Effects of Microplastic on Human Gut Microbiome: Detection of Plastic-Degrading Genes in Human Gut Exposed to Microplastics—Preliminary Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants, Stool Sample Collection and DNA Extraction
2.2. Metagenomic Analysis
2.2.1. Library Preparation and Sequencing
2.2.2. Taxonomic and Functional Data Analysis
2.2.3. Normalized Read Counts
2.3. Statistical Analysis
3. Results
3.1. Coastal and Highland Populations Showed Similar Gut Microbiome Profiles despite Different Microplastic Contamination between the Two Populations
3.2. Microplastic Contamination in the Human Gut Showed No Correlation with the Gut Microbiome Diversity and Richness
3.3. Microplastic Contamination Did Not Correlate with Gut-Health-Related Gene Abundance
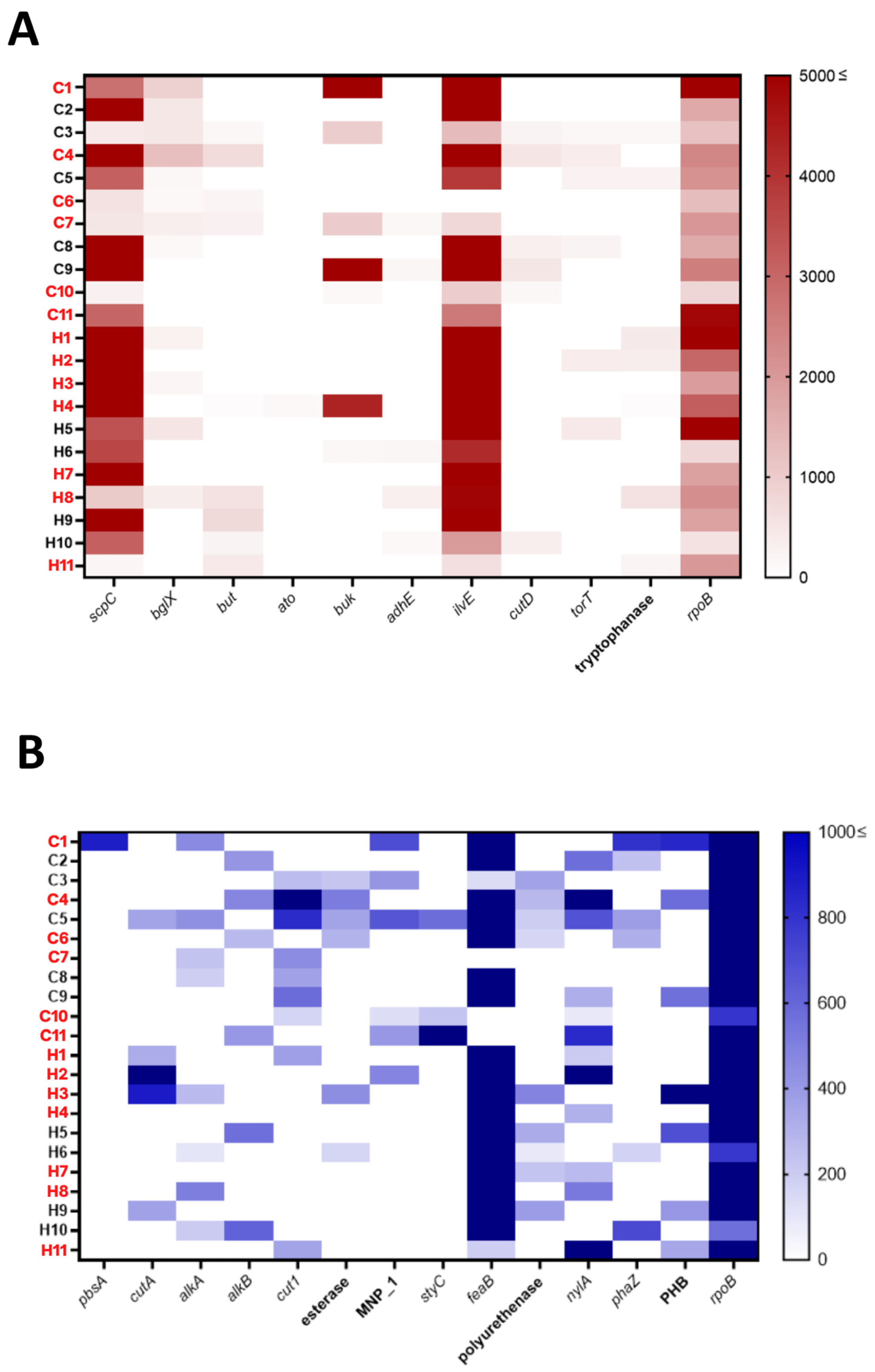
3.4. Genes Encoding Plastic-Degrading Enzymes Were Detected in the Human Gut
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jambeck, J.R.; Geyer, R.; Wilcox, C.; Siegler, T.R.; Perryman, M.; Andrady, A.; Narayan, R.; Law, K.L. Marine pollution. Plastic waste inputs from land into the ocean. Science 2015, 347, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Forum, W.E. The New Plastics Economy: Rethinking the Future of Plastics; Ellen MacArthur Foundation: Geneva, Switzerland, 2016. [Google Scholar]
- Hartmann, N.B.; Huffer, T.; Thompson, R.C.; Hassellov, M.; Verschoor, A.; Daugaard, A.E.; Rist, S.; Karlsson, T.; Brennholt, N.; Cole, M.; et al. Are We Speaking the Same Language? Recommendations for a Definition and Categorization Framework for Plastic Debris. Environ. Sci. Technol. 2019, 53, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.; Lindeque, P.; Fileman, E.; Halsband, C.; Goodhead, R.; Moger, J.; Galloway, T.S. Microplastic ingestion by zooplankton. Environ. Sci. Technol. 2013, 47, 6646–6655. [Google Scholar] [CrossRef] [PubMed]
- Seng, N.; Lai, S.; Fong, J.; Saleh, M.F.; Cheng, C.; Cheok, Z.Y.; Todd, P.A. Early evidence of microplastics on seagrass and macroalgae. Mar. Freshw. Res. 2020, 71, 922–928. [Google Scholar] [CrossRef]
- McGoran, A.R.; Clark, P.F.; Smith, B.D.; Morritt, D. High prevalence of plastic ingestion by Eriocheir sinensis and Carcinus maenas (Crustacea: Decapoda: Brachyura) in the Thames Estuary. Environ. Pollut. 2020, 265, 114972. [Google Scholar] [CrossRef]
- Plee, T.A.; Pomory, C.M. Microplastics in sandy environments in the Florida Keys and the panhandle of Florida, and the ingestion by sea cucumbers (Echinodermata: Holothuroidea) and sand dollars (Echinodermata: Echinoidea). Mar. Pollut. Bull. 2020, 158, 111437. [Google Scholar] [CrossRef]
- Neto, J.G.B.; Rodrigues, F.L.; Ortega, I.; Rodrigues, L.D.S.; Lacerda, A.; Coletto, J.L.; Kessler, F.; Cardoso, L.G.; Madureira, L.; Proietti, M.C. Ingestion of plastic debris by commercially important marine fish in southeast-south Brazil. Environ. Pollut. 2020, 267, 115508. [Google Scholar] [CrossRef]
