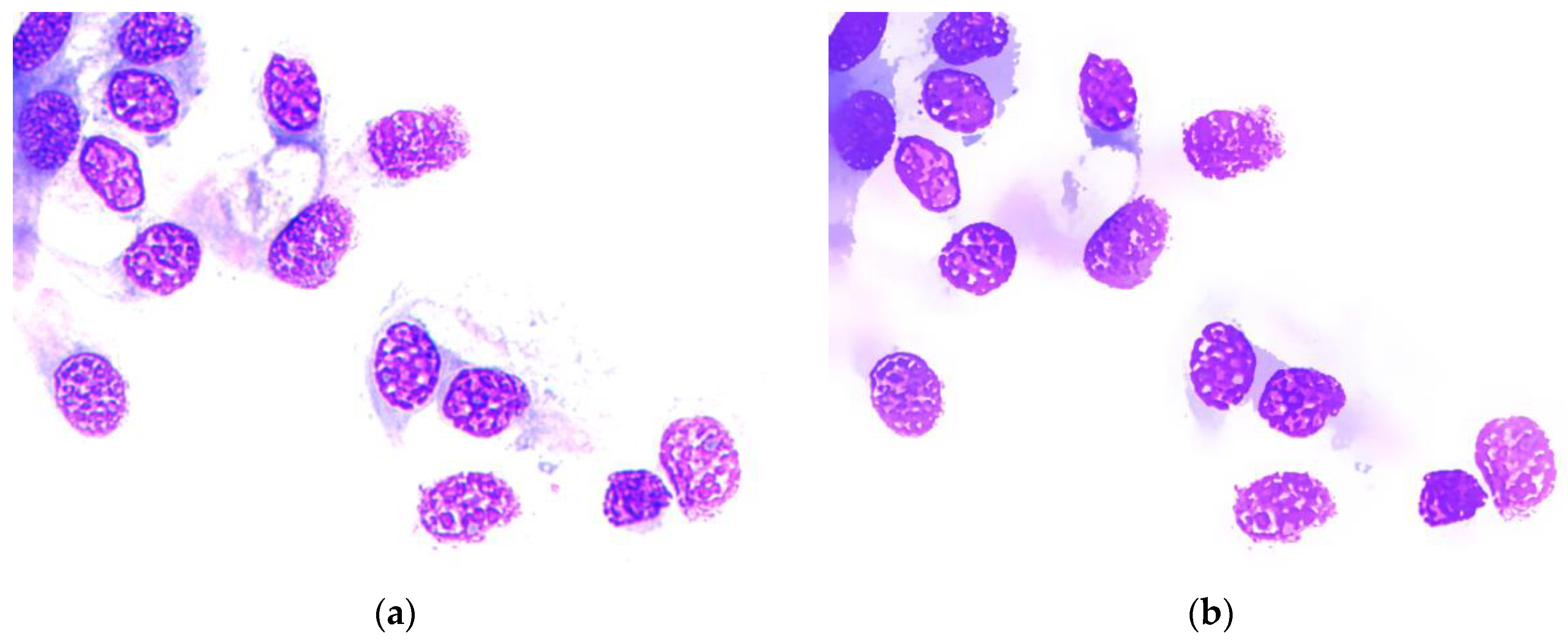
5. Experimental Results
| Accuracy | 0.860 |
| Sensitivity (Recall) | 0.959 |
| Specificity | 0.405 |
| Precision | 0.881 |
| F-score | 0.918 |
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
Appendix B
References
- Dimauro, G.; Caivano, D.; Bevilacqua, V.; Girardi, F.; Napoletano, V. VoxTester, software for digital evaluation of speech changes in Parkinson disease. In Proceedings of the 2016 IEEE International Symposium on Medical Measurements and Applications, MeMeA, Benevento, Italy, 15–18 May 2016; ISBN 9781467391726. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Brunetti, A.; Trotta, G.F.; Dimauro, G.; Elez, K.; Alberotanza, V.; Scardapane, A. A Novel Approach for Hepatocellular Carcinoma Detection and Classification Based on Triphasic CT Protocol. In Proceedings of the IEEE Congress on Evolutionary Computation, San Sebastian, Spain, 5–8 June 2017. [Google Scholar] [CrossRef]
- Rubaiat, S.Y.; Rahman, M.M.; Hasan, M.K. Important Feature Selection & Accuracy Comparisons of Different Machine Learning Models for Early Diabetes Detection. In Proceedings of the 2018 International Conference on Innovation in Engineering and Technology (ICIET), Dhaka, Bangladesh, 27–28 December 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Dimauro, G.; Bevilacqua, V.; Colizzi, L.; Di Pierro, D. TestGraphia, a Software System for the Early Diagnosis of Dysgraphia. IEEE Access 2020, 8, 19564–19575. [Google Scholar] [CrossRef]
- Hasan, M.K.; Aziz, M.H.; Zarif, M.I.I.; Hasan, M.; Hashem, M.M.A.; Guha, S.; Love, R. HeLP ME: Recom-mendations for Non-invasive Hemoglobin Level Prediction in Mobile-phone Environment. JMIR mHealth uHealth 2020, in press. Available online: https://preprints.jmir.org/preprint/16806/accepted (accessed on 20 June 2020).
- Gigantesco, A.; Giuliani, M. Quality of life in mental health services with a focus on psychiatric rehabilitation practice. Annali dell’Istituto Superiore di Sanita 2011, 47, 363–372. [Google Scholar] [CrossRef]
- Dimauro, G.; Caivano, D.; Girardi, F.; Ciccone, M.M. The Patient Centered Electronic Multimedia Health Fascicle-EMHF. In Proceedings of the 2014 IEEE Workshop on Biometric Measurements and Systems for Security and Medical Applications (BIOMS), Rome, Italy, 17 October 2014; ISBN 9781479951758. [Google Scholar] [CrossRef]
- Dimauro, G.; Girardi, F.; Caivano, D.; Colizzi, L. Personal Health E-Record—Toward an enabling Ambient Assisted Living Technology for communication and information sharing between patients and care providers. In Ambient Assisted Living; Springer: Cham, Switzerland, 2018; ISBN 9783030059200. [Google Scholar] [CrossRef]
- Maglietta, R.; Amoroso, N.; Boccardi, M.; Bruno, S.; Chincarini, A.; Frisoni, G.B.; Inglese, P.; Redolfi, A.; Tangaro, S.; Tateo, A.; et al. Automated hippocampal segmentation in 3D MRI using random undersampling with boosting algorithm. Pattern Anal. Appl. 2016, 19, 579–591. [Google Scholar] [CrossRef]
