Soil Is Still an Unknown Biological System
Abstract
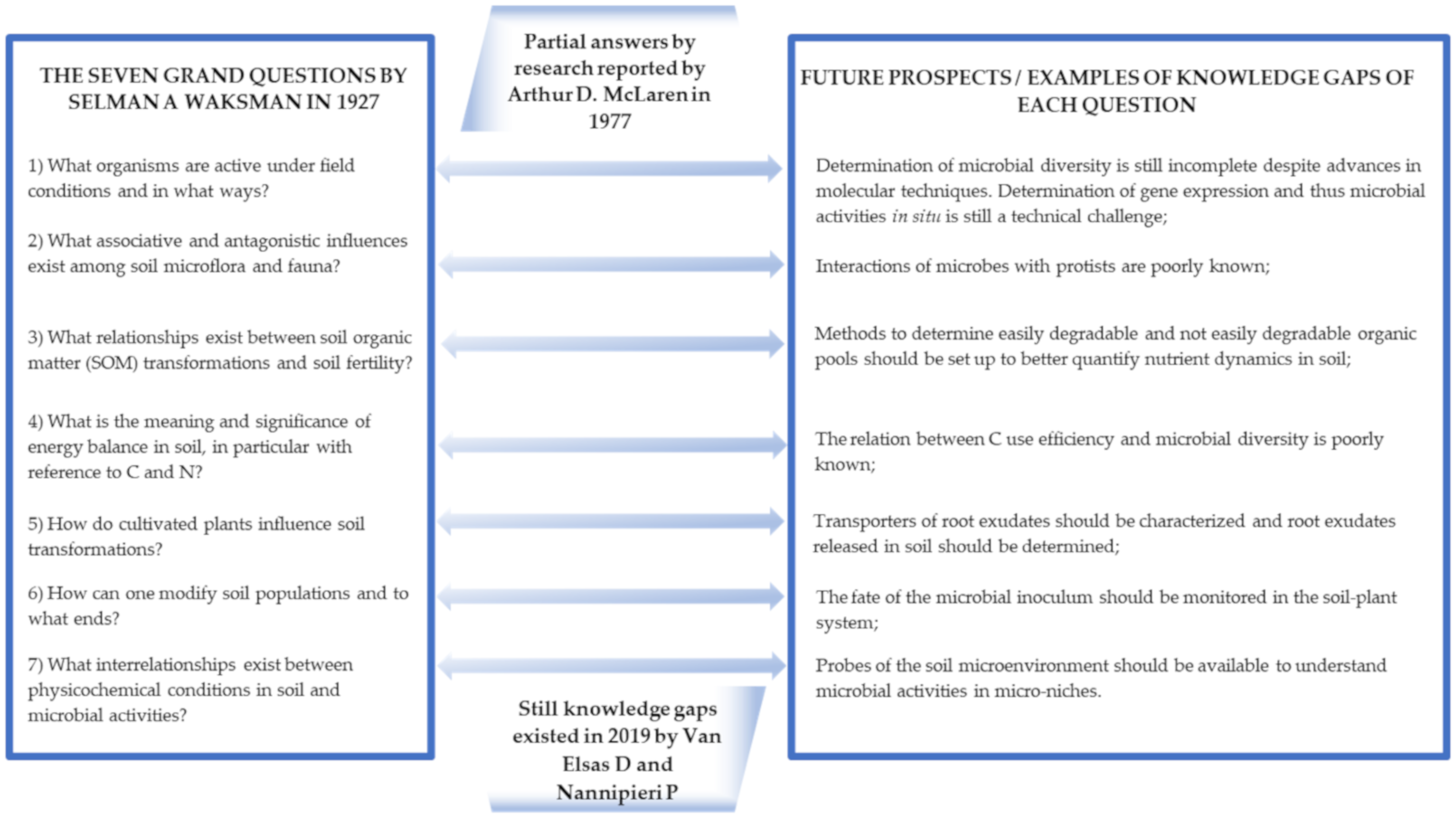
1. Introduction
- What organisms are active under field conditions and in what ways?
- What associative and antagonistic influences exist among soil microflora and fauna?