- Romeo, T.; Pietro, B.; Peda, C.; Consoli, P.; Andaloro, F.; Fossi, M.C. First evidence of presence of plastic debris in stomach of large pelagic fish in the Mediterranean Sea. Mar. Pollut. Bull. 2015, 95, 358–361. [Google Scholar] [CrossRef]
- Goss, H.; Jaskiel, J.; Rotjan, R. Thalassia testudinum as a potential vector for incorporating microplastics into benthic marine food webs. Mar. Pollut. Bull. 2018, 135, 1085–1089. [Google Scholar] [CrossRef]
- Cordova, M.R.; Riani, E.; Shiomoto, A. Microplastics ingestion by blue panchax fish (Aplocheilus sp.) from Ciliwung Estuary, Jakarta, Indonesia. Mar. Pollut. Bull. 2020, 161, 111763. [Google Scholar] [CrossRef]
- Ismail, M.R.; Lewaru, M.W.; Prihadi, D.J. Microplastics Ingestion by Fish in The Pangandaran Bay, Indonesia. 2019. World News Nat. Sci 2019, 23. [Google Scholar]
- Buwono, N.R.; Risjani, Y.; Soegianto, A. Spatio-temporal patterns of occurrence of microplastics in the freshwater fish Gambusia affinis from the Brantas River, Indonesia. Environ. Pollut. 2022, 311, 119958. [Google Scholar] [CrossRef]
- Mason, S.A.; Welch, V.G.; Neratko, J. Synthetic Polymer Contamination in Bottled Water. Front. Chem. 2018, 6, 407. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, E.G.; Li, J.; Chen, Q.; Ma, L.; Zeng, E.Y.; Shi, H. A Review of Microplastics in Table Salt, Drinking Water, and Air: Direct Human Exposure. Environ. Sci. Technol. 2020, 54, 3740–3751. [Google Scholar] [CrossRef]
- Abbasi, S.; Turner, A. Human exposure to microplastics: A study in Iran. J. Hazard. Mater. 2021, 403, 123799. [Google Scholar] [CrossRef]
- Ragusa, A.; Svelato, A.; Santacroce, C.; Catalano, P.; Notarstefano, V.; Carnevali, O.; Papa, F.; Rongioletti, M.C.A.; Baiocco, F.; Draghi, S.; et al. Plasticenta: First evidence of microplastics in human placenta. Environ. Int. 2021, 146, 106274. [Google Scholar] [CrossRef]
- Braun, T.; Ehrlich, L.; Henrich, W.; Koeppel, S.; Lomako, I.; Schwabl, P.; Liebmann, B. Detection of Microplastic in Human Placenta and Meconium in a Clinical Setting. Pharmaceutics 2021, 13, 921. [Google Scholar] [CrossRef]
- Amato-Lourenco, L.F.; Carvalho-Oliveira, R.; Junior, G.R.; Dos Santos Galvao, L.; Ando, R.A.; Mauad, T. Presence of airborne microplastics in human lung tissue. J. Hazard. Mater. 2021, 416, 126124. [Google Scholar] [CrossRef]
- Jenner, L.C.; Rotchell, J.M.; Bennett, R.T.; Cowen, M.; Tentzeris, V.; Sadofsky, L.R. Detection of microplastics in human lung tissue using muFTIR spectroscopy. Sci. Total Environ. 2022, 831, 154907. [Google Scholar] [CrossRef]
- Luqman, A.; Nugrahapraja, H.; Wahyuono, R.A.; Islami, I.; Haekal, M.H.; Fardiansyah, Y.; Putri, B.Q.; Amalludin, F.I.; Rofiqa, E.A.; Götz, F.; et al. Microplastic Contamination in Human Stools, Foods, and Drinking Water Associated with Indonesian Coastal Population. Environments 2021, 8, 138. [Google Scholar] [CrossRef]
