Development of a Deep Learning-Based Algorithm to Detect the Distal End of a Surgical Instrument
Abstract
1. Introduction
2. Materials and Methods
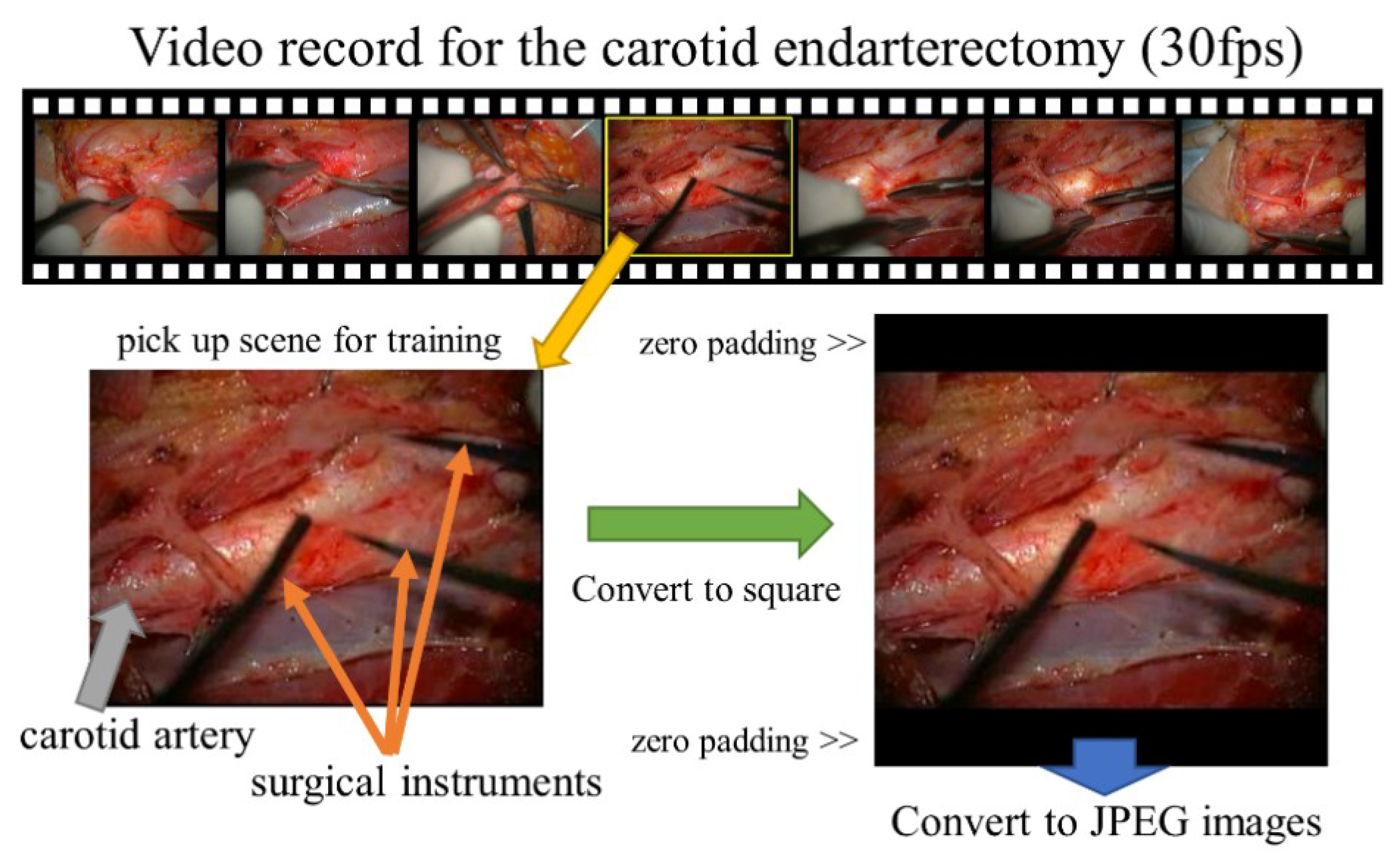
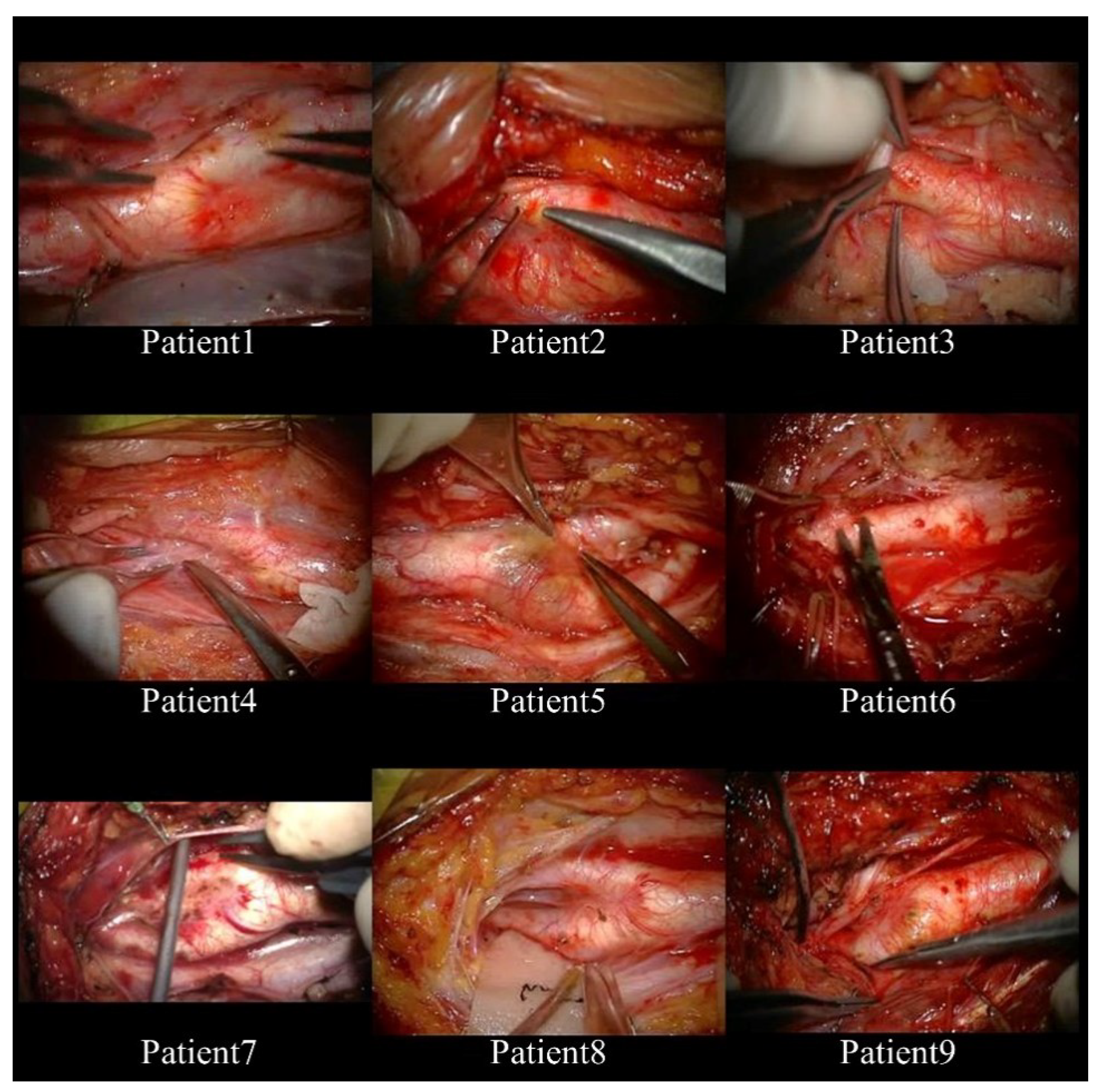
2.1. Subjects and Format of Video Records
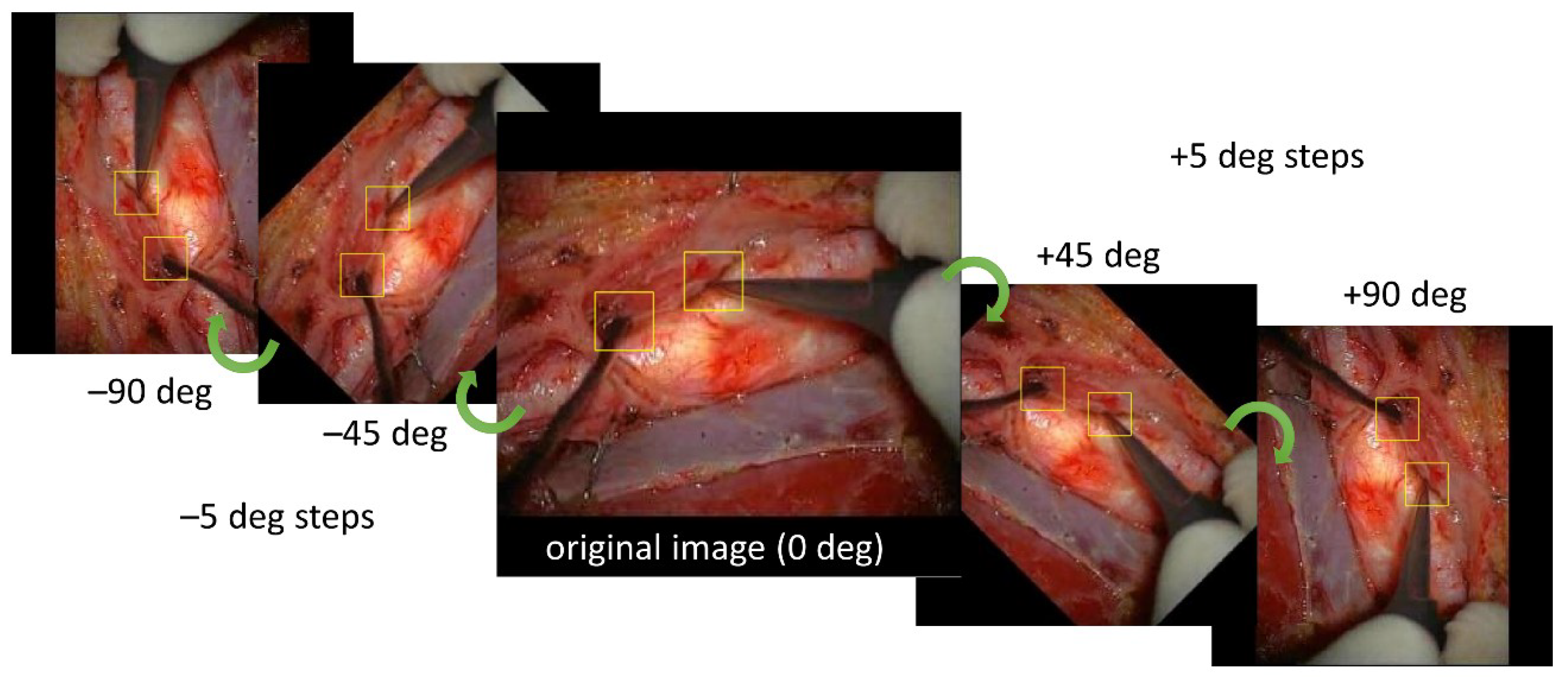
2.2. Preprocessing of Images
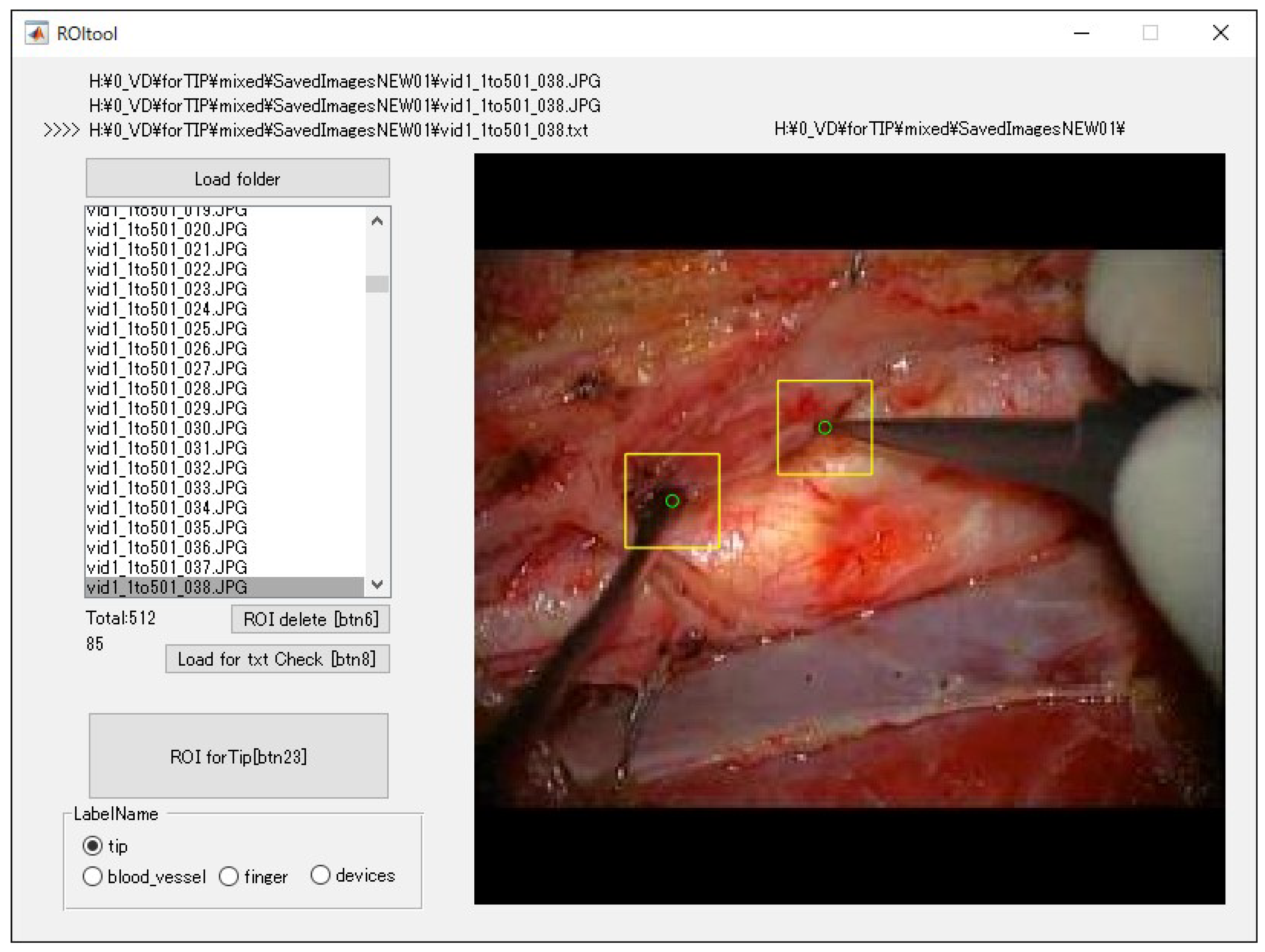
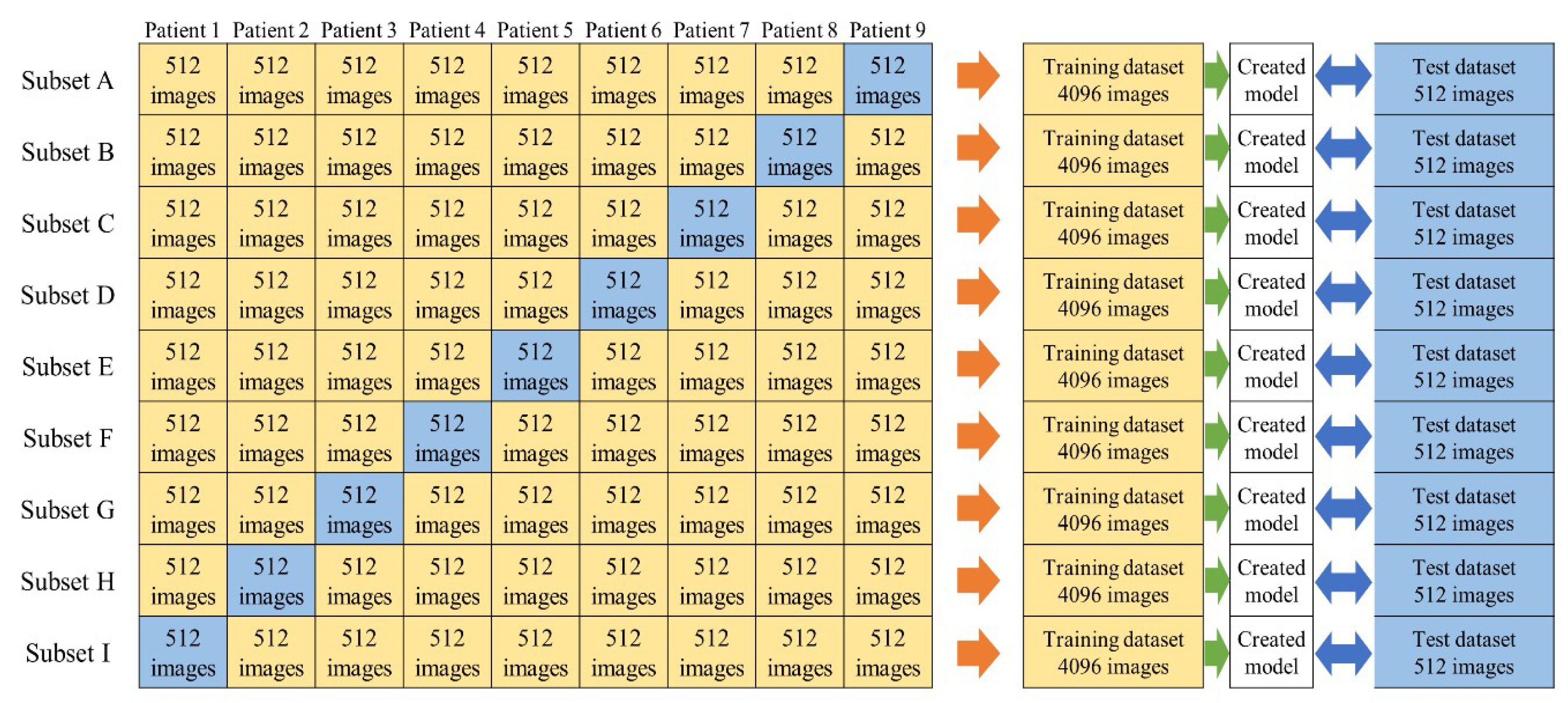
2.3. Dataset
2.4. Training Images for Model Creation
2.5. Evaluation of Created Models
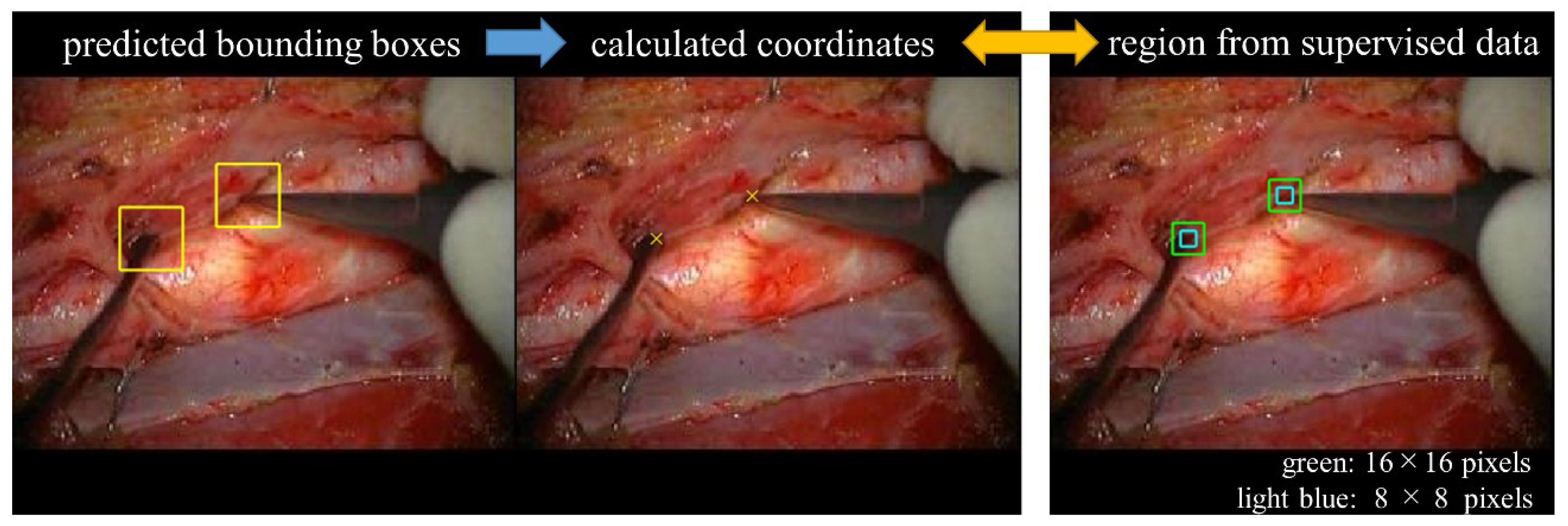
2.6. Algorithm and Evaluation of Distal End Detection
2.7. Statistical Analysis
3. Results
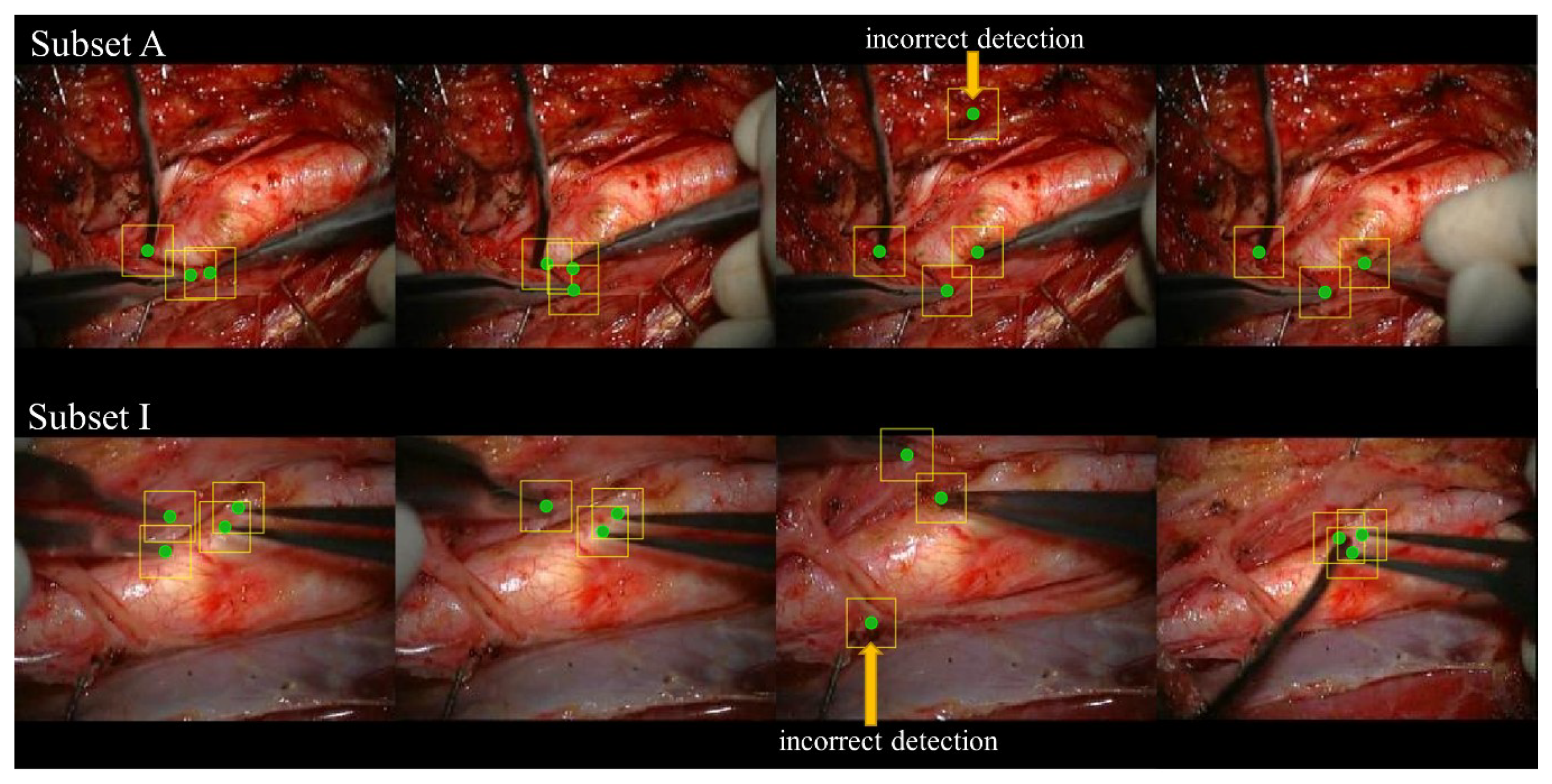
3.1. Bounding Box Detection
3.2. Detection Rate of the Distal End of the Surgical Instrument
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sugimori, H. Classification of computed tomography images in different slice positions using deep learning. J. Healthc. Eng. 2018, 2018, 1753480. [Google Scholar] [CrossRef] [PubMed]
- Sugimori, H. Evaluating the overall accuracy of additional learning and automatic classification system for CT images. Appl. Sci. 2019, 9, 682. [Google Scholar] [CrossRef]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Yasaka, K.; Akai, H.; Kunimatsu, A.; Abe, O.; Kiryu, S. Liver fibrosis: Deep convolutional neural network for staging by using gadoxetic acid–enhanced hepatobiliary phase MR images. Radiology 2017, 287, 146–155. [Google Scholar] [CrossRef]
- Zhang, Q.; Ruan, G.; Yang, W.; Liu, Y.; Zhao, K.; Feng, Q.; Chen, W.; Wu, E.X.; Feng, Y. MRI Gibbs-ringing artifact reduction by means of machine learning using convolutional neural networks. Magn. Reson. Med. 2019, 82, 2133–2145. [Google Scholar] [CrossRef]
