3.1. Results and Predictions on Test Pictures
In order to compare the results between the different models generated, an accuracy indicator was defined as the percentage of pixels correctly classified over the total of the image:
The evaluation is made not only between the best models obtained for SVM and deep-learning, but also between several models generated with different parameters in both cases. For the SVM model, the results were obtained with different dataset sizes: 50, 122, 190 and 251 training images. SVM test accuracy results are presented in
Table 1. For every size, the mean test accuracy of all 50 models iteratively generated with different dataset distributions was calculated, although only the mean test accuracy of the best model is presented in
Table 1. Model number 4, which is the one that was trained with a larger dataset, was found to be the one with the best average accuracy (83.09%) and the best average accuracy for all the iterations of generation of the model (82.53%). Moreover, model 4 was determined as the best model, with the highest accuracy in 64.29% of the test pictures. The best model indicator was defined as the percentage of test pictures that present the best result with each model. This percentage was also reached considering 1% better test accuracy results for the model 4. Quantifying this evolution of percentages reveals small differences between model 4 and the rest in the cases it is not the best, presenting a higher accuracy or up to 1% lower in 89.29% of the test pictures. As expected, the accuracy of the model grows as the number of training examples increases.
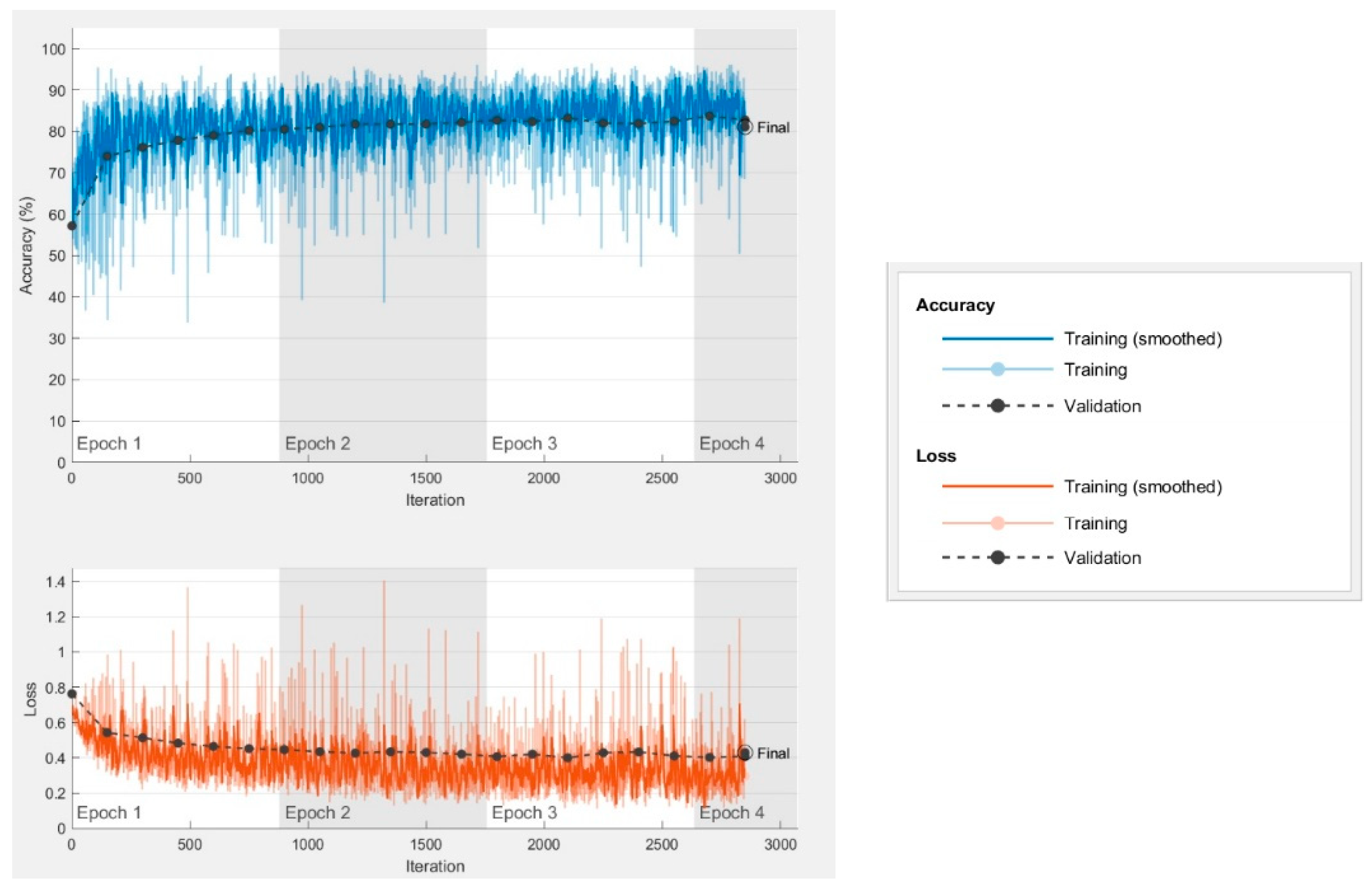
Focusing on the best SVM model obtained, whose learning curve is shown in