- Wibowo, A.T.; Nugrahapraja, H.; Wahyuono, R.A.; Islami, I.; Haekal, M.H.; Fardiansyah, Y.; Sugiyo, P.W.W.; Putro, Y.K.; Fauzia, F.N.; Santoso, H.; et al. Microplastic Contamination in the Human Gastrointestinal Tract and Daily Consumables Associated with an Indonesian Farming Community. Sustainability 2021, 13, 12840. [Google Scholar] [CrossRef]
- Cox, K.D.; Covernton, G.A.; Davies, H.L.; Dower, J.F.; Juanes, F.; Dudas, S.E. Correction to Human Consumption of Microplastics. Environ. Sci. Technol. 2020, 54, 10974. [Google Scholar] [CrossRef] [PubMed]
- Prata, J.C.; da Costa, J.P.; Lopes, I.; Duarte, A.C.; Rocha-Santos, T. Environmental exposure to microplastics: An overview on possible human health effects. Sci. Total Environ. 2020, 702, 134455. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut Microbiota and Immune System Interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Sampson, T.R.; Geschwind, D.H.; Mazmanian, S.K. The Central Nervous System and the Gut Microbiome. Cell 2016, 167, 915–932. [Google Scholar] [CrossRef]
- Ojeda, J.; Avila, A.; Vidal, P.M. Gut Microbiota Interaction with the Central Nervous System throughout Life. J. Clin. Med. 2021, 10, 1299. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Lombard, V.; Henrissat, B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE 2012, 7, e28742. [Google Scholar] [CrossRef]
- Lu, L.; Luo, T.; Zhao, Y.; Cai, C.; Fu, Z.; Jin, Y. Interaction between microplastics and microorganism as well as gut microbiota: A consideration on environmental animal and human health. Sci. Total Environ. 2019, 667, 94–100. [Google Scholar] [CrossRef]
- Qiao, R.; Deng, Y.; Zhang, S.; Wolosker, M.B.; Zhu, Q.; Ren, H.; Zhang, Y. Accumulation of different shapes of microplastics initiates intestinal injury and gut microbiota dysbiosis in the gut of zebrafish. Chemosphere 2019, 236, 124334. [Google Scholar] [CrossRef]
- Lu, L.; Wan, Z.; Luo, T.; Fu, Z.; Jin, Y. Polystyrene microplastics induce gut microbiota dysbiosis and hepatic lipid metabolism disorder in mice. Sci. Total Environ. 2018, 631–632, 449–458. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Gorska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef]
- Luqman, A.; Zabel, S.; Rahmdel, S.; Merz, B.; Gruenheit, N.; Harter, J.; Nieselt, K.; Gotz, F. The Neuromodulator-Encoding sadA Gene Is Widely Distributed in the Human Skin Microbiome. Front. Microbiol. 2020, 11, 573679. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Backhed, F.; Fraser, C.M.; Ringel, Y.; Sanders, M.E.; Sartor, R.B.; Sherman, P.M.; Versalovic, J.; Young, V.; Finlay, B.B. Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host Microbe 2012, 12, 611–622. [Google Scholar] [CrossRef]
- Djouina, M.; Vignal, C.; Dehaut, A.; Caboche, S.; Hirt, N.; Waxin, C.; Himber, C.; Beury, D.; Hot, D.; Dubuquoy, L.; et al. Oral exposure to polyethylene microplastics alters gut morphology, immune response, and microbiota composition in mice. Environ. Res. 2022, 212, 113230. [Google Scholar] [CrossRef]
