Characterizing the Spatial Distribution of Eragrostis Curvula (Weeping Lovegrass) in New Jersey (United States of America) Using Logistic Regression
Abstract
1. Introduction
2. Materials and Methods
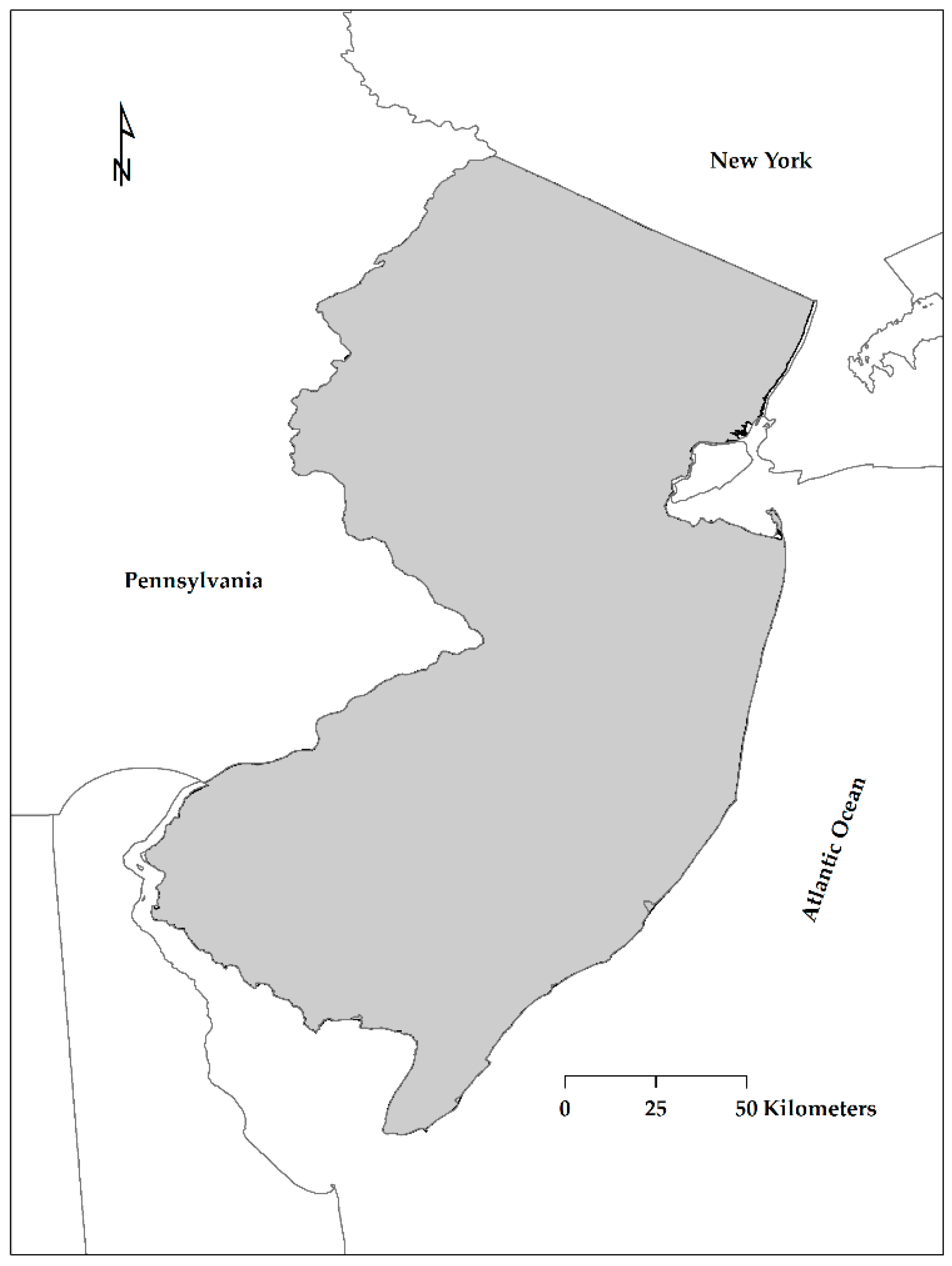
2.1. Study Area and Rationale
2.2. Data and Analysis Tools
3. Results and Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Mack, R.N.; Erneberg, M. The United States Naturalized Flora: Largely the Product of Deliberate Introductions. Ann. Mo. Bot. Gard. 2002, 89, 176–189. [Google Scholar] [CrossRef]
- Mack, R.N.; Lonsdale, W.M. Humans as Global Plant Dispersers: Getting More Than We Bargained For. BioScience 2001, 51, 95–102. [Google Scholar] [CrossRef]
- Mooney, H.A.; Cleland, E.E. The Evolutionary Impact of Invasive Species. Proc. Natl. Acad. Sci. USA 2001, 98, 5446–5451. [Google Scholar] [CrossRef] [PubMed]
- Reichard, S.H.; White, P. Horticulture as a Pathway of Invasive Plant Introductions in the United States. BioScience 2001, 51, 103–113. [Google Scholar] [CrossRef]
- Rossman, A.Y. A Special Issue on Global Movement of Invasive Plants and Fungi. BioScience 2001, 51, 93–94. [Google Scholar] [CrossRef][Green Version]
- Swearingen, J.; Bargeron, C. Invasive Plant Atlas of the United States. 2016. Available online: http://www.invasiveplantatlas.org/ (accessed on 24 April 2016).