- What relationships exist between soil organic matter (SOM) transformations and soil fertility?
- What is the meaning and significance of energy balance in soil, in particular with reference to C and N?
- How do cultivated plants influence soil transformations?
- How can one modify soil populations and to what ends?
- What interrelationships exist between physicochemical conditions in soil and microbial activities?
2. Properties of Soil as a Biological System
2.1. Physicals Structure and Distribution of Organisms in Soil
2.2. Microbial Diversity and Microbial Functions
2.3. The Role of Important Biological Molecules Adsorbed or Entrapped in Soil
2.4. Interactions between Microbes, between Microbes and Plants, and between Microbes and Fauna
2.5. The Main Research Approaches
3. Perspective
4. Conclusions
Funding
Conflicts of Interest
References
- McLaren, A.D. The seven questions of Selman A. Waksman. Soil Biol. Biochem. 1977, 9, 375–376. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Nannipieri, P. The seven grand questions on soil microbiology (Selman A. Waksman, re-examined by Arthur D. McLaren). In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; CRC Press: Baca Raton, CA, USA, 2019; pp. 21–35. [Google Scholar]
- Oades, J.M.; Waters, A.G. Aggregate hierarchy in soils. Aust. J. Soil Res. 1991, 29, 815–828. [Google Scholar] [CrossRef]
- Tisdall, J.M.; Oades, J.M. Organic matter and water-stable aggregates in soils. J. Soil Sci. 1982, 33, 141–163. [Google Scholar] [CrossRef]
- Elliott, E.T.; Coleman, D.C. Let the soil work for us. Ecol. Bull. Natl. Speleol. Soc. 1988, 39, 23–32. [Google Scholar]
- Six, J. A history of research on the link between (micro) aggregates, soil biota, and soil organic matter. Soil Tillage Res. 2004, 79, 7–31. [Google Scholar] [CrossRef]
- Coleman, D.C.; Crossley, D.A. Fundamentals of Soil Ecology; Academic Press: London, UK, 1996. [Google Scholar]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Dini-Andreote, F.; Van Elsas, J.D. The soil microbiome—An overview. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; CRC Press: Baca Raton, CA, USA, 2019; pp. 37–48. [Google Scholar]
- Smith, J.L.; Paul, E.A. The significanceof soil microbial biomass estimations. In Soil Biochemistry; Bollag, J.-M., Stotzky, G., Eds.; Marcel Dekker: New York, NY, USA, 1990; Volume 6, pp. 357–396. [Google Scholar]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.; Benavent-Gonzalez, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Nannipieri, P.; Ascher-Jenull, J.; Ceccherini, M.T.; Pietramellara, G.; Renella, G.; Schloter, M. Beyond microbial diversity for predicting soil functions: A mini review. Pedosphere 2020, 30, 5–17. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Wei, X.; Hu, Y.; Razavi, B.S.; Zhou, J.; Shen, J.; Nannipieri, P.; Wu, J.; Ge, T. Rare taxa of alkaline phosphomonoesterase-harboring microorganisms meadiate soil phosphorus mineralization. Soil Biol. Biochem. 2019, 131, 62–70. [Google Scholar] [CrossRef]
- Jiang, Y.; Song, H.; Lei, Y.; Korpelainen, H.; Li, C. Distinct co-occurrence patterns and driving forces of rare and abundant bacterial subcommunities following a glacial retreat in the eastern Tibetan Plateau. Biol. Fertil. Soils 2019, 55, 351–364. [Google Scholar] [CrossRef]
- Lavelle, P.; Spain, A. Soil Ecology; Kluwer Academic Publisher: Dordrecht, The Netherlands, 2001. [Google Scholar]
- Williamson, K.E.; Srinivasiah, S.; Wommack, K.E. Viruses in soil ecosystems. In Handbook of Soil Science: Perspectives and Processes; Huang, P.M., Li, Y., Summer, M.E., Eds.; CRC Press: Boca Raton, CA, USA, 2012; pp. 24.1–24.10. [Google Scholar]