- Roth, H.R.; Lee, C.T.; Shin, H.-C.; Seff, A.; Kim, L.; Yao, J.; Lu, L.; Summers, R.M. Anatomy-specific classification of medical images using deep convolutional nets. In Proceedings of the 2015 IEEE International Symposium on Biomedical Imaging, New York, NY, USA, 16–19 April 2015; pp. 101–104. [Google Scholar]
- Noguchi, T.; Higa, D.; Asada, T.; Kawata, Y.; Machitori, A.; Shida, Y.; Okafuji, T.; Yokoyama, K.; Uchiyama, F.; Tajima, T. Artificial intelligence using neural network architecture for radiology (AINNAR): Classification of MR imaging sequences. Jpn. J. Radiol. 2018, 36, 691–697. [Google Scholar] [CrossRef]
- Gao, X.W.; Hui, R.; Tian, Z. Classification of CT brain images based on deep learning networks. Comput. Methods Programs Biomed. 2017, 138, 49–56. [Google Scholar] [CrossRef]
- Sugimori, H.; Kawakami, M. Automatic detection of a standard line for brain magnetic resonance imaging using deep learning. Appl. Sci. 2019, 9, 3849. [Google Scholar] [CrossRef]
- Cao, Z.; Duan, L.; Yang, G.; Yue, T.; Chen, Q. An experimental study on breast lesion detection and classification from ultrasound images using deep learning architectures. BMC Med. Imaging 2019, 19, 1–9. [Google Scholar] [CrossRef]
- Ariji, Y.; Yanashita, Y.; Kutsuna, S.; Muramatsu, C.; Fukuda, M.; Kise, Y.; Nozawa, M.; Kuwada, C.; Fujita, H.; Katsumata, A.; et al. Automatic detection and classification of radiolucent lesions in the mandible on panoramic radiographs using a deep learning object detection technique. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 128, 424–430. [Google Scholar] [CrossRef]
