The Influence of Dissolved Organic Carbon on the Microbial Community Associated with Tetraselmis striata for Bio-Diesel Production
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Tetraselmis striata Strain and Culturing
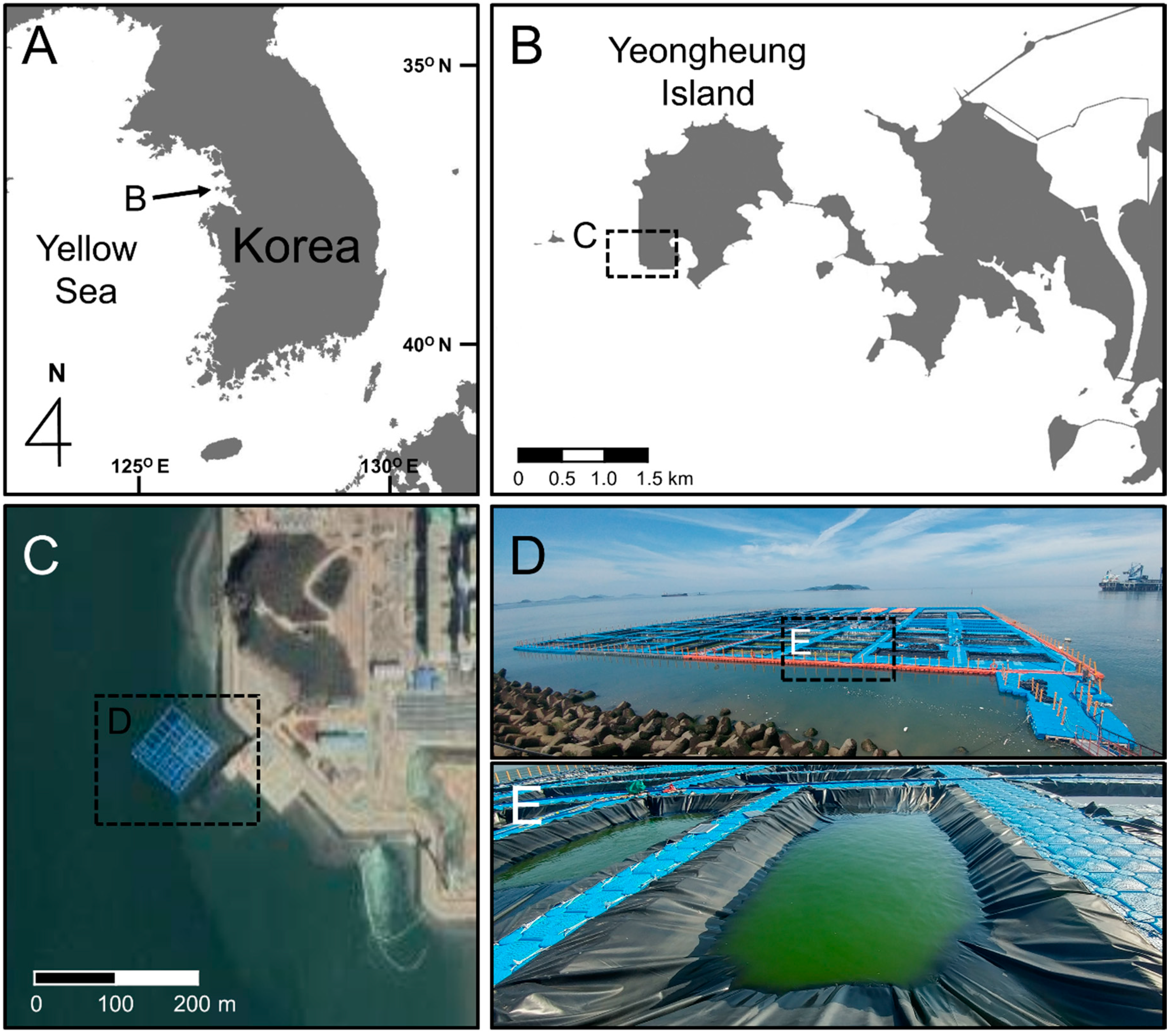
2.2. Sampling of Marine Culturing Field
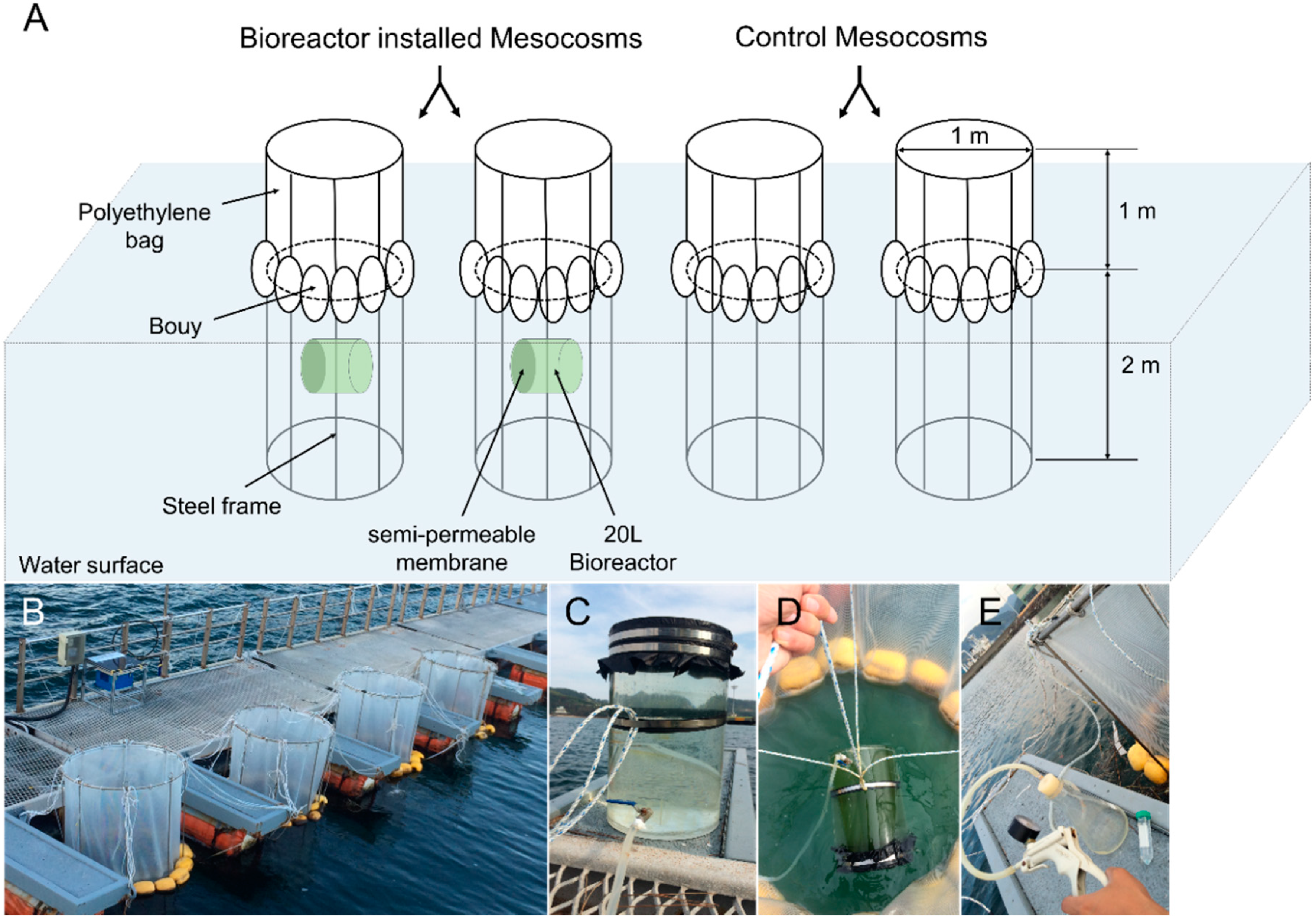
2.3. Mesocosm Design and Sampling
2.4. DOC Analysis
2.5. Bacterial Abundance and Composition
2.6. Preservation and Enumeration of HNFs, Phytoplankton, Ciliates, and Zooplankton
2.7. Nutrients Analyses
3. Results
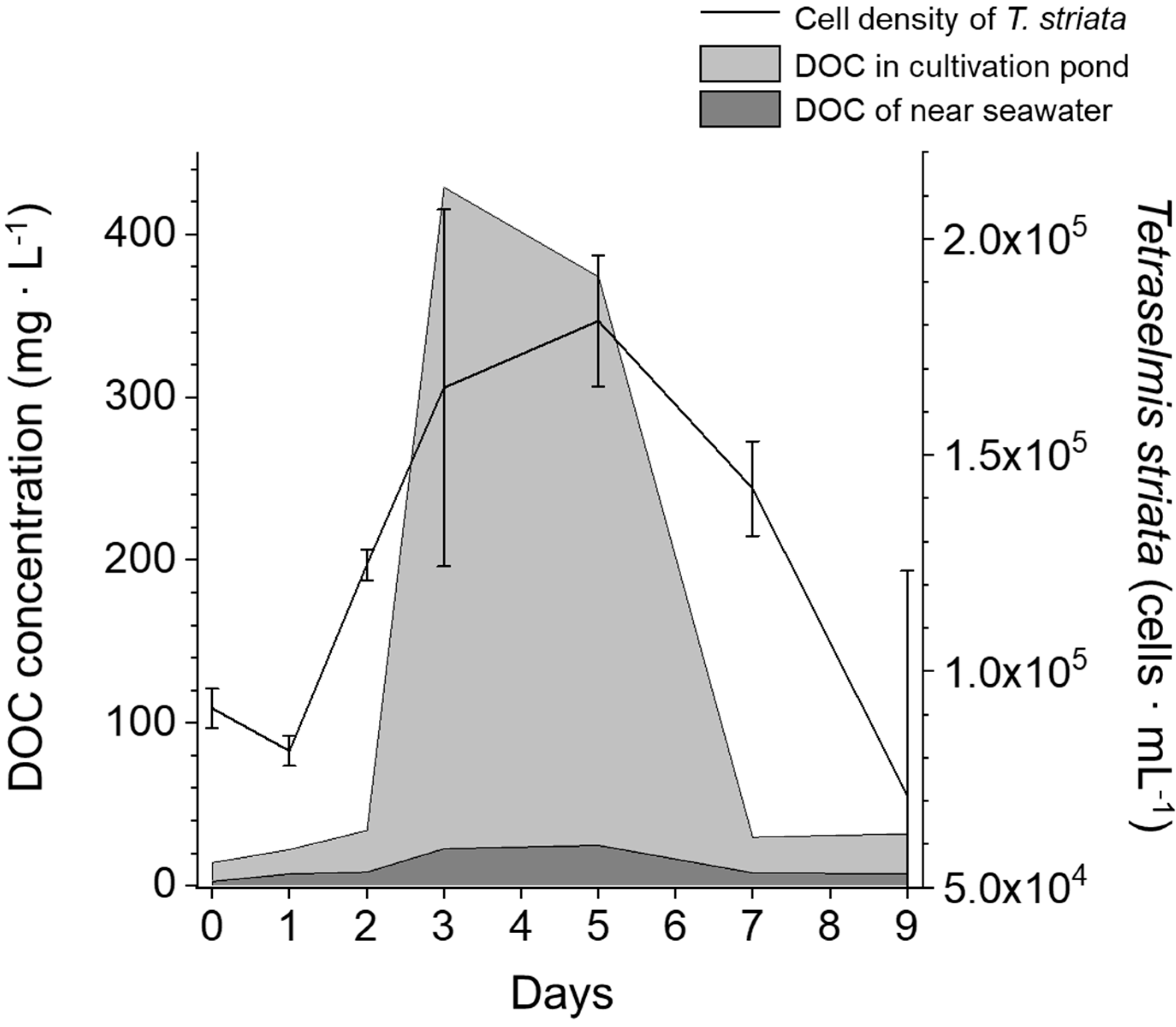
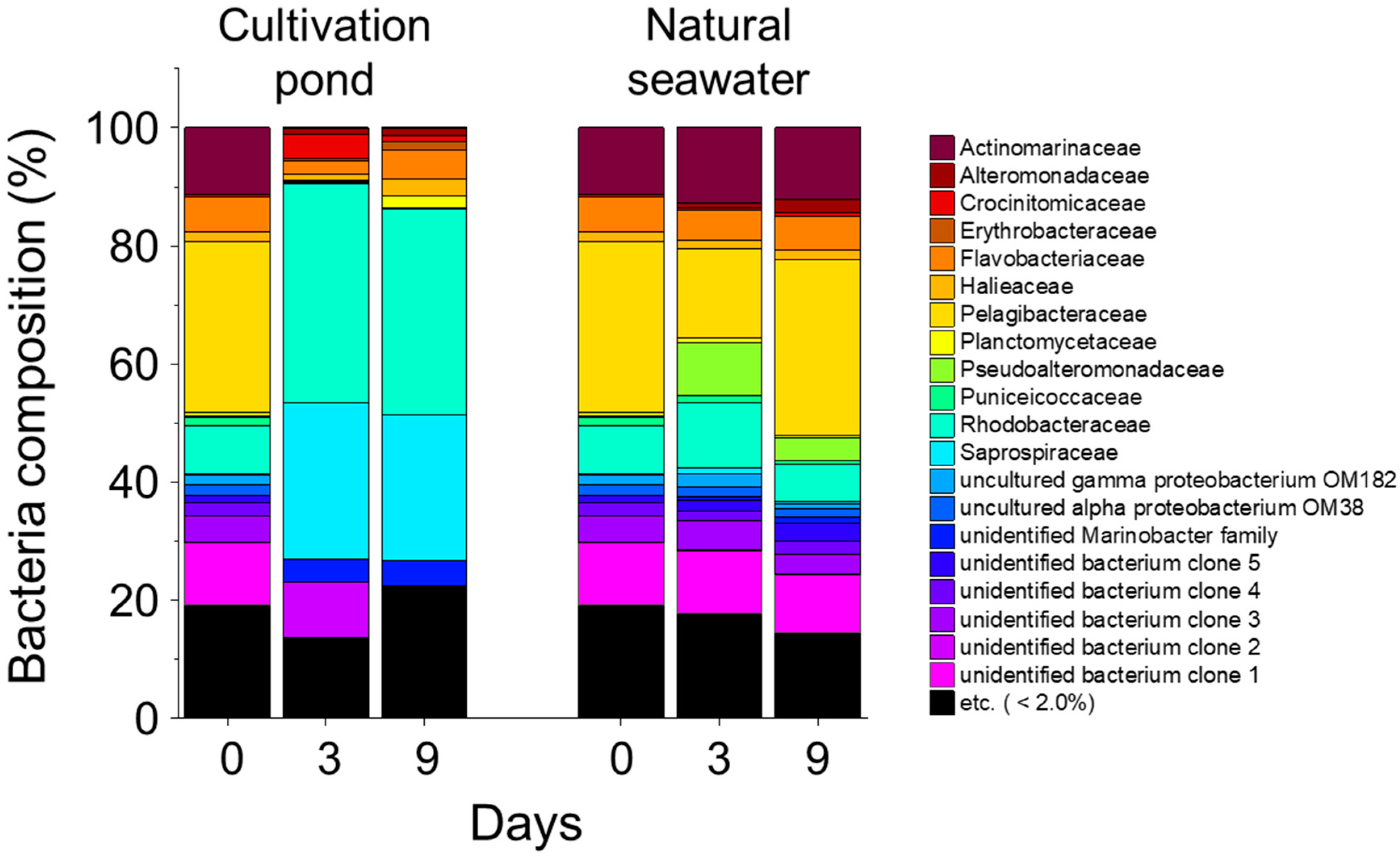
3.1. DOC and Bacteria in Cultivation Pond
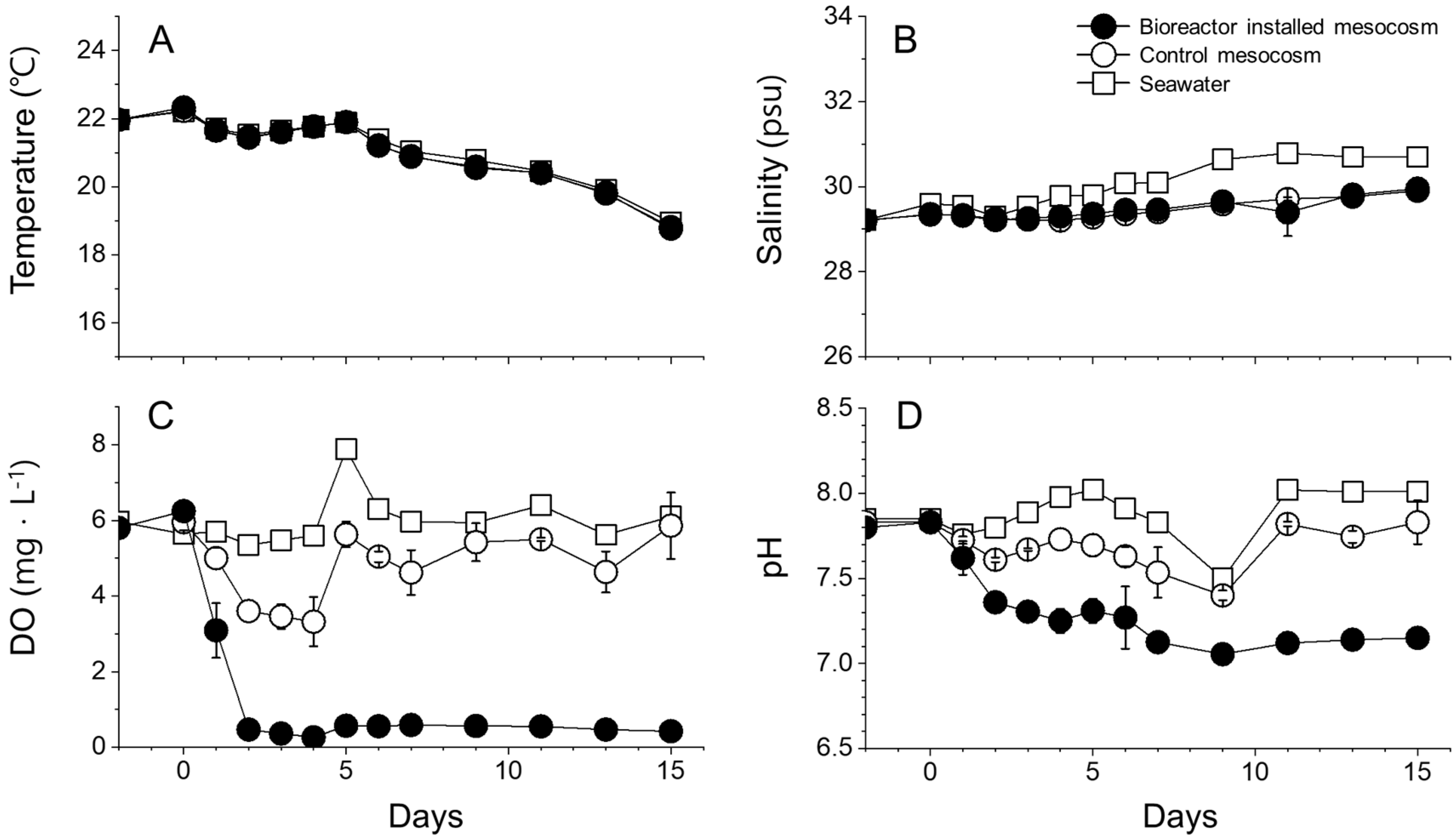
3.2. Environmental Factors during Mesocosm Experiment
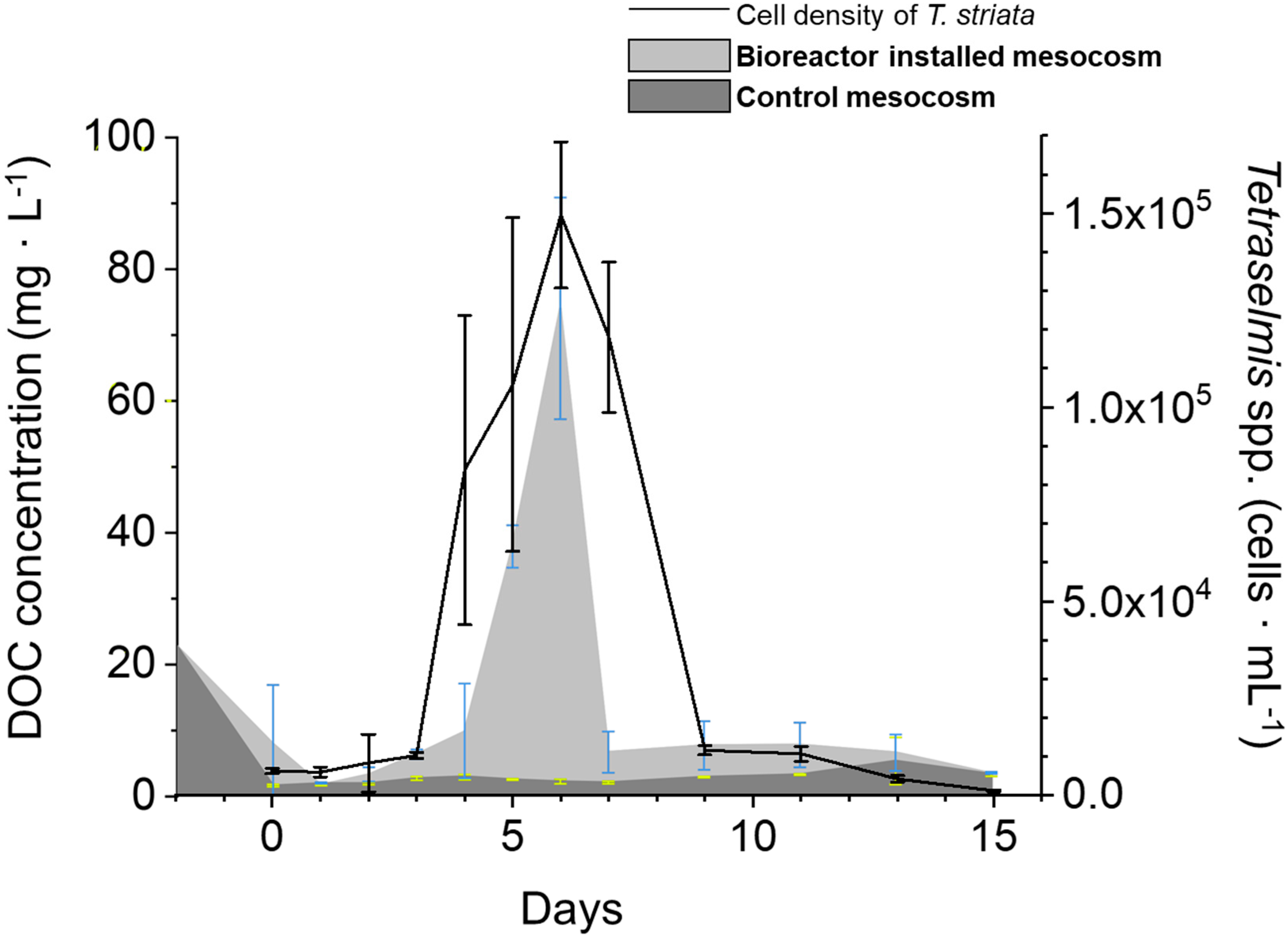
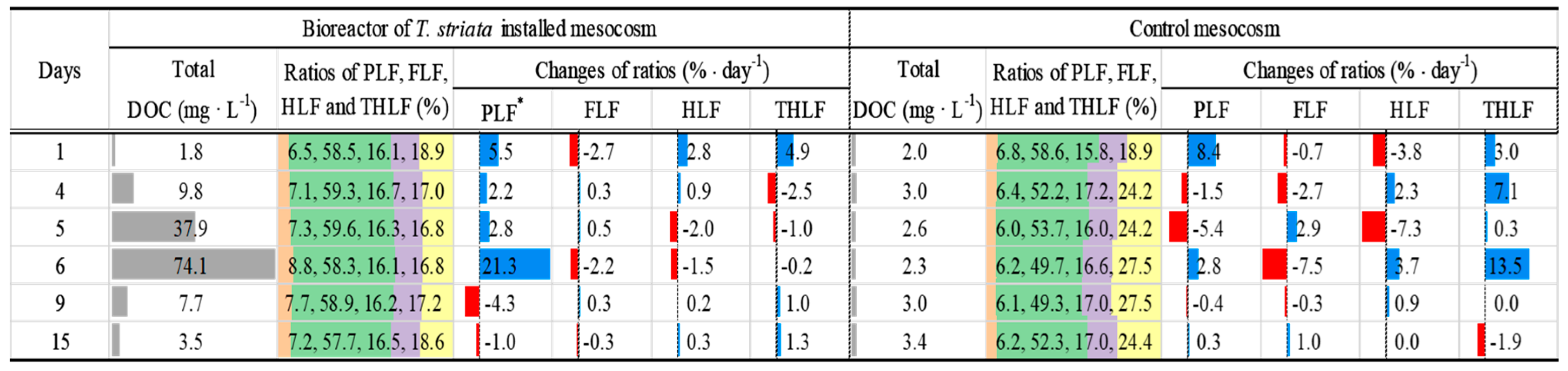
3.3. Abundance of T. striata and DOC Concentration
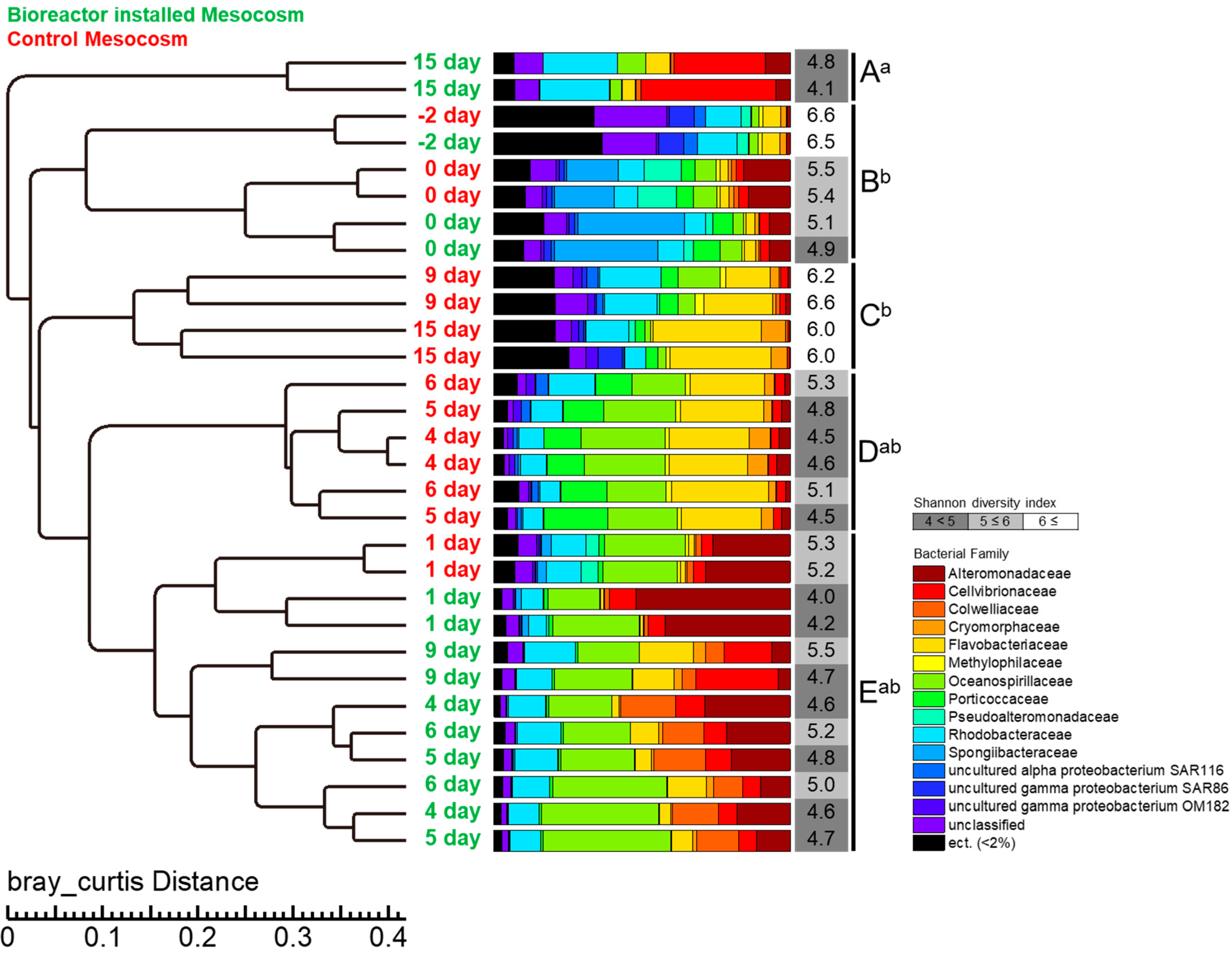
3.4. Bacterial Abundance and Composition
