Allometric Equations for Aboveground Biomass Estimation in Wet Miombo Forests of the Democratic Republic of the Congo Using Terrestrial LiDAR
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Data Collection
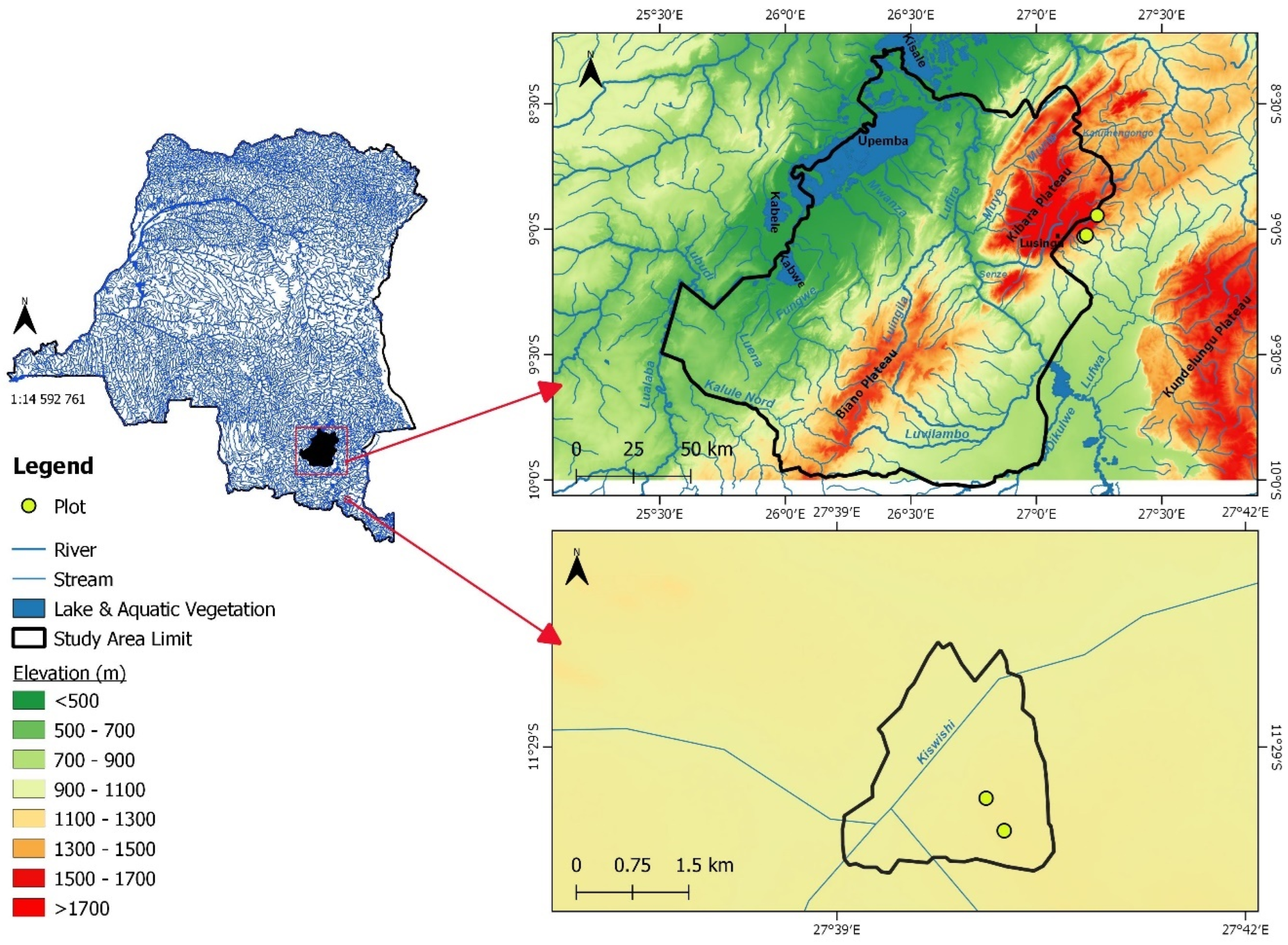
2.1.1. Study Sites
2.1.2. Data Collection
Forest Inventory Data
Terrestrial Laser Scanning Data
Wood Density Data
2.2. Methods
2.2.1. Terrestrial Laser Scanning Data Processing
- -
- An R code developed by Martin-ducup [27] was modified and used to automatically extract individual trees. For each tree, the code builds a cylindrical bounding box centered on that tree (using tree relative coordinates from forest inventory data) and sized so as to englobe the entire tree (using tree DBH from forest inventory data as well as height and crown diameter allometry models, [46]. The bounding box was then used to subset the stand points cloud and export points within the bounding box as a separate CSV file. The file was then imported into the 3Dforest software (Version 052) [16] for automatic ground removal, and into the CloudCompare software (Version V2.12) (Figure 2(C1–C3)) for a careful supervised refinement of the result i.e., the manual removal of points that do not belong to the focal tree.