- Jin, H.; Ma, T.; Sha, X.; Liu, Z.; Zhou, Y.; Meng, X.; Chen, Y.; Han, X.; Ding, J. Polystyrene microplastics induced male reproductive toxicity in mice. J. Hazard. Mater. 2021, 401, 123430. [Google Scholar] [CrossRef]
- Stock, V.; Bohmert, L.; Lisicki, E.; Block, R.; Cara-Carmona, J.; Pack, L.K.; Selb, R.; Lichtenstein, D.; Voss, L.; Henderson, C.J.; et al. Uptake and effects of orally ingested polystyrene microplastic particles in vitro and in vivo. Arch. Toxicol. 2019, 93, 1817–1833. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Q.; Wei, G.; Liang, Q.; Qi, Q. Production in Escherichia coli of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) with differing monomer compositions from unrelated carbon sources. Appl. Environ. Microbiol. 2011, 77, 4886–4893. [Google Scholar] [CrossRef] [PubMed]
- Dellomonaco, C.; Rivera, C.; Campbell, P.; Gonzalez, R. Engineered respiro-fermentative metabolism for the production of biofuels and biochemicals from fatty acid-rich feedstocks. Appl. Environ. Microbiol. 2010, 76, 5067–5078. [Google Scholar] [CrossRef] [PubMed]
- Santiago, B.; MacGilvray, M.; Faustoferri, R.C.; Quivey, R.G., Jr. The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J. Bacteriol. 2012, 194, 2010–2019. [Google Scholar] [CrossRef] [PubMed]
- Madsen, S.M.; Beck, H.C.; Ravn, P.; Vrang, A.; Hansen, A.M.; Israelsen, H. Cloning and inactivation of a branched-chain-amino-acid aminotransferase gene from Staphylococcus carnosus and characterization of the enzyme. Appl. Environ. Microbiol. 2002, 68, 4007–4014. [Google Scholar] [CrossRef] [PubMed][Green Version]
- O’Leary, N.D.; O’Connor, K.E.; Dobson, A.D. Biochemistry, genetics and physiology of microbial styrene degradation. FEMS Microbiol. Rev. 2002, 26, 403–417. [Google Scholar] [CrossRef]
- Danopoulos, E.; Twiddy, M.; West, R.; Rotchell, J.M. A rapid review and meta-regression analyses of the toxicological impacts of microplastic exposure in human cells. J. Hazard. Mater. 2022, 427, 127861. [Google Scholar] [CrossRef]
- Hirt, N.; Body-Malapel, M. Immunotoxicity and intestinal effects of nano- and microplastics: A review of the literature. Part Fibre Toxicol. 2020, 17, 57. [Google Scholar] [CrossRef]
- Espinosa, C.; Garcia Beltran, J.M.; Esteban, M.A.; Cuesta, A. In vitro effects of virgin microplastics on fish head-kidney leucocyte activities. Environ. Pollut. 2018, 235, 30–38. [Google Scholar] [CrossRef]
- Ragusa, A.; Notarstefano, V.; Svelato, A.; Belloni, A.; Gioacchini, G.; Blondeel, C.; Zucchelli, E.; De Luca, C.; D’Avino, S.; Gulotta, A.; et al. Raman Microspectroscopy Detection and Characterisation of Microplastics in Human Breastmilk. Polymers 2022, 14, 2700. [Google Scholar] [CrossRef]