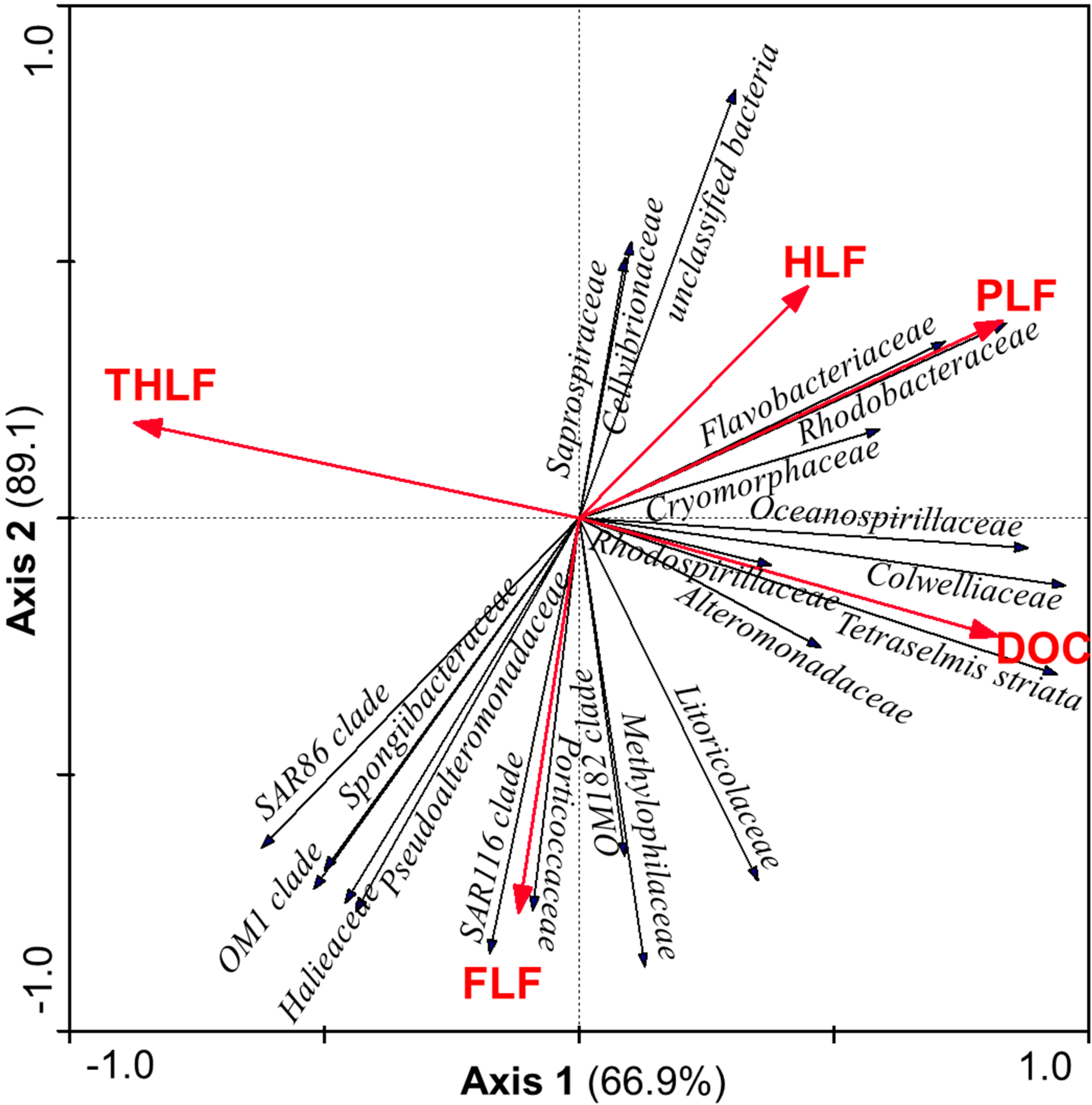
3.5. Relationship between Bacterial Community and DOC
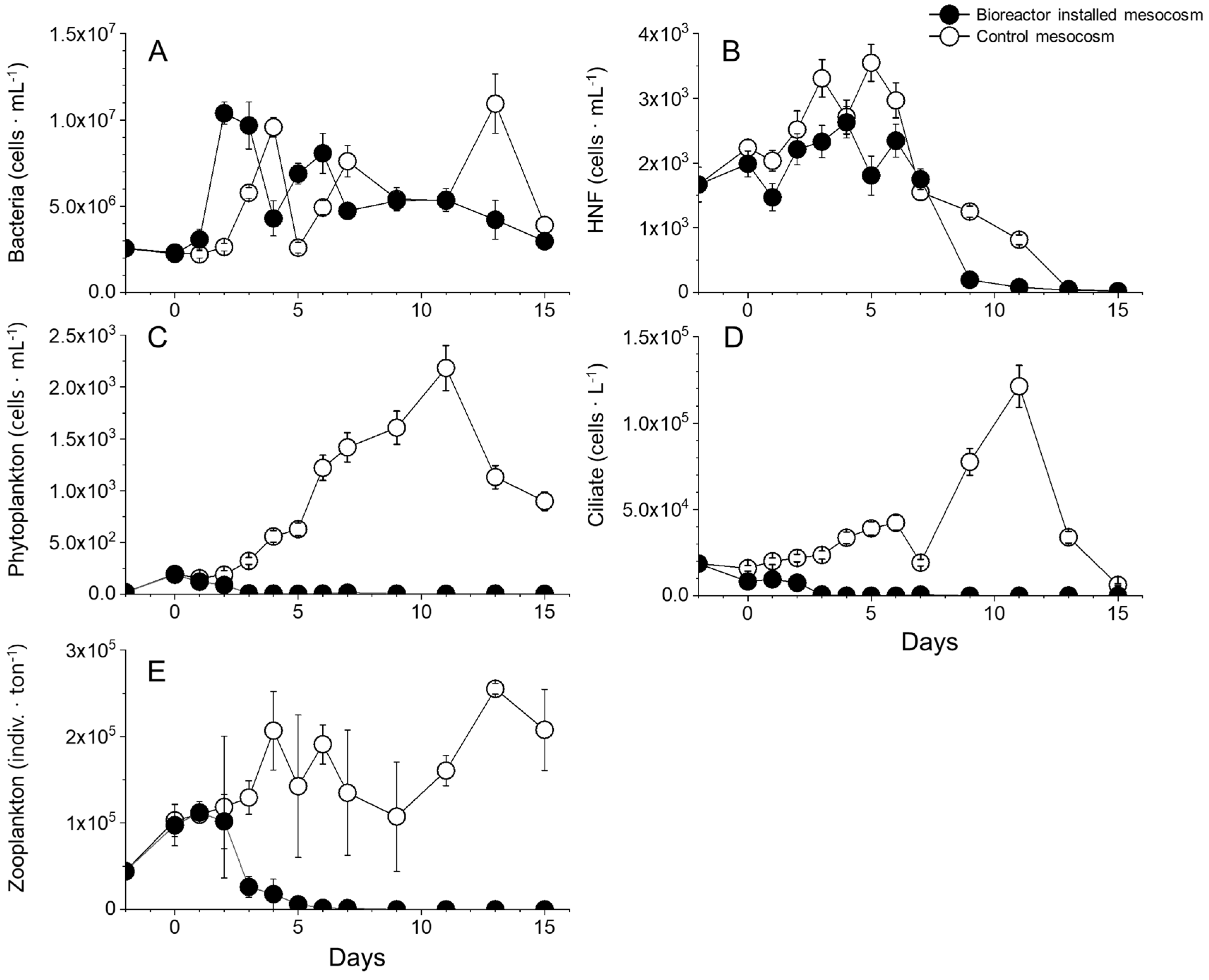
3.6. Other Biotic Factors
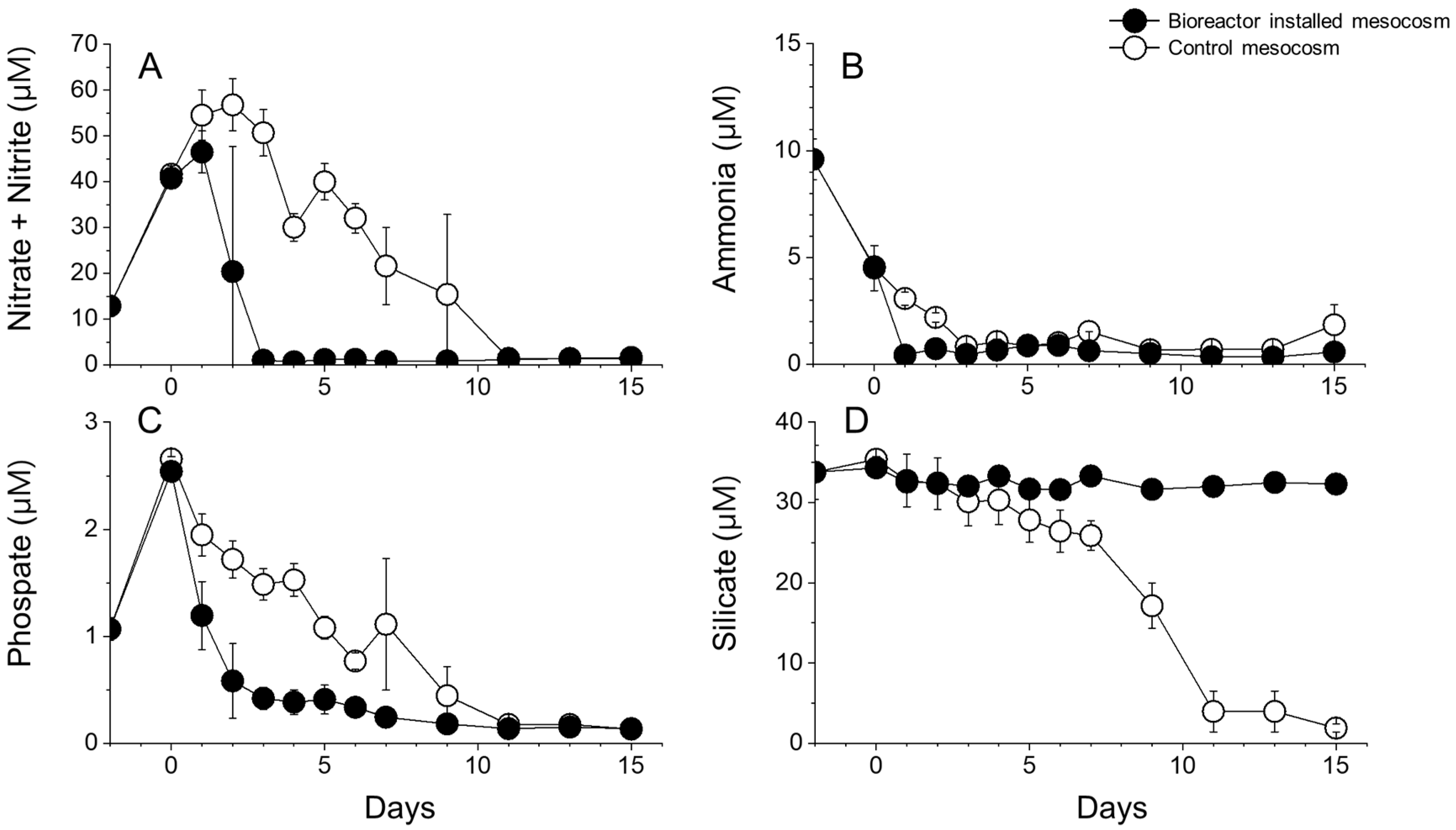
3.7. Nutrient Dynamics
4. Discussion
4.1. DOC Released from T. striata
4.2. Changes in Bacterial Community in Cultivation Pond and Mesocosms
4.3. Microbial Food Web Structure
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nashawi, I.S.; Malallah, A.; Al-Bisharah, M. Forecasting world crude oil production using multicyclic Hubbert model. Energy Fuels 2010, 24, 1788–1800. [Google Scholar] [CrossRef]
- Wijffels, R.H.; Barbosa, M.J. An outlook on microalgal biofuels. Science 2010, 329, 796–799. [Google Scholar] [CrossRef] [PubMed]
- Amaro, H.M.; Guedes, A.C.; Malcata, F.X. Advances and perspectives in using microalgae to produce biodiesel. Appl. Energy 2011, 88, 3402–3410. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Patidar, S.K.; Kim, S.-H.; Kim, J.H.; Park, J.; Park, B.S.; Han, M.