- Celebi, M.E.; Wen, Q.; Hwang, S.; Iyatomi, H.; Schaefer, G. Lesion border detection in dermoscopy images using ensembles of thresholding methods. Skin Res. Technol. 2013, 19, e252–e258. [Google Scholar] [CrossRef]
- Rasche, C. Melanoma Recognition with an Ensemble of Techniques for Segmentation and a Structural Analysis for Classification. arXiv 2018, arXiv:1807.06905. [Google Scholar]
- Dimauro, G.; Simone, L. Novel biased normalized cuts approach for the automatic segmentation of the conjunctiva. Electronics 2020, 9, 997. [Google Scholar] [CrossRef]
- Rasche, C. Fleckmentation: Rapid segmentation using repeated 2-means. IET Image Process. 2019, 13, 1940–1943. [Google Scholar] [CrossRef]
- Piuri, V.; Scotti, F. Morphological classification of blood leucocytes by microscope images. In Proceedings of the 2004 IEEE International Conference on Computational Intelligence for Measurement Systems and Applications, Boston, MA, USA, 14–16 July 2004; pp. 103–108. [Google Scholar] [CrossRef]
- Qiao, G.; Zong, G.; Sun, M.; Wang, J. Automatic neutrophil nucleus lobe counting based on graph representation of region skeleton. Cytom. Part A 2012, 81A, 734–742. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Liu, H.; Wang, J.; Guo, F. A combined spatial-spectral method for auto- mated white blood cells segmentation. Opt. Laser Technol. 2013, 54, 225–231. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Buongiorno, D.; Carlucci, P.; Giglio, F.; Tattoli, G.; Guarini, A.; Sgherza, N.; de Tullio, G.; Minoia, C.; Scattone, A.; et al. A supervised CAD to support telemedicine in hematology. In Proceedings of the 2015 International Joint Conference on Neural Networks, Killarney, Ireland, 12–17 July 2015. [Google Scholar] [CrossRef]
- Zheng, Q.; Milthorpe, B.K.; Jones, A.S. Direct neural network application for automated cell recognition. Cytometry 2004, 57A, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Osowski, S.; Siroi, R.; Markiewicz, T.; Siwek, K. Application of support vector machine and genetic algorithm for improved blood cell recognition. IEEE Trans. Intrum. Meas. 2009, 58, 2159–2168. [Google Scholar] [CrossRef]
- Theera-Umpon, N.; Gader, P.D. System-level training of neural networks for counting white blood cells. IEEE Trans. Syst. Man Cybern. Part C Appl. Rev. 2002, 32, 48–53. [Google Scholar] [CrossRef]
- Bousquet, J.; Schünemann, H.J.; Samolinski, B.; Demoly, P.; Baena-Cagnani, C.E.; Bachert, C.; Bonini, S.; Boulet, L.P.; Bousquet, P.J.; Brozek, J.L.; et al. Allergic Rhinitis and its Impact on Asthma (ARIA): Achievements in 10 years and future needs. World Health Organization Collaborating Center for Asthma and Rhinitis. J. Allergy Clin. Immunol. 2012, 130, 1049–1062. [Google Scholar] [CrossRef]