- Schwabl, P.; Koppel, S.; Konigshofer, P.; Bucsics, T.; Trauner, M.; Reiberger, T.; Liebmann, B. Detection of Various Microplastics in Human Stool: A Prospective Case Series. Ann. Intern. Med. 2019, 171, 453–457. [Google Scholar] [CrossRef]
- Luo, T.; Wang, C.; Pan, Z.; Jin, C.; Fu, Z.; Jin, Y. Maternal Polystyrene Microplastic Exposure during Gestation and Lactation Altered Metabolic Homeostasis in the Dams and Their F1 and F2 Offspring. Environ. Sci. Technol. 2019, 53, 10978–10992. [Google Scholar] [CrossRef]
- Deng, Y.; Yan, Z.; Shen, R.; Wang, M.; Huang, Y.; Ren, H.; Zhang, Y.; Lemos, B. Microplastics release phthalate esters and cause aggravated adverse effects in the mouse gut. Environ. Int. 2020, 143, 105916. [Google Scholar] [CrossRef]
- Jin, Y.; Lu, L.; Tu, W.; Luo, T.; Fu, Z. Impacts of polystyrene microplastic on the gut barrier, microbiota and metabolism of mice. Sci. Total Environ. 2019, 649, 308–317. [Google Scholar] [CrossRef]
- Li, B.; Ding, Y.; Cheng, X.; Sheng, D.; Xu, Z.; Rong, Q.; Wu, Y.; Zhao, H.; Ji, X.; Zhang, Y. Polyethylene microplastics affect the distribution of gut microbiota and inflammation development in mice. Chemosphere 2020, 244, 125492. [Google Scholar] [CrossRef]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Zhang, L.; Yue, Y.; Shi, M.; Tian, M.; Ji, J.; Liao, X.; Hu, X.; Chen, F. Dietary Luffa cylindrica (L.) Roem promotes branched-chain amino acid catabolism in the circulation system via gut microbiota in diet-induced obese mice. Food Chem. 2020, 320, 126648. [Google Scholar] [CrossRef]
- Wang, F.; Wan, Y.; Yin, K.; Wei, Y.; Wang, B.; Yu, X.; Ni, Y.; Zheng, J.; Huang, T.; Song, M.; et al. Lower Circulating Branched-Chain Amino Acid Concentrations Among Vegetarians are Associated with Changes in Gut Microbial Composition and Function. Mol. Nutr. Food Res. 2019, 63, e1900612. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef]
- Luqman, A.; Nega, M.; Nguyen, M.T.; Ebner, P.; Gotz, F. SadA-Expressing Staphylococci in the Human Gut Show Increased Cell Adherence and Internalization. Cell Rep. 2018, 22, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.; Conway, S.; Aminov, R. Commensal gut bacteria: Mechanisms of immune modulation. Trends Immunol. 2005, 26, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Darch, H.; McCafferty, C.P. Gut microbiome effects on neuronal excitability & activity: Implications for epilepsy. Neurobiol. Dis. 2022, 165, 105629. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.B.; Hsiao, E.Y. Roles for the gut microbiota in regulating neuronal feeding circuits. J. Clin. Investig. 2021, 131, e143772. [Google Scholar] [CrossRef]
- Muller, P.A.; Schneeberger, M.; Matheis, F.; Wang, P.; Kerner, Z.; Ilanges, A.; Pellegrino, K.; Del Marmol, J.; Castro, T.B.R.; Furuichi, M.; et al. Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature 2020, 583, 441–446. [Google Scholar] [CrossRef]