-S. Pelagibaca bermudensis promotes biofuel competence of Tetraselmis striata in a broad range of abiotic stressors: Dynamics of quorum-sensing precursors and strategic improvement in lipid productivity. Biotechnol. Biofuels 2018, 11, 102. [Google Scholar] [CrossRef]
- Danquah, M.; Harun, R.; Halim, R.; Forde, G. Cultivation medium design via elemental balancing for Tetraselmis suecica. Chem. Biochem. Eng. Q. 2010, 24, 361–369. [Google Scholar]
- Huang, X.; Huang, Z.; Wen, W.; Yan, J. Effects of nitrogen supplementation of the culture medium on the growth, total lipid content and fatty acid profiles of three microalgae (Tetraselmis subcordiformis, Nannochloropsis oculata and Pavlova viridis). J. Appl. Phycol. 2013, 25, 129–137. [Google Scholar] [CrossRef]
- Moheimani, N.R. Inorganic carbon and pH effect on growth and lipid productivity of Tetraselmis suecica and Chlorella sp (Chlorophyta) grown outdoors in bag photobioreactors. J. Appl. Phycol. 2013, 25, 387–398. [Google Scholar] [CrossRef]
- Kim, G.; Bae, J.; Lee, K. Nitrate repletion strategy for enhancing lipid production from marine microalga Tetraselmis sp. Bioresour. Technol. 2016, 205, 274–279. [Google Scholar] [CrossRef]
- Selvakumar, P.; Umadevi, K. Enhanced lipid and fatty acid content under photoheterotrophic condition in the mass cultures of Tetraselmis gracilis and Platymonas convolutae. Algal Res. 2014, 6, 180–185. [Google Scholar] [CrossRef]
- Teo, C.L.; Atta, M.; Bukhari, A.; Taisir, M.; Yusuf, A.M.; Idris, A. Enhancing growth and lipid production of marine microalgae for biodiesel production via the use of different LED wavelengths. Bioresour. Technol. 2014, 162, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Úbeda-Mínguez, P.; Chileh, T.; Dautor, Y.; García-Maroto, F.; Alonso, D.L. Tools for microalgal biotechnology: Development of an optimized transformation method for an industrially promising microalga—Tetraselmis chuii. J. Appl. Phycol. 2015, 27, 223–232. [Google Scholar] [CrossRef]