- Bernardo, P.C.M.; Charles-Dominique, T.; Barakat, M.; Ortet, P.; Fernandez, E.; Filloux, D.; Hartnady, P.; Rebelo, T.A.; Cousins, S.R.; Mesleard, F.; et al. Geometagenomics illuminates the impact of agriculture on the distribution and prevalence of plant viruses at the ecosystem level. ISME J. 2018, 12, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Juarez, S.; Nunan, N.; Duday, A.-N.; Pouteau, V.; Chenu, C. Soil carbon mineralisation responses to alterations of microbial diversity and soil structure. Biol. Fertil. Soils 2013, 49, 939–948. [Google Scholar] [CrossRef]
- De Graaf, M.A.; Adkins, J.; Kardol, P.; Throop, H.L. A meta-analysis of soil biodiversity impacts on the carbon cycle. Soil 2015, 1, 257–271. [Google Scholar] [CrossRef]
- Bao, Y.; Guo, Z.; Chen, R.; Wu, M.; Li, Z.; Lin, X.; Feng, Y. Functional community composition shows less environmental variability than 4 taxonomic composition in straw-degrading bacteria . Biol. Fertil. Soils 2020, 56. in press. [Google Scholar]
- Singh, B.K.; Quince, C.; Macdonald, C.A.; Khachane, A.; Thomas, N.; Al-Soud, W.A.; Sorensen, S.J.; He, Z.; White, D.; Sinclair, A.; et al. Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environ. Microbiol. 2014, 16, 1–10. [Google Scholar] [CrossRef]
- Burns, R.G. Enzyme activity in soil: Location and a possible role in microbial ecology. Soil Biol. Biochem. 1982, 14, 423–427. [Google Scholar] [CrossRef]
- Nannipieri, P.; Trasar-Cepeda, C.; Dick, R.P. Soil enzyme activity: A brief history and biochemistry as a basis for appropriate interpretations and meta-analysis. Biol. Fertil. Soils 2018, 54, 11–19. [Google Scholar] [CrossRef]
- Pietramellara, G.; Ascher, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: Fate nd ecological relevance. Biol. Fertil. Soils 2009, 45, 219–235. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Nothen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant. Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; de Araujo, W.L.; Trevors, J.T. Microbial interactions. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; CRC Press: Baca Raton, CA, USA, 2019; pp. 141–161. [Google Scholar]
- Bonkowsky, M.; Clarholm, M. Stimulation of plant growth through interactions of bacteria and protozoa: Testing the auxiliary microbial loop hypothesis. Acta Protozool. 2012, 51, 237–247. [Google Scholar]
- Bonkowski, M.; Dumack, K.; Fiore-Donno, A.M. The protists in soil—A token of untold eukaryotc diversity. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; CRC Press: Baca Raton, CA, USA, 2019; pp. 125–140. [Google Scholar]
- Samad, A.; Brader, G.; Pfaffenbichler, N.; Sessitsch, A. Plant-associated bacteria and the rhizosphere. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; CRC Press: Baca Raton, CA, USA, 2019; pp. 163–178. [Google Scholar]
- Cooper, J.E. Early interactions between legumes and rhizobia: Disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 2007, 103, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.M.; Perotto, S.; Bonfante, P. Mycorrhizal fungi: A fungal community at the interface between soil and roots. In The Rhizosphere. Biochemistry and Organic Substances at the Soil-Plant Interface, 3rd ed.; Pinton, R., Varanini, Z., Nannipieri, P., Eds.; CRC Press: Boca Raton, CA, USA, 2007; pp. 201–236. [Google Scholar]
- De Ridder-Duine, A.S.; Kowalchuk, G.A.; Gunnewiek, P.J.A.K.; Smant, W.; van Veen, J.A.; de Boer, W. Rhizosphere bacterial community composition in natural stands of Carex arenaria (sand sedge) is determined by bulk soil community composition. Soil Biol. Biochem. 2005, 37, 349–357. [Google Scholar] [CrossRef]
- Yech, Y.K.; Dennis, P.G.; Paungfoo-Lohienne, C.; Weber, L.; Brackin, R.; Ragan, M.A.; Schmidt, S.; Hugenholtz, P. Evolutionary concervation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat. Commun. 2017, 8, 215–224. [Google Scholar]