Figure 4, a case of high bias is observed, which may indicate an underfitting problem. The model does not fit the data sufficiently, it lacks information and requires more parameters to reduce the error. Specifically, method limitations were observed with regard to the clustering and SVM structure. Clustering for data preparation and SVM training are strictly conducted by the colour space parameters of the pixels, which seem to be insufficient for the segmentation. The problem does not apparently be related to the selection of channels or colour spaces, neither with the absence to add one of them, but with the nature of the method. Additional procedures are required for a segmentation that performs an analysis based on regions and textures.
In the case of the deep-learning model, trainings were performed with different dataset configurations, network architecture parameters and training options, which are presented in
Table A1 and
Table A2. The results for each model are shown in
Table 2 and
Table 3. Mean test accuracy of all 30 models generated iteratively with different dataset distributions was calculated, as well as the mean test accuracy of the best model. According to the results, model 13, with an accuracy of 85.05%, was found to be the best. Model 15 has a very similar accuracy (84.90%), obtained with a double value of validation patience parameter. Furthermore, the comparison of accuracy between models 11 (83.07%) and 10 (84.67%) demonstrates that the application of the image enhancement pre-process of CLAHE did not improve the results.
Valuable information is obtained from the training curves, as presented in
Figure 5 for model 13. A high variability in accuracy is observed during training due to the differences presented between training images. As previously stated, the objective was to generate a segmentation model capable of working with pictures that included complex backgrounds and regions with problematic lighting. These pictures with characteristics that are more difficult to discriminate are responsible for the fact that poor results are frequently produced during training.