- Lehan, N.E.; Murphy, J.R.; Thorburn, L.P.; Bradley, B.A. Accidental Introductions Are an Important Source of Invasive Plants in the Continental United States. Am. J. Bot. 2013, 100, 1287–1293. [Google Scholar] [CrossRef]
- Allen, J.M.; Leininger, T.J.; Hurd, J.D.; Civco, D.L.; Gelfand, A.E.; Silander, J.A. Socioeconomics Drive Woody Invasive Plant Richness in New England, USA through Forest Fragmentation. Landsc. Ecol. 2013, 28, 1671–1686. [Google Scholar] [CrossRef]
- Staudhammer, C.L.; Escobedo, F.J.; Holt, N.; Young, L.J.; Brandeis, T.J.; Zipperer, W. Predictors, Spatial Distribution, and Occurrence of Woody Invasive Plants in Subtropical Urban Ecosystems. J. Environ. Manag. 2015, 155, 97–105. [Google Scholar] [CrossRef]
- Richardson, D.M.; Rejmánek, M. Trees and Shrubs as Invasive Alien Species-a Global Review: Global Review of Invasive Trees & Shrubs. Divers. Distrib. 2011, 17, 788–809. [Google Scholar] [CrossRef]
- Vicente, J.R.; Fernandes, R.F.; Randin, C.F.; Broennimann, O.; Gonçalves, J.; Marcos, B.; Pôças, I.; Alves, P.; Guisan, A.; Honrado, J.P. Will Climate Change Drive Alien Invasive Plants into Areas of High Protection Value? An Improved Model-Based Regional Assessment to Prioritise the Management of Invasions. J. Environ. Manag. 2013, 131, 185–195. [Google Scholar] [CrossRef]
- New Jersey Invasive Species Council. New Jersey Strategic Management Plan for Invasive Species. Available online: https://www.nj.gov/dep/njisc/docs/Final%20NJ%20Strategic%20Management%20Plan%20for%20Invasive%20Species%2011.09.pdf (accessed on 24 April 2016).
- Brown, A. Ballast Plants in and Near New York City. Bull. Torrey Bot. Club 1880, 7, 122–126. [Google Scholar] [CrossRef]
- Martindale, I.C. The Introduction of Foreign Plants. Bot. Gaz. 1876, 2, 55–58. [Google Scholar] [CrossRef]
- Snyder, D.; Kaufman, S.R. An Overview of Nonindigenous Plant Species in New Jersey; New Jersey Department of Environmental Protection, Division of Parks and Forestry, Office of Natural Lands Management, Natural Heritage Program: Trenton, NJ, USA, 2004.
- New Jersey Invasive Species Strike Team. New Jersey Invasive Species Strike Team. Available online: http://njisst.org (accessed on 15 September 2016).
- Beck, K.G.; Zimmerman, K.; Schardt, J.D.; Stone, J.; Lukens, R.R.; Reichard, S.; Randall, J.; Cangelosi, A.A.; Cooper, D.; Thompson, J.P. Invasive Species Defined in a Policy Context: Recommendations from the Federal Invasive Species Advisory Committee. Invasive Plant Sci. Manag. 2008, 1, 414–421. [Google Scholar] [CrossRef]
- National Invasive Species Council. 2008–2012 National Invasive Species Management Plan. Available online: https://www.doi.gov/sites/doi.gov/files/migrated/invasivespecies/upload/2008-2012-National-Invasive-Species-Management-Plan.pdf (accessed on 24 April 2016).