- Andoh, A. Physiological Role of Gut Microbiota for Maintaining Human Health. Digestion 2016, 93, 176–181. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Uchida, H.; Shigeno-Akutsu, Y.; Nomura, N.; Nakahara, T.; Nakajima-Kambe, T. Cloning and sequence analysis of poly(tetramethylene succinate) depolymerase from Acidovorax delafieldii strain BS-3. J. Biosci. Bioeng. 2002, 93, 245–247. [Google Scholar] [CrossRef]
- Hanlon, S.P.; Hill, T.K.; Flavell, M.A.; Stringfellow, J.M.; Cooper, R.A. 2-phenylethylamine catabolism by Escherichia coli K-12: Gene organization and expression. Microbiology 1997, 143 Pt 2, 513–518. [Google Scholar] [CrossRef][Green Version]
- Oelschlagel, M.; Heiland, C.; Schlomann, M.; Tischler, D. Production of a recombinant membrane protein in an Escherichia coli strain for the whole cell biosynthesis of phenylacetic acids. Biotechnol. Rep. 2015, 7, 38–43. [Google Scholar] [CrossRef]


| Type of Microplastic | Significant Correlated Taxa | Spearman’s Correlation Coefficient |
|---|---|---|
| High-density Polyethylene (Hdpe) | Genus | |
| Bacteroides | −0.821 | |
| Polypropylene (Pp) | Genus | |
| Roseburia | 0.900 | |
| Clostridium | 1.000 | |
| Prevotellamassilia | 0.900 | |
| Species | ||
| Prevotella copri | −1.000 | |
| Prevotellamassilia timonensis | 0.900 | |
| Polystyrene (Ps) | Genus | |
| Roseburia | −1.000 | |
| Clostridium | −1.000 | |
| Species | ||
| Prevotella copri | 1.000 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nugrahapraja, H.; Sugiyo, P.W.W.; Putri, B.Q.; Ni’matuzahroh; Fatimah; Huang, L.; Hafza, N.; Götz, F.; Santoso, H.; Wibowo, A.T.; et al. Effects of Microplastic on Human Gut Microbiome: Detection of Plastic-Degrading Genes in Human Gut Exposed to Microplastics—Preliminary Study. Environments 2022, 9, 140. https://doi.org/10.3390/environments9110140
Nugrahapraja H, Sugiyo PWW, Putri BQ, Ni’matuzahroh, Fatimah, Huang L, Hafza N, Götz F, Santoso H, Wibowo AT, et al. Effects of Microplastic on Human Gut Microbiome: Detection of Plastic-Degrading Genes in Human Gut Exposed to Microplastics—Preliminary Study. Environments. 2022; 9(11):140. https://doi.org/10.3390/environments9110140
Chicago/Turabian StyleNugrahapraja, Husna, Pramudya Wisnu Wicaksono Sugiyo, Balqis Qonita Putri, Ni’matuzahroh, Fatimah, Li Huang, Nourhane Hafza, Friedrich Götz, Heri Santoso, Anjar Tri Wibowo, and et al. 2022. "Effects of Microplastic on Human Gut Microbiome: Detection of Plastic-Degrading Genes in Human Gut Exposed to Microplastics—Preliminary Study" Environments 9, no. 11: 140. https://doi.org/10.3390/environments9110140
APA StyleNugrahapraja, H., Sugiyo, P. W. W., Putri, B. Q., Ni’matuzahroh, Fatimah, Huang, L., Hafza, N., Götz, F., Santoso, H., Wibowo, A. T., & Luqman, A. (2022). Effects of Microplastic on Human Gut Microbiome: Detection of Plastic-Degrading Genes in Human Gut Exposed to Microplastics—Preliminary Study. Environments, 9(11), 140. https://doi.org/10.3390/environments9110140