- Vuttipongchaikij, S. Genetic manipulation of microalgae for improvement of biodiesel production. Genom. Genet. 2012, 5, 130–148. [Google Scholar]
- Kim, Z.-H.; Park, H.; Lee, C.-G. Seasonal assessment of biomass and fatty acid productivity by Tetraselmis sp. in the ocean using semi-permeable membrane photobioreactors. J. Microbiol. Biotechnol. 2016, 26, 1098–1102. [Google Scholar] [CrossRef] [PubMed]
- Kim, Z.-H.; Park, H.; Ryu, Y.-J.; Shin, D.-W.; Hong, S.-J.; Tran, H.-L.; Lim, S.-M.; Lee, C.-G. Algal biomass and biodiesel production by utilizing the nutrients dissolved in seawater using semi-permeable membrane photobioreactors. J. Appl. Phycol. 2015, 27, 1763–1773. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, F. Effects of temperature and temperature shift on docosahexaenoic acid production by the marine microalge Crypthecodinium cohnii. J. Am. Oil Chem. Soc. 2000, 77, 613–617. [Google Scholar] [CrossRef]
- Jorquera, O.; Kiperstok, A.; Sales, E.A.; Embirucu, M.; Ghirardi, M.L. Comparative energy life-cycle analyses of microalgal biomass production in open ponds and photobioreactors. Bioresour. Technol. 2010, 101, 1406–1413. [Google Scholar] [CrossRef]
- Wang, J.; Sommerfeld, M.R.; Lu, C.; Hu, Q. Combined effect of initial biomass density and nitrogen concentration on growth and astaxanthin production of Haematococcus pluvialis (Chlorophyta) in outdoor cultivation. Algae 2013, 28, 193–202. [Google Scholar] [CrossRef]
- Zhu, L.; Hiltunen, E.; Antila, E.; Zhong, J.; Yuan, Z.; Wang, Z. Microalgal biofuels: Flexible bioenergies for sustainable development. Renew. Sustain. Energy Rev. 2014, 30, 1035–1046. [Google Scholar] [CrossRef]
- Broome, S.; Craft, C.; Struck, S.; San Clements, M. Effects of Shading from Bridges on Estuarine Wetlands; Final Report; US Department of Transportation Research and Special Programs Administration: NC, USA, 2005. Available online: http://www.ncdot.gov/doh/preconstruct/tpb/research/download/2001-12finalreport.pdf (accessed on June 2019).