- Richardson, D.M.; Pyšek, P.; Carlton, J.T. A Compendium of Essential Concepts and Terminology in Invasion Ecology. In Fifty Years of Invasion Ecology; Richardson, D.M., Ed.; Wiley-Blackwell: Oxford, UK, 2010; pp. 409–420. [Google Scholar] [CrossRef]
- Richardson, D.M.; Pysek, P.; Rejmanek, M.; Barbour, M.G.; Panetta, F.D.; West, C.J. Naturalization and Invasion of Alien Plants: Concepts and Definitions. Divers. Distrib. 2000, 6, 93–107. [Google Scholar] [CrossRef]
- Daehler, C.C. Performance Comparisons of Co-Occurring Native and Alien Invasive Plants: Implications for Conservation and Restoration. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 183–211. [Google Scholar] [CrossRef]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; With, K.A.; Baughman, S.; Cabin, R.J.; Cohen, J.E.; Ellstrand, N.C.; et al. The Population Biology of Invasive Species. Annu. Rev. Ecol. Syst. 2001, 32, 305–332. [Google Scholar] [CrossRef]
- Davis, M.A.; Grime, J.P.; Thompson, K. Fluctuating Resources in Plant Communities: A General Theory of Invasibility. J. Ecol. 2000, 88, 528–534. [Google Scholar] [CrossRef]
- Funk, J.L. Differences in Plasticity between Invasive and Native Plants from a Low Resource Environment. J. Ecol. 2008, 96, 1162–1173. [Google Scholar] [CrossRef]
- Funk, J.L.; Vitousek, P.M. Resource-Use Efficiency and Plant Invasion in Low-Resource Systems. Nature 2007, 446, 1079–1081. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Davidson, A. Adaptive Phenotypic Plasticity and Plant Water Use. Funct. Plant Biol. 2010, 37, 117–127. [Google Scholar] [CrossRef]
- Pyšek, P.; Richardson, D.M. Invasive Species, Environmental Change and Management, and Health. Annu. Rev. Environ. Resour. 2010, 35, 25–55. [Google Scholar] [CrossRef]
- Pyšek, P.; Richardson, D.M. Traits Associated with Invasiveness in Alien Plants: Where Do We Stand? In Biological Invasions; Nentwig, W., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 193, pp. 97–125. [Google Scholar] [CrossRef]
- Werner, C.; Zumkier, U.; Beyschlag, W.; Máguas, C. High Competitiveness of a Resource Demanding Invasive Acacia under Low Resource Supply. Plant Ecol. 2010, 206, 83–96. [Google Scholar] [CrossRef]
- Whitney, K.D.; Gabler, C.A. Rapid Evolution in Introduced Species, ‘Invasive Traits’ and Recipient Communities: Challenges for Predicting Invasive Potential: Evolution and Invasion Predictions. Divers. Distrib. 2008, 14, 569–580. [Google Scholar] [CrossRef]
- Firn, J. African lovegrass in Australia: A valuable pasture species or embarrassing invader? Tropical Grasslands 2009, 43, 86–97. [Google Scholar]
- Han, Y.; Buckley, Y.M.; Firn, J. An Invasive Grass Shows Colonization Advantages over Native Grasses under Conditions of Low Resource Availability. Plant Ecol. 2012, 213, 1117–1130. [Google Scholar] [CrossRef]
- United States Congress, Office of Technology Assessment. Harmful Non-Indigenous Species in the United States; OTA-F-565; U.S. Government Printing Office: Washington, DC, USA, 1993; p. 391. Available online: https://govinfo.library.unt.edu/ota/Ota_1/DATA/1993/9325.PDF (accessed on 15 September 2016).
- Vaz, A.S.; Alcaraz-Segura, D.; Campos, J.C.; Vicente, J.R.; Honrado, J.P. Managing Plant Invasions through the Lens of Remote Sensing: A Review of Progress and the Way Forward. Sci. Total Environ. 2018, 642, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Bazzichetto, M.; Malavasi, M.; Barták, V.; Acosta, A.T.R.; Moudrý, V.; Carranza, M.L. Modeling Plant Invasion on Mediterranean Coastal Landscapes: An Integrative Approach Using Remotely Sensed Data. Landsc. Urban Plan. 2018, 171, 98–106. [Google Scholar] [CrossRef]
- Iacona, G.; Price, F.D.; Armsworth, P.R. Predicting the Presence and Cover of Management Relevant Invasive Plant Species on Protected Areas. J. Environ. Manag. 2016, 166, 537–543. [Google Scholar] [CrossRef]
- Barbosa, F.G. The Future of Invasive African Grasses in South America under Climate Change. Ecol. Inform. 2016, 36, 114–117. [Google Scholar] [CrossRef]
- Gucker, C.L. Eragrostis Curvula. In Fire Effects Information System; USDA Forest Service, Rocky Mountain Research Station, Fire Sciences Laboratory: Missoula, MT, USA, 2009. Available online: https://www.fs.fed.us/database/feis/plants/graminoid/eracur/all.html (accessed on 28 May 2016).