- Ge, C.; Gu, I.Y.H.; Jakola, A.S.; Yang, J. Deep learning and multi-sensor fusion for glioma classification using multistream 2D convolutional networks. In Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Honolulu, HI, USA, 17–22 July 2018; Volume 2018, pp. 5894–5897. [Google Scholar]
- Dalmiş, M.U.; Litjens, G.; Holland, K.; Setio, A.; Mann, R.; Karssemeijer, N.; Gubern-Mérida, A. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med. Phys. 2017, 44, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Duong, M.T.; Rudie, J.D.; Wang, J.; Xie, L.; Mohan, S.; Gee, J.C.; Rauschecker, A.M. Convolutional neural network for automated FLAIR lesion segmentation on clinical brain MR imaging. Am. J. Neuroradiol. 2019, 40, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Bentley, P.; Rueckert, D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. NeuroImage Clin. 2017, 15, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Khalili, N.; Lessmann, N.; Turk, E.; Claessens, N.; de Heus, R.; Kolk, T.; Viergever, M.A.; Benders, M.J.N.L.; Išgum, I. Automatic brain tissue segmentation in fetal MRI using convolutional neural networks. Magn. Reson. Imaging 2019, 64, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Lama, S.; Gan, L.S.; Maddahi, Y.; Zareinia, K.; Sutherland, G.R. Forces of tool-tissue interaction to assess surgical skill level. JAMA Surg. 2018, 153, 234–242. [Google Scholar] [CrossRef]
- Birkmeyer, J.D.; Finks, J.F.; O’Reilly, A.; Oerline, M.; Carlin, A.M.; Nunn, A.R.; Dimick, J.; Banerjee, M.; Birkmeyer, N.J.O. Surgical skill and complication rates after bariatric surgery. N. Engl. J. Med. 2013, 369, 1434–1442. [Google Scholar] [CrossRef]