- Kennish, M.J.; Haag, S.M.; Sakowicz, G.P. Seagrass demographic and spatial habitat characterization in Little Egg Harbor, New Jersey, using fixed transects. J. Coast. Res. 2008, 55, 148–170. [Google Scholar] [CrossRef]
- Seiyaboh, E.; Inyang, I.; Gijo, A. Environmental impact of Tombia bridge construction across Nun river in central Niger delta, Nigeria. Int. J. Eng. Sci. 2013, 2, 32–41. [Google Scholar]
- Livanou, E.; Lagaria, A.; Psarra, S.; Lika, K. A DEB-based approach of modeling dissolved organic matter release by phytoplankton. J. Sea Res. 2019, 143, 140–151. [Google Scholar] [CrossRef]
- Thomas, J.P. Release of dissolved organic matter from natural populations of marine phytoplankton. Mar. Biol. 1971, 11, 311–323. [Google Scholar] [CrossRef]
- Thornton, D.C. Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 2014, 49, 20–46. [Google Scholar] [CrossRef]
- Crump, B.C.; Kling, G.W.; Bahr, M.; Hobbie, J.E. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 2003, 69, 2253–2268. [Google Scholar] [CrossRef]
- Eiler, A.; Langenheder, S.; Bertilsson, S.; Tranvik, L.J. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol. 2003, 69, 3701–3709. [Google Scholar] [CrossRef]
- Park, J.; Park, B.S.; Wang, P.; Patidar, S.K.; Kim, J.H.; Kim, S.-H.; Han, M.-S. Phycospheric native bacteria Pelagibaca bermudensis and Stappia sp. ameliorate biomass productivity of Tetraselmis striata (KCTC1432BP) in co-cultivation system through mutualistic interaction. Front. Plant Sci. 2017, 8, 289. [Google Scholar] [CrossRef]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; González, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014, 12, 686. [Google Scholar] [CrossRef]
- Ederington, M.C.; McManus, G.B.; Harvey, H.R. Trophic transfer of fatty acids, sterols, and a triterpenoid alcohol between bacteria, a ciliate, and the copepod Acartia tonsa. Limnol. Oceanogr. 1995, 40, 860–867. [Google Scholar] [CrossRef]
- Wang, P.; Joo, J.-H.; Park, B.S.; Kim, J.-H.; Kim, J.H.; Han, M.-S. Relationship between dissolved organic carbon and bacterial community in the coastal waters of Incheon, Korea. Oceanol. Hydrobiol. Stud. 2017, 46, 50–61. [Google Scholar] [CrossRef]
- Guillard, R.R. Culture of Phytoplankton for Feeding Marine Invertebrates, Culture of Marine Invertebrate Animals; Springer: Berlin, Germany, 1975; pp. 29–60. [Google Scholar]
- Sournia, A. Phytoplankton, manual. In Monographs on Oceanographic Methodology; UNESCO: Paris, France, 1978. [Google Scholar]
- Hur, J.; Hwang, S.-J.; Shin, J.-K. Using synchronous fluorescence technique as a water quality monitoring tool for an urban river. Water Air Soil Pollut. 2008, 191, 231–243. [Google Scholar] [CrossRef]
- Hur, J.; Lee, B.-M.; Shin, H.-S. Microbial degradation of dissolved organic matter (DOM) and its influence on phenanthrene—DOM interactions. Chemosphere 2011, 85, 1360–1367. [Google Scholar] [CrossRef] [PubMed]
- Porter, K.; Feig, Y. The use of DAPI for identification and enumeration of bacteria and blue-green algae. Limnol. Oceanogr. 1980, 25, 943. [Google Scholar] [CrossRef]
- Park, B.S.; Wang, P.; Kim, J.H.; Kim, J.-H.; Gobler, C.J.; Han, M.-S. Resolving the intra-specific succession within Cochlodinium polykrikoides populations in southern Korean coastal waters via use of quantitative PCR assays. Harmful Algae 2014, 37, 133–141. [Google Scholar] [CrossRef]
- Lane, D. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Turner, S.; Pryer, K.M.; Miao, V.P.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef]
- Wishart, D. Note: An algorithm for hierarchical classifications. Biometrics 1969, 256, 165–170. [Google Scholar] [CrossRef]
- Ter Braak, C.J. The analysis of vegetation-environment relationships by canonical correspondence analysis. Vegetatio 1987, 69, 69–77. [Google Scholar] [CrossRef]
- Saraceni, C.; Ruggiu, D. Techniques for sampling water and phytoplankton. In A Manual on Methods for Measuring Primary Production in Aquatic Environments; Blackwell Scientific: Philadelphia, PA, USA, 1969; pp. 5–7. [Google Scholar]
- Borja, V.M. Marine Phytoplankton of the Western Pacific; Kouseisha Kouseikaku: Tokyo, Japan, 2012. [Google Scholar]
- Makoto, O.; Tsutomu, I. Methods in Marine Zooplankton Ecology; John Wiley: Hoboken, NJ, USA, 1984. [Google Scholar]
- McNichol, A.P.; Aluwihare, L.I. The power of radiocarbon in biogeochemical studies of the marine carbon cycle: Insights from studies of dissolved and particulate organic carbon (DOC and POC). Chem. Rev. 2007, 107, 443–466. [Google Scholar] [CrossRef]
- Maranón, E.; Cermeno, P.; Fernández, E.; Rodríguez, J.; Zabala, L. Significance and mechanisms of photosynthetic production of dissolved organic carbon in a coastal eutrophic ecosystem. Limnol. Oceanogr. 2004, 49, 1652–1666. [Google Scholar] [CrossRef]
- Caballero, M.A.; Jallet, D.; Shi, L.; Rithner, C.; Zhang, Y.; Peers, G. Quantification of chrysolaminarin from the model diatom Phaeodactylum tricornutum. Algal Res. 2016, 20, 180–188. [Google Scholar] [CrossRef]
- Hildebrand, M.; Manandhar-Shrestha, K.; Abbriano, R. Effects of chrysolaminarin synthase knockdown in the diatom Thalassiosira pseudonana: Implications of reduced carbohydrate storage relative to green algae. Algal Res. 2017, 23, 66–77. [Google Scholar] [CrossRef]
- Ball, S.G.; Morell, M.K. From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 2003, 54, 207–233. [Google Scholar] [CrossRef]
- Benner, R.; Pakulski, J.D.; McCarthy, M.; Hedges, J.I.; Hatcher, P.G. Bulk chemical characteristics of dissolved organic matter in the ocean. Science 1992, 255, 1561–1564. [Google Scholar] [CrossRef]
- Engel, A.; Harlay, J.; Piontek, J.; Chou, L. Contribution of combined carbohydrates to dissolved and particulate organic carbon after the spring bloom in the northern Bay of Biscay (North-Eastern Atlantic Ocean). Cont. Shelf Res. 2012, 45, 42–53. [Google Scholar] [CrossRef]
- Myklestad, S.M.; Børsheim, K.Y. Dynamics of carbohydrates in the Norwegian Sea inferred from monthly profiles collected during 3 years at 66 N, 2 E. Mar. Chem. 2007, 107, 475–485. [Google Scholar] [CrossRef]
- Sannigrahi, P.; Ingall, E.D.; Benner, R. Cycling of dissolved and particulate organic matter at station Aloha: Insights from 13C NMR spectroscopy coupled with elemental, isotopic and molecular analyses. Deep Sea Res. Part I Oceanogr. Res. Pap. 2005, 52, 1429–1444. [Google Scholar] [CrossRef]
- Ogawa, H.; Tanoue, E. Dissolved organic matter in oceanic waters. J. Oceanogr. 2003, 59, 129–147. [Google Scholar] [CrossRef]
- Lancelot, C.; Fasham, M.; Legendre, L.; Radach, G.; Scott, M.; Kirchman, D.L. Dissolved Organic Matter in Biogeochemical Models of the Ocean, Towards a Model of Ocean Biogeochemical Processes; Springer: Berlin, Germany, 1993; pp. 209–225. [Google Scholar]
- Norrman, B.; LiZweifel, U.; Hopkinson, C.; Fry, B. Production and utilization of dissolved organic carbon during an experimental diatom bloom. Oceanogr. Lit. Rev. 1996, 5, 451–452. [Google Scholar] [CrossRef]