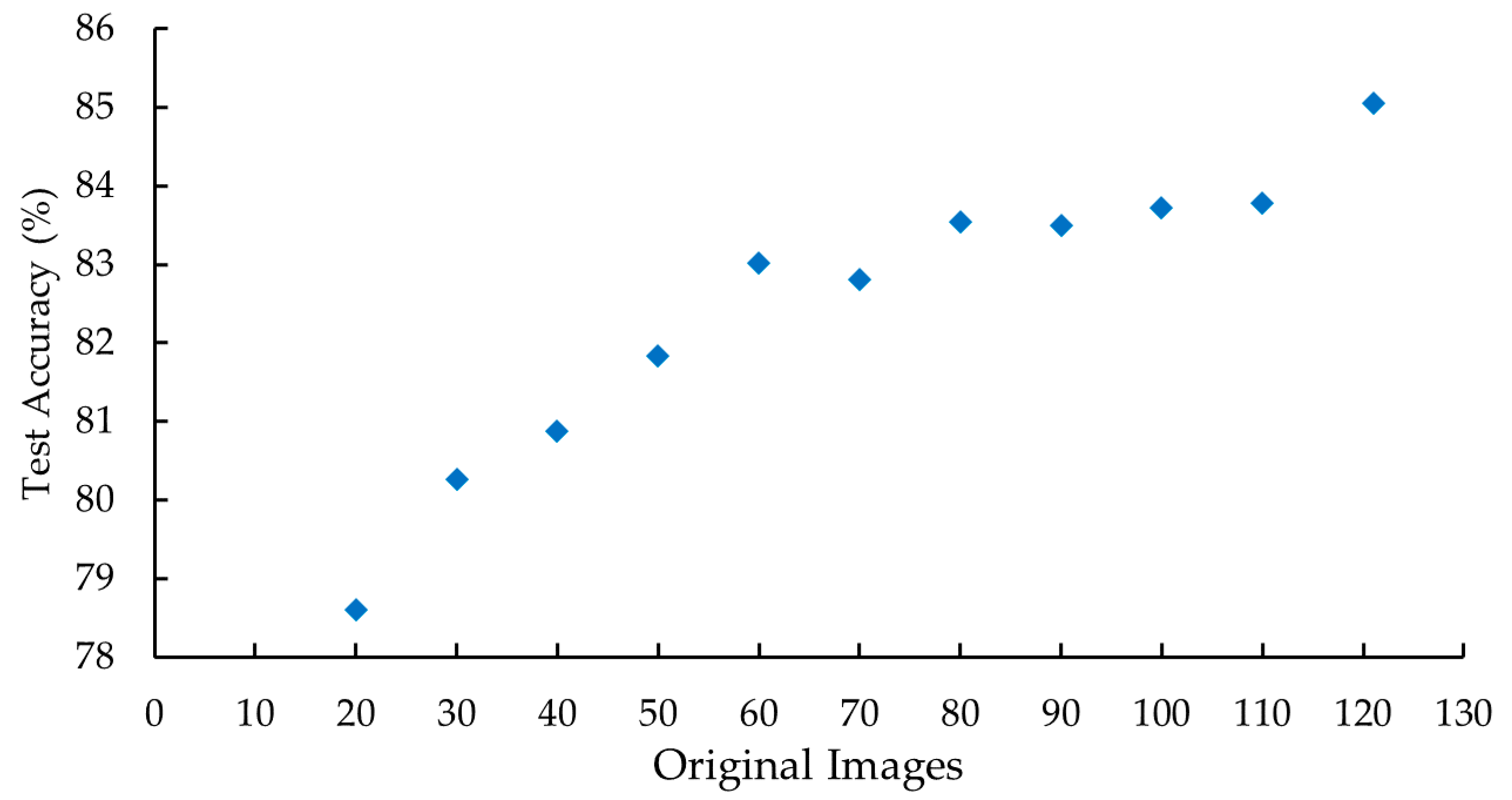
Once the best model was chosen, training was executed employing the same parameters with different dataset sizes in order to analyse the evolution of the test accuracy. The aim was to predict the possibility of model improvement in case we were to add new training pictures. The dataset size varied between 20 and 121 original images, i.e., 160 and 968 images after applying data augmentation, with 10 original images steps. The test accuracy indicator obtained from each case evidenced an enhancement as the dataset size increased, as shown in
Figure 6. This trend suggests that by increasing the size of the training set, a more accurate model could be achieved.
To perform a comparison between the best models found for SVM and deep-learning, the following indicators were defined.
where tp is the number of true positives, fp the number of false positives and fn the number of false negatives.