- Peterson, P.M. Eragrostis. In Flora of North America North of Mexico; Magnoliophyta: Commelinidae (in Part): Poaceae, Part 2; Barkworth, M.E., Capels, K.M., Long, S., Piep, M.B., Eds.; Oxford University Press: New York, NY, USA, 2003; Volume 25, pp. 65–105. [Google Scholar]
- Skerman, P.J.; Cameron, D.G.; Riveros, F. Tropical Forage Legumes, 2nd ed.; Food and Agriculture Organization of the United Nations: Rome, Italy, 1988. [Google Scholar]
- Fairbrothers, D.E. Naturalization of Eragrostis Curvula (Weeping Lovegrass) in New Jersey. Bull. Torrey Bot. Club 1960, 87, 216. [Google Scholar] [CrossRef]
- New Jersey Department of Transportation (NJDOT). Statewide Capital Investment Strategy Fy 2013–2022. Available online: http://www.njleg.state.nj.us/OPI/Reports_to_the_Legislature/transportation_capital_investment_strategy_FY2013_2022.pdf (accessed on 15 September 2016).
- Colom, M.R.; Vazzana, C. Photosynthesis and PSII Functionality of Drought-Resistant and Drought-Sensitive Weeping Lovegrass Plants. Environ. Exp. Bot. 2003, 49, 135–144. [Google Scholar] [CrossRef]
- Mack, M.C.; D’Antonio, C.M. Impacts of Biological Invasions on Disturbance Regimes. Trends Ecol. Evol. 1998, 13, 195–198. [Google Scholar] [CrossRef]
- D’Antonio, C.M.; Vitousek, P.M. Biological Invasions by Exotic Grasses, the Grass/Fire Cycle, and Global Change. Annu. Rev. Ecol. Syst. 1992, 23, 63–87. [Google Scholar] [CrossRef]
- Skerman, P.J.; Riveros, F. Tropical Grasses; FAO Plant Production and Protection Series; Food and Agriculture Organization of the United Nations; UNIPUB: Rome, Italy; Laham, MD, USA, 1990. [Google Scholar]
- Cox, J.R.; Martin, M.H. Effects of Planting Depth and Soil Texture on the Emergence of Four Lovegrasses. J. Range Manag. 1984, 37, 204–205. [Google Scholar] [CrossRef]
- Cox, J.R.; Martin-R, M.H.; Ibarra-F, F.A.; Fourie, J.H.; Rethman, J.F.G.; Wilcox, D.G. The Influence of Climate and Soils on the Distribution of Four African Grasses. J. Range Manag. 1988, 41, 127–139. [Google Scholar] [CrossRef]
- Dahl, B.E.; Cotter, P.F.; Wester, D.B.; Britton, C.M. Grass Seeding in West Texas. Noxious Brush Weed Control Range Wildl. Manag. 1986, 17, 8–15. [Google Scholar]
- Crider, F.J. Three Introduced Lovegrasses for Soil Conservation; U.S. Dept. of Agriculture: Washington, DC, USA, 1945.
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression: Hosmer/Applied Logistic Regression; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2000. [Google Scholar] [CrossRef]
- Zaiontz, C. Real Statistics Using Excel. Available online: http://www.real-statistics.com (accessed on 10 February 2017).
- Ludlum, D.M. The New Jersey Weather Book; Rutgers University Press: New Brunswick, NJ, USA, 1983. [Google Scholar]
- Office of the New Jersey State Climatologist. Climate Information. Available online: https://climate.rutgers.edu/stateclim/?section=njcp&target=NJCoverview (accessed on 10 February 2017).
- US Census Bureau 2016. Annual Estimates of the Resident Population: 1 April 2010 to 1 July 2016. Available online: https://factfinder.census.gov/faces/tableservices/jsf/pages/productview.xhtml?src=bkmk (accessed on 10 February 2017).
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning: With Applications in R; Springer: New York, NY, USA, 2013; Available online: https://www.academia.edu/36691506/An_Introduction_to_Statistical_Learning_Springer_Texts_in_Statistics_An_Introduction_to_Statistical_Learning (accessed on 7 September 2017).
- Ekström, M.; Esseen, P.-A.; Westerlund, B.; Grafström, A.; Jonsson, B.G.; Ståhl, G. Logistic Regression for Clustered Data from Environmental Monitoring Programs. Ecol. Inform. 2018, 43, 165–173. [Google Scholar] [CrossRef]
- Goslee, S.C.; Beck, K.G.; Peters, D.P.C. Distribution of Russian Knapweed in Colorado: Climate and Environmental Factors. J. Range Manag. 2003, 56, 206. [Google Scholar] [CrossRef]
- Mansourian, S.; Darbandi, E.I.; Rashed Mohassel, M.H.; Rastgoo, M.; Kanouni, H. Comparison of Artificial Neural Networks and Logistic Regression as Potential Methods for Predicting Weed Populations on Dryland Chickpea and Winter Wheat Fields of Kurdistan Province, Iran. Crop Prot. 2017, 93, 43–51. [Google Scholar] [CrossRef]
- Pearce, J.; Ferrier, S. Evaluating the Predictive Performance of Habitat Models Developed Using Logistic Regression. Ecol. Model. 2000, 133, 225–245. [Google Scholar] [CrossRef]
- Reese, G.C.; Wilson, K.R.; Hoeting, J.A.; Flather, C.H. Factors Affecting Species Distribution Predictions: A Simulation Modeling Experiment. Ecol. Appl. 2005, 15, 554–564. [Google Scholar] [CrossRef]
- New Jersey Office of GIS Open Data. New Jersey Road Centerlines. Available online: http://njogis-newjersey.opendata.arcgis.com/datasets/new-jersey-road-centerlines (accessed on 24 April 2016).