- Elek, R.N.; Haidegger, T. Robot-assisted minimally invasive surgical skill assessment—Manual and automated platforms. Acta Polytech. Hung. 2019, 16, 141–169. [Google Scholar]
- Jo, K.; Choi, Y.; Choi, J.; Chung, J.W. Robust real-time detection of laparoscopic instruments in robot surgery using convolutional neural networks with motion vector prediction. Appl. Sci. 2019, 9, 2865. [Google Scholar] [CrossRef]
- Zhao, Z.; Cai, T.; Chang, F.; Cheng, X. Real-time surgical instrument detection in robot-assisted surgery using a convolutional neural network cascade. Healthc. Technol. Lett. 2019, 6, 275–279. [Google Scholar] [CrossRef]
- Bouget, D.; Allan, M.; Stoyanov, D.; Jannin, P. Vision-based and marker-less surgical tool detection and tracking: A review of the literature. Med. Image Anal. 2017, 35, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Bouget, D.; Benenson, R.; Omran, M.; Riffaud, L.; Schiele, B.; Jannin, P. Detecting surgical tools by modelling local appearance and global shape. IEEE Trans. Med. Imaging 2015, 34, 2603–2617. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Nakamura, T.; Ito, Y.; Tokairin, K.; Kazumata, K.; Nakayama, N.; Houkin, K. A pilot study on measuring tissue motion during carotid surgery using video-based analyses for the objective assessment of surgical performance. World J. Surg. 2019, 43, 2309–2319. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, Z.; Voros, S.; Cheng, X. Real-time tracking of surgical instruments based on spatio-temporal context and deep learning. Comput. Assist. Surg. 2019, 24, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, 1–20. [Google Scholar] [CrossRef]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the 2018 International Interdisciplinary Ph.D. Workshop (IIPhDW), Świnoujście, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar]
- Mash, R.; Borghetti, B.; Pecarina, J. Improved aircraft recognition for aerial refueling through data augmentation in convolutional neural networks. In Proceedings of the Advances in Visual Computing; Springer International Publishing: Cham, Switzerland, 2016; pp. 113–122. [Google Scholar]
- Taylor, L.; Nitschke, G. Improving deep learning using generic data augmentation. arXiv 2017, arXiv:1708.06020. Available online: https://arxiv.org/abs/1708.06020 (accessed on 1 April 2020). [Google Scholar]
- Eaton-rosen, Z.; Bragman, F. Improving data augmentation for medical image segmentation. In Proceedings of the 1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlands, 4–6 July 2018; pp. 1–3. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6517–6525. [Google Scholar]
- Lalonde, R.; Zhang, D.; Shah, M. ClusterNet: Detecting small objects in large scenes by exploiting spatio-temporal information. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2018, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4003–4012. [Google Scholar]
- Chen, K.; Wang, J.; Yang, S.; Zhang, X.; Xiong, Y.; Loy, C.C.; Lin, D. Optimizing video object detection via a scale-time lattice. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2018, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7814–7823. [Google Scholar]