The deep-learning model has a better average result on accuracy, recall and F
1 score, whereas the SVM model presents a better average precision, as shown in
Table 4. This means that the deep-learning model is less restrictive, generating a lower number of false negatives, which leads to a 5.16% higher recall. However, as it is less restrictive, more false positives are also found, which lowers the precision to 2.19%. Taking into account the accuracy and F
1 score, it can be determined that the best model is the deep learning one. Moreover, the percentages of the test pictures that have the best result with each model have also been obtained for each indicator and presented in the ‘Best model’ row of
Table 4. In this case, the best result is achieved by the SVM model except for recall. However, as it can be seen in the following row, the percentages of best model vary significantly in favour of the deep-learning model if it is considered to be the best model with a higher result or up to 3% lower. In contrast, if we proceed in the same way by favouring the SVM model with the same percentage, the results are not improved so substantially. These improvements derived from the 3% favouring in each case are summarised in the last row of
Table 4. The enhancement in the percentage of test pictures that obtain their best result for each model, on average for all indicators, would be 15.18% for SVM and 42.86% for deep learning. These results serve as an argument to define the deep-learning model as a priority when it comes to improving it in future work, since a small improvement in the indicators (3%) would lead to a significant improvement in the comparative results with the SVM model, in terms of percentage of best model (42.86%).
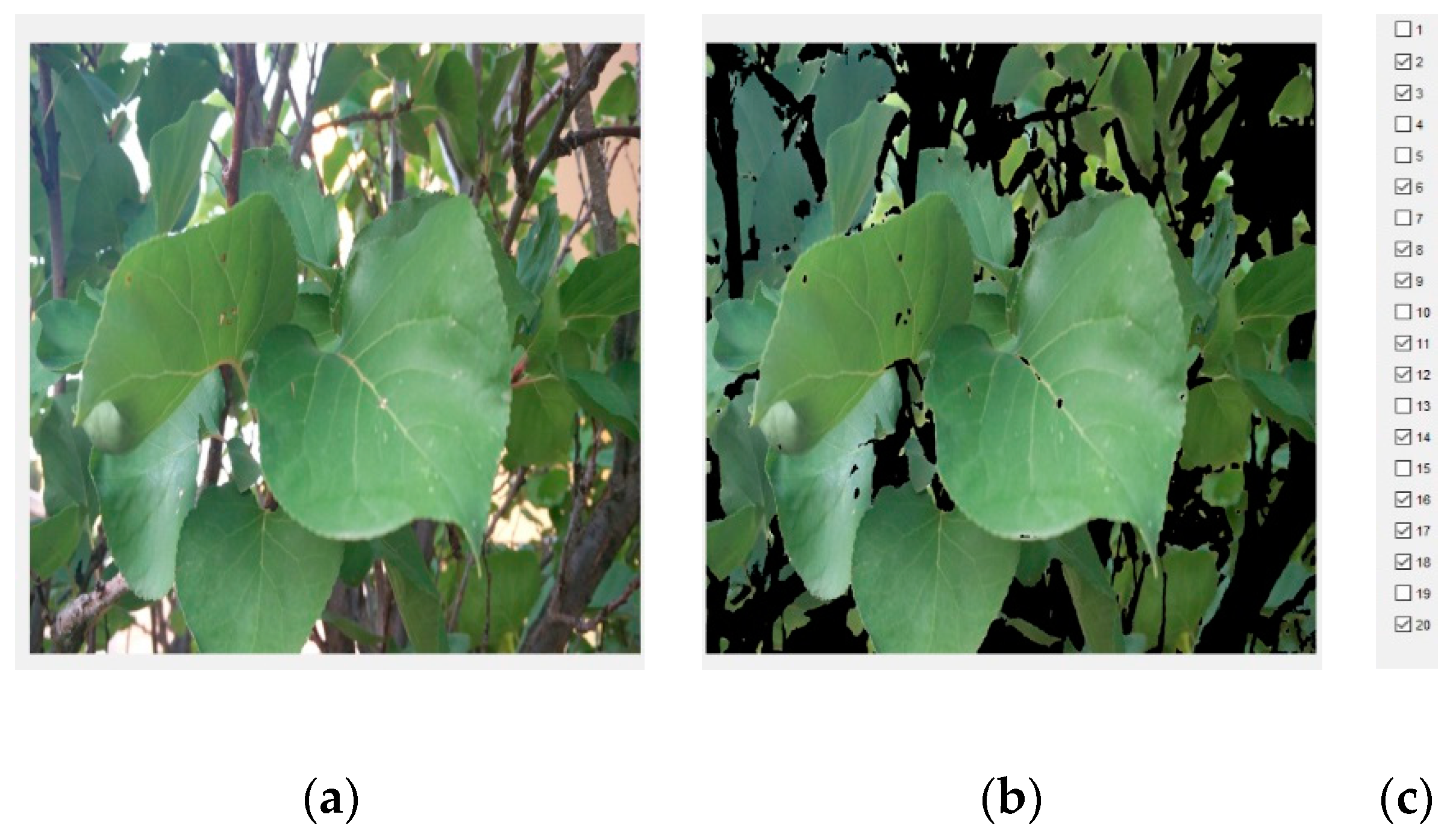
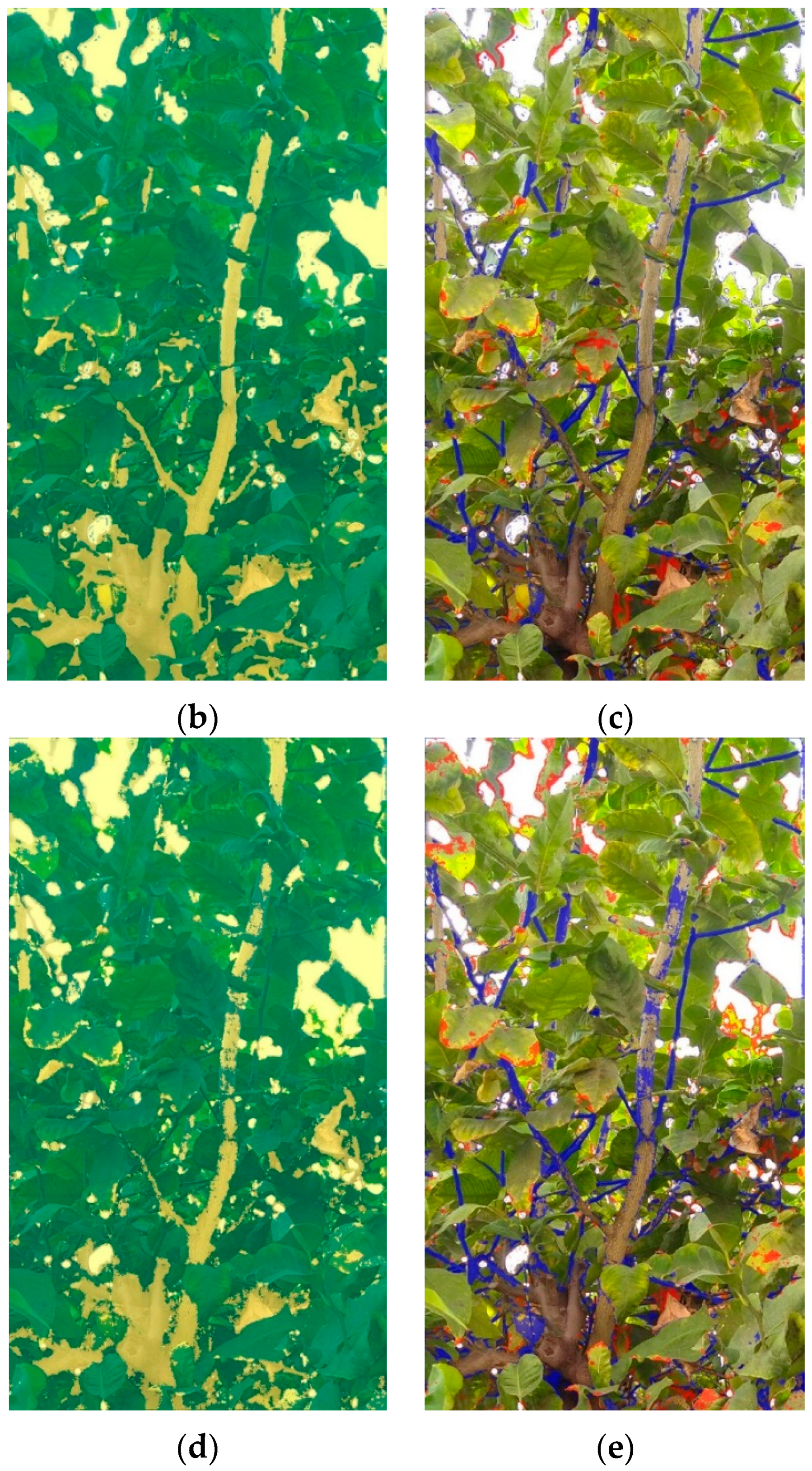
In