- Dimauro, G.; Girardi, F.; Gelardi, M.; Bevilacqua, V.; Caivano, D. Rhino-Cyt: A System for Supporting the Rhinologist in the Analysis of Nasal Cytology. Lect. Notes Comput. Sci. 2018, 619–630. [Google Scholar] [CrossRef]
- Dimauro, G.; Ciprandi, G.; Deperte, F.; Girardi, F.; Ladisa, E.; Latrofa, S.; Gelardi, M. Nasal cytology with deep learning techniques. Int. J. Med. Inform. 2019, 122, 13–19. [Google Scholar] [CrossRef]
- Dimauro, G.; Deperte, F.; Maglietta, R.; Bove, M.; La Gioia, F.; Renò, V.; Simone, L.; Gelardi, M. A Novel Approach for Biofilm Detection Based on a Convolutional Neural Network. Electronics 2020, 9, 881. [Google Scholar] [CrossRef]
- Merenda, M.; Porcaro, C.; Iero, D. Edge Machine Learning for AI-Enabled IoT Devices: A Review. Sensors 2020, 20, 2533. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning in intelligent embedded systems. In WOES’99, Proceedings of the Workshop on Embedded Systems on Workshop on Embedded Systems, Cambridge, MA, USA, 29–31 March 1999; USENIX Association: Berkeley, CA, USA, 1999; p. 9. [Google Scholar]
- Haigh, K.Z.; Mackay, A.M.; Cook, M.R.; Lin, L.G. Machine Learning for Embedded Systems: A Case Study; Technical Report; BBN Technologies: Cambridge, MA, USA, 2015. [Google Scholar]
- Chen, J.; Ran, X. Deep Learning With Edge Computing: A Review. Proc. IEEE 2019, 107, 1655–1674. [Google Scholar] [CrossRef]
- Sze, V.; Chen, Y.H.; Emer, J.; Suleiman, A.; Zhang, Z. Hardware for machine learning: Challenges and opportunities. In Proceedings of the 2017 IEEE Custom Integrated Circuits Conference (CICC), Austin, TX, USA, 30 April–3 May 2017; pp. 1–8. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Valueva, M.; Valuev, G.; Semyonova, N.; Lyakhov, P.; Chervyakov, N.; Kaplun, D.; Bogaevskiy, D. Construction of Residue Number System Using Hardware Efficient Diagonal Function. Electronics 2019, 8, 694. [Google Scholar] [CrossRef]
- Dimauro, G.; Impedovo, S.; Pirlo, G.; Salzo, A. RNS architectures for the implementation of the ‘diagonal function’. Inf. Process. Lett. 2000, 73, 189–198. [Google Scholar] [CrossRef]
- Dimauro, G.; Impedovo, S.; Modugno, R.; Pirlo, G.; Stefanelli, R. Residue-to-binary conversion by the “quotient function”. In IEEE Transactions on Circuits and Systems II: Analog and Digital Signal Processing; IEEE: Piscataway, NJ, USA, 2003; Volume 50, pp. 488–493. [Google Scholar] [CrossRef]
- Gelardi, M. Atlas of Nasal Cytology for the Differential Diagnosis of Nasal Diseases; Edi. Ermes: Milano, Italy, 2012; ISBN 9781467530354. [Google Scholar]
- Gelardi, M.; Iannuzzi, L.; Quaranta, N.; Landi, M.; Passalacqua, G. Nasal cytology-Pratical aspects and clinical relevance. Clin. Exp. Allergy 2016, 46, 785–792. [Google Scholar] [CrossRef]