- Hasan, S.M.K.; Linte, C.A. U-NetPlus: A modified encoder-decoder u-net architecture for semantic and instance segmentation of surgical instruments from laparoscopic images. In Proceedings of the 41th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Berlin, Germany, 23–27 July 2019; pp. 7205–7211. [Google Scholar]
- Ni, Z.-L.; Bian, G.-B.; Xie, X.-L.; Hou, Z.-G.; Zhou, X.-H.; Zhou, Y.-J. RASNet: Segmentation for tracking surgical instruments in surgical videos using refined attention segmentation network. In Proceedings of the 41th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Berlin, Germany, 23–27 July 2019; pp. 5735–5738. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2014, Columbus, OH, USA, 24–27 June 2014; pp. 580–587. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single shot multibox detector. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 21–37. [Google Scholar]
- Oikawa, K.; Kato, T.; Oura, K.; Narumi, S.; Sasaki, M.; Fujiwara, S.; Kobayashi, M.; Matsumoto, Y.; Nomura, J.i.; Yoshida, K.; et al. Preoperative cervical carotid artery contrast-enhanced ultrasound findings are associated with development of microembolic signals on transcranial Doppler during carotid exposure in endarterectomy. Atherosclerosis 2017, 260, 87–93. [Google Scholar] [CrossRef]
- Cao, Z.; Simon, T.; Wei, S.E.; Sheikh, Y. Realtime multi-person 2D pose estimation using part affinity fields. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1302–1310. [Google Scholar]
- Mathis, A.; Mamidanna, P.; Cury, K.M.; Abe, T.; Murthy, V.N.; Mathis, M.W.; Bethge, M. DeepLabCut: Markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 2018, 21, 1281–1289. [Google Scholar] [CrossRef]