Figure 7, an example of the image segmentations made by the models for a test picture is shown. The segmentation mask of the model is overlaid on the original picture, assigning the green and yellow colours to “leaf” and “non-leaf” classes, respectively. The mask of the model’s errors is overlaid on the original picture, assigning blue to the false positives (they are not leaves, but have been classified as such) and red to the false negatives (they are leaves, but have not been classified as such).
3.2. Prediction, Training and Data Preparation Time
Not only performance but time cost of the models should be accounted since economic and technical restrictions have to be considered. Prediction, training and data preparation times were analysed in order to evaluate the feasibility of the methods and to underscore the differences between them.
The prediction time of the model for image segmentation is crucial to determine the feasibility of its implementation in a future field application. Additionally, it must be noted that the computing power in this case would be significantly lower if it were not performed remotely. The average times for the segmentation of the test pictures executed by SVM models are presented in
Table 5. According to results, SVM prediction time increases with the number of training images. The larger the dataset, the more complex and heavier the model is, so the prediction requires a higher computational cost. In the case of deep-learning models, prediction times are affected by the encoder depth parameter, which defines the number of network layers.
Table 5 summarizes the average prediction times for trained deep-learning models with the same encoder depth values.
From the average prediction times of the test pictures in the best SVM and deep-learning models, which are shown in
Table 6, it is appreciated that the SVM model takes approximately 42 times more prediction time. The SVM model is simpler, but requires individual prediction of each of the pixels that make up the picture. In contrast, the deep-learning model with the SegNet network architecture based on the encoder-decoder structure is more agile in prediction.