- US Census Bureau. 2014 TIGER/Line Shapefiles (Machine-Readable Data Files)/Prepared by the U.S. Census Bureau, 2014 Economic and Statistics. 2016. Available online: https://www.census.gov/geographies/mapping-files/time-series/geo/tiger-line-file.2014.html (accessed on 24 April 2016).
- US Census Bureau 2016. Primary and Secondary Roads. Available online: https://www.census.gov/geo/maps-data/data/tiger.html (accessed on 24 April 2016).
- U.S. Department of Agriculture (USDA). National Geospatial Center of Excellence (NGCE)|NRCS. Available online: https://www.nrcs.usda.gov/wps/portal/nrcs/main/national/ngce/ (accessed on 24 April 2016).
- Soil Survey Staff. Natural Resources Conservation Service, United States Department of Agriculture. Web Soil Survey. Available online: https://websoilsurvey.nrcs.usda.gov/ (accessed on 12 April 2016).
- PRISM Climate Group, Oregon State U. Available online: http://prism.oregonstate.edu/ (accessed on 7 September 2017).
- U.S. Geological Survey, The National Map, 3DEP Products and Services: The National Map, 3D Elevation Program Web Page. Available online: https://www.usgs.gov/core-science-systems/ngp/3dep (accessed on 12 April 2016).
- Wasserstein, R.L.; Lazar, N.A. The ASA Statement on p -Values: Context, Process, and Purpose. Am. Stat. 2016, 70, 129–133. [Google Scholar] [CrossRef]