- Papandreou, G.; Zhu, T.; Chen, L.C.; Gidaris, S.; Tompson, J.; Murphy, K. Personlab: Person pose estimation and instance segmentation with a bottom-up, part-based, geometric embedding model. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 282–299. [Google Scholar]
- Allan, M.; Shvets, A.; Kurmann, T.; Zhang, Z.; Duggal, R.; Su, Y.-H.; Rieke, N.; Laina, I.; Kalavakonda, N.; Bodenstedt, S.; et al. 2017 robotic instrument segmentation challenge. arXiv 2019, arXiv:1902.06426. Available online: https://arxiv.org/abs/1902.06426 (accessed on 1 April 2020). [Google Scholar]
- Bernal, J.; Tajkbaksh, N.; Sanchez, F.J.; Matuszewski, B.J.; Chen, H.; Yu, L.; Angermann, Q.; Romain, O.; Rustad, B.; Balasingham, I.; et al. Comparative validation of polyp detection methods in video colonoscopy: Results from the MICCAI 2015 endoscopic vision challenge. IEEE Trans. Med. Imaging 2017, 36, 1231–1249. [Google Scholar] [CrossRef] [PubMed]
| Without Data Augmentation | With Data Augmentation | |||||
|---|---|---|---|---|---|---|
| AP | LAMR | FPS [fps] | AP | LAMR | FPS [fps] | |
| Subset A | 0.3895 | 0.7275 | 43.2 | 0.8258 | 0.4234 | 41.9 |
| Subset B | 0.4649 | 0.6361 | 43.5 | 0.6929 | 0.4382 | 28.0 |
| Subset C | 0.4958 | 0.6216 | 44.0 | 0.6866 | 0.4206 | 28.3 |
| Subset D | 0.2960 | 0.7263 | 44.1 | 0.6689 | 0.4124 | 28.0 |
| Subset E | 0.4150 | 0.6515 | 43.9 | 0.8639 | 0.2006 | 28.1 |
| Subset F | 0.3104 | 0.7367 | 43.8 | 0.8533 | 0.4117 | 28.7 |
| Subset G | 0.6216 | 0.5288 | 43.4 | 0.8603 | 0.1821 | 28.3 |
| Subset H | 0.3336 | 0.7159 | 41.5 | 0.8055 | 0.3841 | 28.7 |
| Subset I | 0.5179 | 0.6124 | 42.7 | 0.9083 | 0.2585 | 28.7 |