- Ajani, P.A.; Kahlke, T.; Siboni, N.; Carney, R.; Murray, S.A.; Seymour, J.R. The microbiome of the cosmopolitan diatom Leptocylindrus reveals significant spatial and temporal variability. Front. Microbiol. 2018, 9, 2758. [Google Scholar] [CrossRef]
- Garcés, E.; Vila, M.; Reñé, A.; Alonso-Sáez, L.; Anglès, S.; Lugliè, A.; Masó, M.; Gasol, J.M. Natural bacterioplankton assemblage composition during blooms of Alexandrium spp.(Dinophyceae) in NW Mediterranean coastal waters. Aquat. Microb. Ecol. 2007, 46, 55–70. [Google Scholar]
- Green, D.H.; Llewellyn, L.E.; Negri, A.P.; Blackburn, S.I.; Bolch, C.J. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 2004, 47, 345–357. [Google Scholar] [CrossRef]
- Jasti, S.; Sieracki, M.E.; Poulton, N.J.; Giewat, M.W.; Rooney-Varga, J.N. Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 2005, 71, 3483–3494. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.L.; Mikulski, C.M.; Barnhorst, A.; Doucette, G.J. Comparative analysis of bacterioplankton assemblages from Karenia brevis bloom and nonbloom water on the west Florida shelf (Gulf of Mexico, USA) using 16S rRNA gene clone libraries. FEMS Microbiol. Ecol. 2010, 73, 468–485. [Google Scholar] [CrossRef]
- Lafay, B.; Ruimy, R.; De Traubenberg, C.R.; Breittmayer, V.; Gauthier, M.J.; Christen, R. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int. J. Syst. Evol. Microbiol. 1995, 45, 290–296. [Google Scholar] [CrossRef]
- Yang, C.; Li, Y.; Zhou, B.; Zhou, Y.; Zheng, W.; Tian, Y.; Van Nostrand, J.D.; Wu, L.; He, Z.; Zhou, J. Illumina sequencing-based analysis of free-living bacterial community dynamics during an Akashiwo sanguine bloom in Xiamen sea, China. Sci. Rep. 2015, 5, 8476. [Google Scholar] [CrossRef]
- Sañudo-Wilhelmy, S.A.; Gómez-Consarnau, L.; Suffridge, C.; Webb, E.A. The role of B vitamins in marine biogeochemistry. Annu. Rev. Mar. Sci. 2014, 6, 339–367. [Google Scholar] [CrossRef]
- Floodgate, G.; Hayes, P. The Adansonian taxonomy of some yellow pigmented marine bacteria. Microbiology 1963, 30, 237–244. [Google Scholar] [CrossRef]
- Mudarris, M.; Austin, B. Systemic disease in turbot Scophthalmus maximus caused by a previously unrecognised Cytophaga-like bacterium. Dis. Aquat. Org. 1989, 6, 161–166. [Google Scholar] [CrossRef]
- Williams, T.J.; Wilkins, D.; Long, E.; Evans, F.; DeMaere, M.Z.; Raftery, M.J.; Cavicchioli, R. The role of planktonic F lavobacteria in processing algal organic matter in coastal E ast A ntarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 2013, 15, 1302–1317. [Google Scholar] [CrossRef]
- DeLong, E.F.; Franks, D.G.; Alldredge, A.L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 1993, 38, 924–934. [Google Scholar] [CrossRef]
- Pinhassi, J.; Sala, M.M.; Havskum, H.; Peters, F.; Guadayol, O.; Malits, A.; Marrasé, C. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 2004, 70, 6753–6766. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Barbeyron, T.; Martin, R.; Portetelle, D.; Michel, G.; Vandenbol, M. The cultivable surface microbiota of the brown alga Ascophyllum nodosum is enriched in macroalgal-polysaccharide-degrading bacteria. Front. Microbiol. 2015, 6, 1487. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P. The family colwelliaceae. In The Prokaryotes: Gammaproteobacteria; Springer: Berlin, Germany, 2014; pp. 179–195. [Google Scholar]
- Michels, M.H.; Vaskoska, M.; Vermuë, M.H.; Wijffels, R.H. Growth of Tetraselmis suecica in a tubular photobioreactor on wastewater from a fish farm. Water Res. 2014, 65, 290–296. [Google Scholar] [CrossRef]
- Powell, N.; Shilton, A.; Chisti, Y.; Pratt, S. Towards a luxury uptake process via microalgae–defining the polyphosphate dynamics. Water Res. 2009, 43, 4207–4213. [Google Scholar] [CrossRef]
- Powell, N.; Shilton, A.N.; Pratt, S.; Chisti, Y. Factors influencing luxury uptake of phosphorus by microalgae in waste stabilization ponds. Environ. Sci. Technol. 2008, 42, 5958–5962. [Google Scholar] [CrossRef]
- Yao, C.; Jiang, J.; Cao, X.; Liu, Y.; Xue, S.; Zhang, Y. Phosphorus enhances photosynthetic storage starch production in a green microalga (Chlorophyta) Tetraselmis subcordiformis in nitrogen starvation conditions. J. Agric. Food Chem. 2018, 66, 10777–10787. [Google Scholar] [CrossRef]
- Legrand, C.; Rengefors, K.; Fistarol, G.O.; Graneli, E. Allelopathy in phytoplankton-biochemical, ecological and evolutionary aspects. Phycologia 2003, 42, 406–419. [Google Scholar] [CrossRef]
- Imai, I.; Ishida, Y.; Sakaguchi, K.; Hata, Y. Algicidal marine bacteria isolated from northern Hiroshima Bay, Japan. Fish. Sci. 1995, 61, 628–636. [Google Scholar] [CrossRef]
- Imai, I.; Kimura, S. Resistance of the fish-killing dinoflagellate Cochlodinium polykrikoides against algicidal bacteria isolated from the coastal sea of Japan. Harmful Algae 2008, 7, 360–367. [Google Scholar] [CrossRef]
- Kang, Y.-H.; Jung, S.W.; Joo, J.-H.; Han, M.-S. Use of immobilized algicidal bacteria to control natural freshwater diatom blooms. Hydrobiologia 2012, 683, 151–162. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, J.H.; Song, Y.H.; Chang, D.S. Isolation and characterization of the marine bacterium, Alteromonas sp. SR-14 inhibiting the growth of diatom, Chaetoceros species. Korean J. Fish. Aquat. Sci. 1999, 32, 155–159. [Google Scholar]
- Lee, B.-K.; Katano, T.; Kitamura, S.-I.; Oh, M.-J.; Han, M.-S. Monitoring of algicidal bacterium, Alteromonas sp. Strain A14 in its application to natural Cochlodinium polykrikoides blooming seawater using fluorescence in situ hybridization. J. Microbiol. 2008, 46, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Su, J.Q.; Yang, X.R.; Zheng, T.L.; Tian, Y.; Jiao, N.Z.; Cai, L.Z.; Hong, H.S. Isolation and characterization of a marine algicidal bacterium against the toxic dinoflagellate Alexandrium tamarense. Harmful Algae 2007, 6, 799–810. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-H.; Kim, J.H.; Baek, S.H.; Kim, J.-H.; Ajani, P.A.; Park, B.S.; Han, M.-S. The Influence of Dissolved Organic Carbon on the Microbial Community Associated with Tetraselmis striata for Bio-Diesel Production. Appl. Sci. 2020, 10, 3601. https://doi.org/10.3390/app10103601
Kim S-H, Kim JH, Baek SH, Kim J-H, Ajani PA, Park BS, Han M-S. The Influence of Dissolved Organic Carbon on the Microbial Community Associated with Tetraselmis striata for Bio-Diesel Production. Applied Sciences. 2020; 10(10):3601. https://doi.org/10.3390/app10103601
Chicago/Turabian StyleKim, Sae-Hee, Jin Ho Kim, Seung Ho Baek, Joo-Hwan Kim, Penelope A. Ajani, Bum Soo Park, and Myung-Soo Han. 2020. "The Influence of Dissolved Organic Carbon on the Microbial Community Associated with Tetraselmis striata for Bio-Diesel Production" Applied Sciences 10, no. 10: 3601. https://doi.org/10.3390/app10103601
APA StyleKim, S.-H., Kim, J. H., Baek, S. H., Kim, J.-H., Ajani, P. A., Park, B. S., & Han, M.-S. (2020). The Influence of Dissolved Organic Carbon on the Microbial Community Associated with Tetraselmis striata for Bio-Diesel Production. Applied Sciences, 10(10), 3601. https://doi.org/10.3390/app10103601