- Physiographic Provinces of New Jersey. Available online: https://rucore.libraries.rutgers.edu/rutgers-lib/18149/ (accessed on 28 September 2019).
- Coastal Plain Province (U.S. National Park Service). Available online: https://www.nps.gov/articles/coastalplain.htm (accessed on 28 September 2019).
- Clarke, S.; French, K. Germination Response to Heat and Smoke of 22 Poaceae Species from Grassy Woodlands. Aust. J. Bot. 2005, 53, 445. [Google Scholar] [CrossRef]
- U.S. Department of Agriculture (USDA). Field Guide for Managing Lehmann and Weeping Lovegrasses in the Southwest. Available online: https://www.fs.usda.gov/Internet/FSE_DOCUMENTS/stelprdb5410118.pdf (accessed on 24 April 2016).
- Taylor, C.M.; Hastings, A. Finding Optimal Control Strategies for Invasive Species: A Density-Structured Model for Spartina Alterniflora: Optimal Control Strategies for an Invasive Grass. J. Appl. Ecol. 2004, 41, 1049–1057. [Google Scholar] [CrossRef]
- Giljohann, K.M.; Hauser, C.E.; Williams, N.S.G.; Moore, J.L. Optimizing Invasive Species Control across Space: Willow Invasion Management in the Australian Alps: Spatial Allocation of Invasive Species Control. J. Appl. Ecol. 2011, 48, 1286–1294. [Google Scholar] [CrossRef]
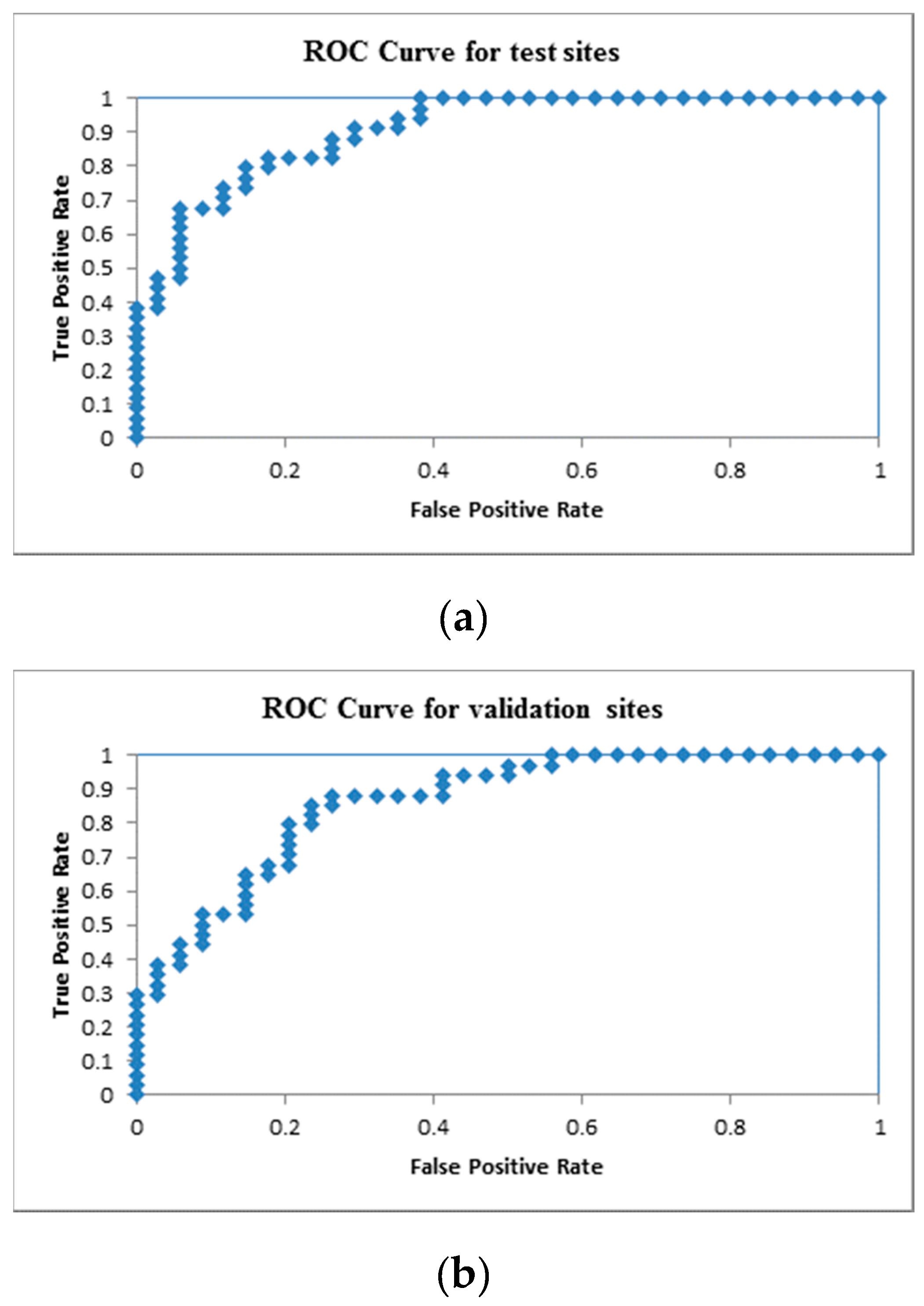
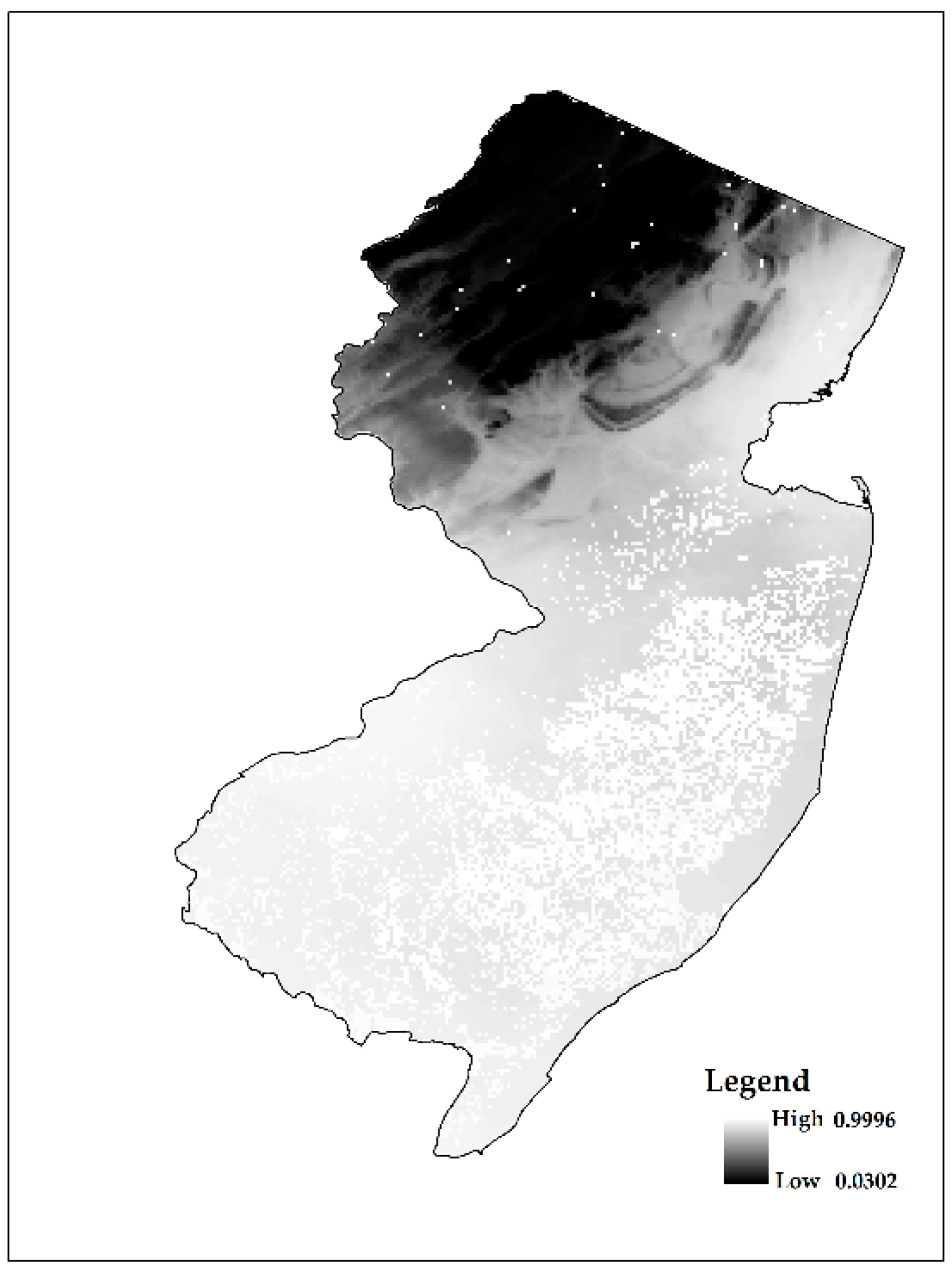
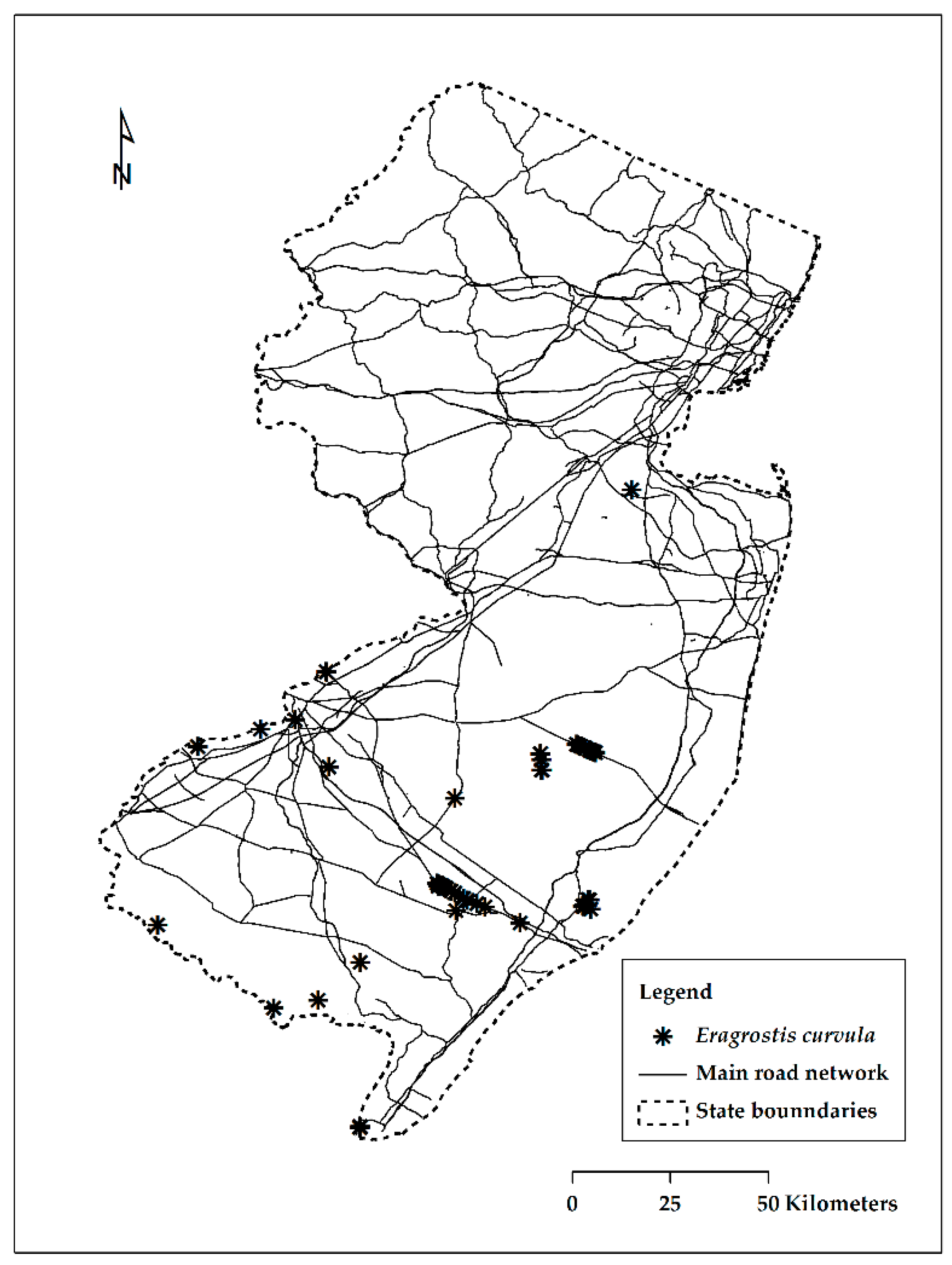
| Number of Cases | Suc-Obs | Fail-Obs | Total Predicted |