- -
- -

2.2.2. Computation of Reference Aboveground Biomass Estimates
2.2.3. Statistical Analyses
Flora Analysis
AGB Model Calibration
Model Cross-Validation
Comparison of Local and Literature-Based Model Predictions
3. Results
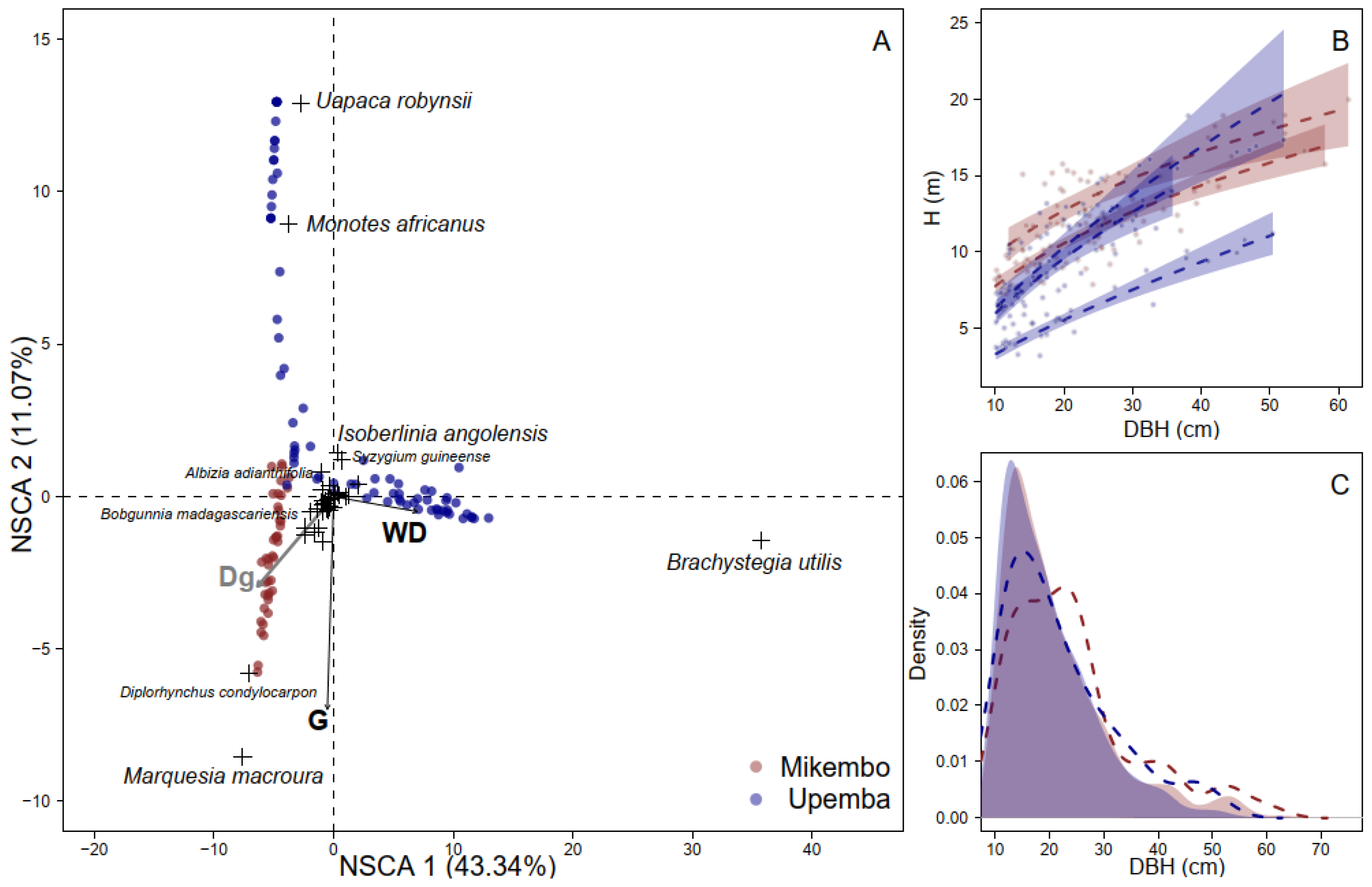
3.1. Floristic Structure
3.2. Calibrating and Validating Allometry Models
3.3. Comparing Local Allometry Equations with State of Art
4. Discussion
4.1. Floristic Communities
4.2. Allometric Models
4.3. Comparison with Models from State of Art
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodman, R.; Herold, M. Why Maintaining Tropical Forests is Essential and Urgent for a Stable Climate; Center for Global Development Working Paper, no 385; Center for Global Development: Washington, DC, USA, 2014. [Google Scholar] [CrossRef]
- Chave, J.; Davies, S.J.; Phillips, O.L.; Lewis, S.L.; Sist, P.; Schepaschenko, D.; Armston, J.; Baker, T.R.; Coomes, D.; Disney, M.; et al. Ground Data are Essential for Biomass Remote Sensing Missions. Surv. Geophys. 2019, 40, 863–880. [Google Scholar] [CrossRef]
- Calders, K.; Verbeeck, H.; Burt, A.; Origo, N.; Nightingale, J.; Malhi, Y.; Wilkes, P.; Raumonen, P.; Bunce, R.G.H.; Disney, M. Laser scanning reveals potential underestimation of biomass carbon in temperate forest. Ecol. Solut. Evid. 2022, 3, e12197. [Google Scholar] [CrossRef]
- IPCC SESSION OF THE IPCC 8–12 May 2019, Kyoto, Japan Decisions adopted by the Panel Decision IPCC-XLIX-1. Adoption of the Provisional Agenda. 2019, 1–17. Available online: https://www.ipcc.ch/site/assets/uploads/2019/05/IPCC-49_decisions_adopted.pdf (accessed on 28 June 2025).