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; van Thernaat, E.V.L.; Schulze-Lefert, P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2014, 111, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Martinez, C.; Bulgarelli, D. Tracing the evolutionary routes of plant-microbiota interactions. Curr. Opin. Microbiol. 2019, 49, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Fizpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly of ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef]
- Stozky, G. Influence of soil mineral colloids and metabolic processes, growth adhesion, and ecology of microbes and viruses. In Interactions of Soil Minerals with Natural Organics and Microbes; Huang, M., Schnitzer, M., Eds.; Soil Science Society of America: Madison, WI, USA, 1986; pp. 305–428. [Google Scholar]
- Nannipieri, P.; Giagnoni, L.; Renella, G. Metaproteomics of soil microbial communities. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; CRC Press: Baca Raton, CA, USA, 2019; pp. 257–268. [Google Scholar]
- Bao, Y.Y.; Dolfing, J.; Wang, B.Z.; Chen, R.R.; Huang, M.S.; Li, Z.P.; Lin, X.G.; Feng, Y.Z. Bacterial communities involved directly or indirectly in the anaerobic degradation of cellulose. Biol. Fertil. Soils 2019, 55, 201–211. [Google Scholar] [CrossRef]
- Dias, A.C.F.; Dini-Andreote, F.; Hannula, S.E.; Dini-Andreote, F.; de Cassia Pereira e Silva, M.; Salles, J.F.; de Boer, W.; van Veen, J.; van Elsas, J.D. Different selective effects on rhizosphere bacteria exerted by genetically modified versus conventional potato lines. PLoS ONE 2013, 8, e67948. [Google Scholar] [CrossRef] [PubMed]
- Loeppmann, S.; Semenov, M.; Kuzyakov, Y.; Blagodatskaya, E. Shift from dormancy to microbial growth revealed by RNA:DNA ratio. Soil Biol. Biochem. 2018, 85, 603–612. [Google Scholar] [CrossRef]
- Jenkinson, D.S.; Powlson, D.S. The effect of biocidal treatments on metabolism in soil. V. A method for measuring soil biomass. Soil Biol. Biochem. 1976, 8, 209–213. [Google Scholar] [CrossRef]
- Kästner, M.; Miltner, A. SOM and microbes—What is left from microbial life. In The Future of Soil Carbon; Garcia, C., Nannipieri, P., Hernandez, T., Eds.; Academic Press: London, UK, 2018; pp. 125–163. [Google Scholar]
- Gamalero, E.; Glick, B.R. Plant growth-promoting rhizobacteria in agricultural and stressed soil. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; CRC Press: Baca Raton, CA, USA, 2019; pp. 361–380. [Google Scholar]
- Rilling, J.I.; Acuna, J.J.; Nannipieri, P.; Cassan, F.; Maruyama, F.; Jorquera, M.A. Current opinion and perspectives on the methods for trackimg and monitoring plant growth-promoting bacteria. Soil Biol. Biochem. 2018, 130, 205–219. [Google Scholar] [CrossRef]
| Soil | Vegetation | MBC (kg·ha−1) | MBN (kg·ha−1) |
|---|---|---|---|
| Sandy loam | Pasture | 280 | 40 |
| Silt loam | Cereal-grass | 288 | 48 |
| Clay | Pasture | 750 | 100 |
| Silt | Pasture | 800 | 309 |
| Clay loam | Cereals | 1200 | 240 |
| Clay loam | Wheat | 1940 | 385 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nannipieri, P. Soil Is Still an Unknown Biological System. Appl. Sci. 2020, 10, 3717. https://doi.org/10.3390/app10113717
Nannipieri P. Soil Is Still an Unknown Biological System. Applied Sciences. 2020; 10(11):3717. https://doi.org/10.3390/app10113717
Chicago/Turabian StyleNannipieri, Paolo. 2020. "Soil Is Still an Unknown Biological System" Applied Sciences 10, no. 11: 3717. https://doi.org/10.3390/app10113717
APA StyleNannipieri, P. (2020). Soil Is Still an Unknown Biological System. Applied Sciences, 10(11), 3717. https://doi.org/10.3390/app10113717