The time needed for models training is not a determining parameter to consider in the comparison between models, since it is a machine processing time and it is performed only once. However, it is interesting to take this into account as excessively high times could be a problem for future training with a greater number of pictures.
Table 7 shows how the training time of SVM models rises as the dataset increases, as expected. In the case of the deep-learning model, the training time depends on the number of training images that compose the dataset, added to the validation frequency and validation patience parameters that define the stop criteria, as well as the learning rate and the regularization value.
Based on the results of the best SVM and deep-learning models, which are indicated in
Table 8, the SVM model takes approximately 67 times less training time. The SVM model is simpler and its training does not require the computational capacity that the deep-learning model does.
The time it takes to prepare the data for training is a key factor in the process. This can be a bottleneck and the most determinant procedure, as the resulting model will be as good as the data we are training with. The data preparation times are then compared for both methods in
Table 9. In the case of SVM, the computation time of the clustering and the time of manual classification of the clusters by means of the GUI are taken into account. Instead, for the deep-learning model it is necessary to add to the time for mask generation in the previous process, the manual editing time to adjust it perfectly. It is obtained that for each picture the preparation time of the SVM model is significantly lower (27 times) than that of the deep learning one. The manual editing time of the ground-truth mask represents the biggest stumbling block in this process. If these times are considered for all the pictures used in both cases, the total time is approximately 10 hours for SVM (251 pictures) and 126 hours for deep learning (121 pictures). The deep learning model required 116 hours more time for half of the training images.