- Gelardi, M. Citologia Nasale. Available online: http://www.citologianasale.eu/citologia.htm (accessed on 20 June 2020).
- Paulista, U.E.; Em, P.D.E.P.; Biológicas, C. The Electrical Engineering Handbook; CRC Press: London, UK, 1997; ISBN 978-0133354492. [Google Scholar]
- Covington, M.A. Overview of image processing. In Digital SLR Astrophotography; Cambridge University Press: Cambridge, UK, 2009; pp. 145–164. ISBN 978-0-511-37853-9. [Google Scholar]
- Dimauro, G.; Guarini, A.; Caivano, D.; Girardi, F.; Pasciolla, C.; Iacobazzi, A. Detecting clinical signs of anaemia from digital images of the palpebral conjunctiva. IEEE Access 2019, 7, 113488–113498. [Google Scholar] [CrossRef]
- Dimauro, G.; Baldari, L.; Caivano, D.; Colucci, G.; Girardi, F. Automatic Segmentation of Relevant Sections of the Conjunctiva for Non-Invasive Anemia Detection. In Proceedings of the 2018 3rd International Conference on Smart and Sustainable Technologies (SpliTech), Split, Croatia, 26–29 June 2018; pp. 1–5. [Google Scholar]
- Hasan, M.K.; Haque, M.; Sakib, N.; Love, R.; Ahamed, S.I. Smartphone-based Human Hemoglobin Level Measurement Analyzing Pixel Intensity of a Fingertip Video on Different Color Spaces. Smart Health 2018, 5–6, 26–39. [Google Scholar] [CrossRef]
- Shih, F.Y. Image Processing and Mathematical Morphology: Fundamentals and Applications; CRC Press: Boca Raton, FL, USA, 2017; ISBN 9781315218557. [Google Scholar]
- Bankman, I. Handbook of Medical Image Processing and Analysis; Elsevier: Amsterdam, The Netherlands, 2008; p. 1393. ISBN 9780123739049. [Google Scholar]
- Dimauro, G. A new image quality metric based on human visual system. In Proceedings of the 2012 IEEE International Conference on Virtual Environments Human-Computer Interfaces and Measurement Systems (VECIMS) Proceedings, Tianjin, China, 2–4 July 2012; pp. 69–73. [Google Scholar] [CrossRef]
- Dimauro, G.; Altomare, N.; Scalera, M. PQMET: A digital image quality metric based on human visual system. In Proceedings of the 4th International Conference on Image Processing Theory, Tools and Applications (IPTA), Paris, France, 14–17 October 2014; pp. 1–6. [Google Scholar] [CrossRef]
- Kaur, D.; Kaur, Y. Various Image Segmentation Techniques: A Review. Int. J. Comput. Sci. Mob. Comput. 2014, 3, 809–814. [Google Scholar]
- Comaniciu, D.; Meer, P. Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef]
- Fukunaga, K.; Hostetler, L.D. The Estimation of the Gradient of a Density Function, with Applications in Pattern Recognition. IEEE Trans. Inf. Theory 1975, 21, 32–40. [Google Scholar] [CrossRef]
- Cheng, Y. Mean Shift, Mode Seeking, and Clustering. IEEE Trans. Pattern Anal. Mach. Intell. 1995, 17, 790–799. [Google Scholar] [CrossRef]
- Nedrich, M. Mean Shift Clustering. Available online: https://spin.atomicobject.com/2015/05/26/mean-shift-clustering/ (accessed on 20 June 2020).