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Dassot, M.; Constant, T.; Fournier, M. The use of terrestrial LiDAR technology in forest science: Application fields, benefits and challenges. Ann. For. Sci. 2011, 68, 959–974. [Google Scholar] [CrossRef]
- Dassot, M.; Colin, A.; Santenoise, P.; Fournier, M.; Constant, T. Terrestrial laser scanning for measuring the solid wood volume, including branches, of adult standing trees in the forest environment. Comput. Electron. Agric. 2012, 89, 86–93. [Google Scholar] [CrossRef]
- Demol, M.; Verbeeck, H.; Gielen, B.; Armston, J.; Burt, A.; Disney, M.; Duncanson, L.; Hackenberg, J.; Kükenbrink, D.; Lau, A.; et al. Estimating forest above-ground biomass with terrestrial laser scanning: Current status and future directions. Methods Ecol. Evol. 2022, 13, 1628–1639. [Google Scholar] [CrossRef]
- Demol, M.; Calders, K.; Verbeeck, H.; Gielen, B. Forest above-ground volume assessments with terrestrial laser scanning: A ground-truth validation experiment in temperate, managed forests. Ann. Bot. 2021, 128, 805–819. [Google Scholar] [CrossRef]
- Disney, M.; Burt, A.; Wilkes, P.; Armston, J.; Duncanson, L. New 3D measurements of large redwood trees for biomass and structure. Sci. Rep. 2020, 10, 16721. [Google Scholar] [CrossRef]
- Malhi, Y.; Jackson, T.; Patrick Bentley, L.; Lau, A.; Shenkin, A.; Herold, M.; Calders, K.; Bartholomeus, H.; Disney, M.I. New perspectives on the ecology of tree structure and tree communities through terrestrial laser scanning. Interface Focus. 2018, 8, 20170052. [Google Scholar] [CrossRef]
- Åkerblom, M.; Kaitaniemi, P. Terrestrial laser scanning: A new standard of forest measuring and modelling? Ann. Bot. 2021, 128, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Markku, Å.; Raumonen, P.; Kaasalainen, M.; Casella, E.; Markku, Å.; Raumonen, P.; Kaasalainen, M.; Casella, E. Analysis of Geometric Primitives in Quantitative Structure Models of Tree Stems. Remote Sens. 2015, 7, 4581–4603. [Google Scholar] [CrossRef]
- Hackenberg, J.; Spiecker, H.; Calders, K.; Disney, M.; Raumonen, P. SimpleTree—An efficient open-source tool to build tree models from TLS clouds. Forests 2015, 6, 4245–4294. [Google Scholar] [CrossRef]
- Raumonen, P.; Kaasalainen, M.; Akerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast automatic precision tree models from terrestrial laser scanner data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef]
- Trochta, J.; Kruček, M.; Vrška, T.; Kraâl, K. 3D Forest: An application for descriptions of three-dimensional forest structures using terrestrial LiDAR. PLoS ONE 2017, 12, e0176871. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Momo Takoudjou, S.; Casella, E. LeWoS: A universal leaf-wood classification method to facilitate the 3D modelling of large tropical trees using terrestrial LiDAR. Methods Ecol. Evol. 2020, 11, 376–389. [Google Scholar] [CrossRef]
- Wilkes, P.; Disney, M.; Armston, J.; Bentley, L.; Brede, B.; Burt, A.; Calders, K.; Chavana-Bryant, C.; Clewley, D.; Duncanson, L.; et al. TLS2trees: A scalable tree segmentation pipeline for TLS data. bioRxiv 2022, 24. [Google Scholar] [CrossRef]
- Bogdanovich, E.; Perez-Priego, O.; El-Madany, T.S.; Guderle, M.; Pacheco-Labrador, J.; Levick, S.R.; Moreno, G.; Carrara, A.; Martín, M.P.; Migliavacca, M. Using terrestrial laser scanning for characterizing tree structural parameters and their changes under different management in a Mediterranean open woodland. For. Ecol. Manag. 2021, 486, 118945. [Google Scholar] [CrossRef]
- Burt, A.; Disney, M.; Calders, K. Extracting individual trees from lidar point clouds using treeseg. Methods Ecol. Evol. 2019, 10, 438–445. [Google Scholar] [CrossRef]
- Hackenberg, J.; Wassenberg, M.; Spiecker, H.; Sun, D. Non-Destructive Method for Biomass Prediction Combining TLS Derived Tree Volume and Wood Density. Forests 2015, 6, 1274–1300. [Google Scholar] [CrossRef]