|---|---|---|---|
| Suc-Pred | TP | FP | PP |
| Fail-Pred | FN | TN | PN |
| Total | OP | ON | TO |
| Suc-Obs Presence E. Curvula | Fail-Obs Absence E. Curvula | Total | |
|---|---|---|---|
| Suc-Pred | 28 | 7 | 35 |
| Fail-Pred | 6 | 27 | 33 |
| Total | 34 | 34 | 68 |
| Accuracy | 0.823529 | 0.794118 | 0.808824 |
| Suc-Obs Presence E. Curvula | Fail-Obs Absence E. Curvula | Total | |
|---|---|---|---|
| Suc-Pred | 27 | 8 | 35 |
| Fail-Pred | 7 | 26 | 33 |
| Total | 34 | 34 | 68 |
| Accuracy | 0.794118 | 0.764706 | 0.779412 |
| Datasets Training Set Model | Validation Set Model | |
|---|---|---|
| LL0 | −47.134 | −47.134 |
| LL1 | −24.8012 | −31.7043 |
| Chi Square | 44.66558 | 30.85932 |
| df | 4 | 4 |
| p-value | 4.67 × 10−9 | 3.27 × 10−6 |
| alpha | 0.05 | 0.05 |
| significance | yes | yes |
| Datasets Training Set Model | Validation Set Model | |
|---|---|---|
| Hosmer-Lemeshow | 46.37123 | 59.09845 |
| df | 66 | 66 |
| p-value | 0.968234 | 0.713769 |
| alpha | 0.05 | 0.05 |
| significance | no | no |
| Coeff b | s.e. | Wald | p-value | exp(b) | Lower | Upper | |
|---|---|---|---|---|---|---|---|
| Intercept | −22.3593 | 23.46228 | 0.908192 | 0.340595 | 1.95 × 10−10 | ||
| Mean annual temperature | 3.415998 | 1.238602 | 7.60627 | 0.005817 | 30.44733 | 2.686886 | 345.0239 |