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1996, 9, 62–66. [Google Scholar] [CrossRef]
- Sahir, S. Canny Edge Detection Step by Step in Python. Available online: https://towardsdatascience.com/canny-edge-detection-step-by-step-in-python-computer-vision-b49c3a2d8123 (accessed on 20 June 2020).
- Canny, J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 8, 679–698. [Google Scholar] [CrossRef]
- Dougherty, E.; Lotufo, R.A. Hands-on Morphological Image Processing; SPIE Press Book: Bellingham, DC, USA, 2003; ISBN 9780819447203. [Google Scholar]
- Efford, N. Morphological Image Processing, in Digital Image Processing: A Practical Introduction Using Java; Pearson Education: Harrow, UK, 2000; ISBN 978-0201596236. [Google Scholar]
- Dimauro, G.; Bevilacqua, V.; Fina, P.R.; Buongiorno, D.; Brunetti, A.; Latrofa, S.; Cassano, M.; Gelardi, M. Comparative Analysis of Rhino-Cytological Specimens with Image Analysis and Deep Learning Techniques. Electronics 2020, 9, 952. [Google Scholar] [CrossRef]
- Renò, V.; Sciancalepore, M.; Dimauro, G.; Maglietta, R.; Cassano, M.; Gelardi, M. A novel approach for the automatic estimation of Ciliated cells Beating Frequency. Electronics 2020, 9, 1002. [Google Scholar] [CrossRef]





| Confusion Matrix | ||
|---|---|---|
| True Condition | ||
| Predicted | positive | negative |
| Positive | 1224 | 166 |
| Negative | 52 | 113 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimauro, G.; Di Pierro, D.; Deperte, F.; Simone, L.; Fina, P.R. A Smartphone-Based Cell Segmentation to Support Nasal Cytology. Appl. Sci. 2020, 10, 4567. https://doi.org/10.3390/app10134567
Dimauro G, Di Pierro D, Deperte F, Simone L, Fina PR. A Smartphone-Based Cell Segmentation to Support Nasal Cytology. Applied Sciences. 2020; 10(13):4567. https://doi.org/10.3390/app10134567
Chicago/Turabian StyleDimauro, Giovanni, Davide Di Pierro, Francesca Deperte, Lorenzo Simone, and Pio Raffaele Fina. 2020. "A Smartphone-Based Cell Segmentation to Support Nasal Cytology" Applied Sciences 10, no. 13: 4567. https://doi.org/10.3390/app10134567
APA StyleDimauro, G., Di Pierro, D., Deperte, F., Simone, L., & Fina, P. R. (2020). A Smartphone-Based Cell Segmentation to Support Nasal Cytology. Applied Sciences, 10(13), 4567. https://doi.org/10.3390/app10134567