- Momo Takoudjou, S.; Ploton, P.; Sonké, B.; Hackenberg, J.; Griffon, S.; De Coligny, F.; Kamdem, N.G.; Libalah, M.; Mofack, G.I.; Le Moguédec, G.; et al. Using terrestrial laser scanning data to estimate large tropical trees biomass and calibrate allometric models: A comparison with traditional destructive approach. Methods Ecol. Evol. 2018, 9, 905–916. [Google Scholar] [CrossRef]
- Raumonen, P.; Casella, E.; Calders, K.; Murphy, S.; Åkerblom, M.; Kaasalainen, M. Massive-scale tree modelling from TLS data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, 2, 189–196. [Google Scholar] [CrossRef]
- Woodgate, W.; Jones, S.D.; Suarez, L.; Hill, M.J.; Armston, J.D.; Wilkes, P.; Soto-Berelov, M.; Haywood, A.; Mellor, A. Understanding the variability in ground-based methods for retrieving canopy openness, gap fraction, and leaf area index in diverse forest systems. Agric. For. Meteorol. 2015, 205, 83–95. [Google Scholar] [CrossRef]
- Abd Rahman, M.Z.; Abu Bakar, M.A.; Razak, K.A.; Rasib, A.W.; Kanniah, K.D.; Wan Kadir, W.H.; Omar, H.; Faidi, A.; Kassim, A.R.; Abd Latif, Z. Non-destructive, laser-based individual tree aboveground biomass estimation in a tropical rainforest. Forests 2017, 8, 86. [Google Scholar] [CrossRef]
- Phalla Thuch Ota Tetsuji Mizoue Nobuya et, a.l. The importance of tree height in estimating individual tree biomass while considering errors in measurements and allometric models. AGRIVITA J. Agric. Sci. 2017, 40, 131–140. [Google Scholar]
- Martin-Ducup, O.; Mofack, G.; Wang, D.; Raumonen, P.; Ploton, P.; Sonké, B.; Barbier, N.; Couteron, P.; Pélissier, R. Evaluation of automated pipelines for tree and plot metric estimation from TLS data in tropical forest areas. Ann. Bot. 2021, 128, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Trochta, J.; Král, K.; Janík, D.; Adam, D. Arrangement of terrestrial laser scanner positions for area-wide stem mapping of natural forests. Can. J. For. Res. 2013, 43, 355–363. [Google Scholar] [CrossRef]
- Timberlake, J.; Chidumayo, E. Division report. IEEE Control Syst. Mag. 2004, 3, 25. [Google Scholar] [CrossRef]
- Ribeiro, N.S.; Silva de Miranda, P.L.; Timberlake, J. Biogeography and ecology of miombo woodlands. In Miombo Woodlands in a Changing Environment: Securing the Resilience and Sustainability of People and Woodlands; Springer International Publishing: Cham, Switzerland, 2020; pp. 9–53. [Google Scholar]
- Tarimo, B.; Dick, Ø.B.; Gobakken, T.; Totland, Ø. Spatial distribution of temporal dynamics in anthropogenic fires in miombo savanna woodlands of Tanzania. Carbon Balance Manag. 2015, 10, 18. [Google Scholar] [CrossRef]
- Campbell, B. The Miombo in Transition: Woodlands and Welfare in Africa; CIFOR, Center for International Forestry Research (CIFOR): Bogor, Indonesia, 1996; 273p. [Google Scholar]
- Ilunga Muledi, J.; Bauman, D.; Drouet, T.; Vleminckx, J.; Jacobs, A.; Lejoly, J.; Meerts, P.; Shutcha, M.N. Fine-scale habitats influence tree species assemblage in a miombo forest. J. Plant Ecol. 2016, 10, rtw104. [Google Scholar] [CrossRef]
- Bauman, D.; Raspe, O.; Meerts, P.; Degreef, J.; Ilunga Muledi, J.; Drouet, T. Multiscale assemblage of an ectomycorrhizal fungal community: The influence of host functional traits and soil properties in a 10-ha miombo forest. FEMS Microbiol. Ecol. 2016, 92, 151. [Google Scholar] [CrossRef]
- Chirwa, P.; Syampungani, S.; Geldenhuys, C.J. The ecology and management of the Miombo woodlands for sustainable livelihoods in southern Africa: The case for non-timber forest products. South. For. J. For. Sci. 2008, 70, 237–245. [Google Scholar] [CrossRef]
- Des, E.; Unis, E.; Washington, S.N.W. Parcs et Réserves de la République Démocratique du Congo Parcs et réserves de la République Démocratique du Congo. 2015; 1–149. Available online: https://papaco.org/wp-content/uploads/2015/09/RAPPAM-RDC-impression-110629-A4.pdf (accessed on 28 June 2025).