| mean ± SD | 0.4272 ± 0.108 | 0.6619 ± 0.0703 | 43.3 ± 0.8 | 0.7962 ± 0.0897 | 0.3488 ± 0.1036 | 29.9 ± 4.5 |
| Detection Rate of the Distal End of the Surgical Instrument | ||
|---|---|---|
| Within the Center of 8 × 8 Pixels | Within the Center of 16 × 16 Pixels | |
| Subset A | 0.5388 | 0.9660 |
| Subset B | 0.4916 | 0.9241 |
| Subset C | 0.5826 | 0.9604 |
| Subset D | 0.5033 | 0.9560 |
| Subset E | 0.6430 | 0.9706 |
| Subset F | 0.6742 | 0.9796 |
| Subset G | 0.8210 | 0.9828 |
| Subset H | 0.6419 | 0.9733 |
| Subset I | 0.5934 | 0.9750 |
| mean ± SD | 0.6100 ± 0.1014 | 0.9653 ± 0.0177 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugimori, H.; Sugiyama, T.; Nakayama, N.; Yamashita, A.; Ogasawara, K. Development of a Deep Learning-Based Algorithm to Detect the Distal End of a Surgical Instrument. Appl. Sci. 2020, 10, 4245. https://doi.org/10.3390/app10124245
Sugimori H, Sugiyama T, Nakayama N, Yamashita A, Ogasawara K. Development of a Deep Learning-Based Algorithm to Detect the Distal End of a Surgical Instrument. Applied Sciences. 2020; 10(12):4245. https://doi.org/10.3390/app10124245
Chicago/Turabian StyleSugimori, Hiroyuki, Taku Sugiyama, Naoki Nakayama, Akemi Yamashita, and Katsuhiko Ogasawara. 2020. "Development of a Deep Learning-Based Algorithm to Detect the Distal End of a Surgical Instrument" Applied Sciences 10, no. 12: 4245. https://doi.org/10.3390/app10124245
APA StyleSugimori, H., Sugiyama, T., Nakayama, N., Yamashita, A., & Ogasawara, K. (2020). Development of a Deep Learning-Based Algorithm to Detect the Distal End of a Surgical Instrument. Applied Sciences, 10(12), 4245. https://doi.org/10.3390/app10124245