| Soil classes | 2.015138 | 0.64378 | 9.797938 | 0.001747 | 7.501764 | 2.124109 | 26.49415 |
| Precipitation | −0.02105 | 0.01294 | 2.645039 | 0.033874 | 0.979175 | 0.954653 | 1.004326 |
| Distance away from road | −6.2 × 10−5 | 3.12 × 10−5 | 3.880523 | 0.048849 | 0.999938 | 0.999877 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngoy, K.I.; Shebitz, D. Characterizing the Spatial Distribution of Eragrostis Curvula (Weeping Lovegrass) in New Jersey (United States of America) Using Logistic Regression. Environments 2019, 6, 125. https://doi.org/10.3390/environments6120125
Ngoy KI, Shebitz D. Characterizing the Spatial Distribution of Eragrostis Curvula (Weeping Lovegrass) in New Jersey (United States of America) Using Logistic Regression. Environments. 2019; 6(12):125. https://doi.org/10.3390/environments6120125
Chicago/Turabian StyleNgoy, Kikombo Ilunga, and Daniela Shebitz. 2019. "Characterizing the Spatial Distribution of Eragrostis Curvula (Weeping Lovegrass) in New Jersey (United States of America) Using Logistic Regression" Environments 6, no. 12: 125. https://doi.org/10.3390/environments6120125
APA StyleNgoy, K. I., & Shebitz, D. (2019). Characterizing the Spatial Distribution of Eragrostis Curvula (Weeping Lovegrass) in New Jersey (United States of America) Using Logistic Regression. Environments, 6(12), 125. https://doi.org/10.3390/environments6120125