- Batumike, M.J.; Kampunzu, A.B.; Cailteux, J.H. Petrology and geochemistry of the Neoproterozoic Nguba and Kundelungu Groups, Katangan Supergroup, southeast Congo: Implications for provenance, paleoweathering and geotectonic setting. J. Afr. Earth Sci. 2006, 44, 97–115. [Google Scholar] [CrossRef]
- KayaMuyumba D._2015. Available online: https://doaj.org/article/f6853ed0ff8a4912a100ec46f1aa2ce6 (accessed on 28 June 2025).
- Phillips, O.; Baker, T.; Feldpausch, T.; Brienen, R. RAINFOR Field Manual for Plot Establishment and Remeasurement. 2009. Available online: https://forestplots.net/upload/manualsenglish/rainfor_field_manual_en.pdf (accessed on 28 June 2025).
- SEOSAW partnership. A network to understand the changing socio-ecology of the southern African woodlands (SEOSAW): Challenges, benefits, and methods. Plants People Planet 2021, 3, 249–267. [Google Scholar] [CrossRef]
- Wang, Z. Real-Time Updated free Station as a Georeferencing Method in Terrestrial Laser Scanning. 2011. Available online: http://www.diva-portal.org/smash/get/diva2:423230/FULLTEXT01.pdf (accessed on 28 June 2025).
- Fayolle, A.; Ngomanda, A.; Mbasi, M.; Barbier, N.; Bocko, Y.; Boyemba, F.; Couteron, P.; Fonton, N.; Kamdem, N.; Katembo, J.; et al. A regional allometry for the Congo basin forests based on the largest ever destructive sampling. For. Ecol. Manag. 2018, 430, 228–240. [Google Scholar] [CrossRef]
- Henry, M.; Besnard, A.; Asante, W.A.; Eshun, J.; Adu-Bredu, S.; Valentini, R.; Bernoux, M.; Saint-André, L. Wood density, phytomass variations within and among trees, and allometric equations in a tropical rainforest of Africa. For. Ecol. Manag. 2010, 260, 1375–1388. [Google Scholar] [CrossRef]
- Vieilledent, G.; Vaudry, R.; Andriamanohisoa, S.F.; Rakotonarivo, O.S.; Randrianasolo, H.Z.; Razafindrabe, H.N.; Rakotoarivony, C.B.; Ebeling, J.; Rasamoelina, M. A universal approach to estimate biomass and carbon stock in tropical forests using generic allometric models. Ecol. Appl. 2012, 22, 572–583. [Google Scholar] [CrossRef]
- Muledi, J.; Bauman, D.; Jacobs, A.; Meerts, P.; Shutcha, M.; Drouet, T. Tree growth, recruitment, and survival in a tropical dry woodland: The importance of soil and functional identity of the neighbourhood. For. Ecol. Manag. 2020, 460, 117894. [Google Scholar] [CrossRef]
- Martinez Cano, I.; Muller-Landau, H.C.; Wright, S.J.; Bohlman, S.A.; Pacala, S.W. Interspecific variation in tropical tree height and crown allometries in relation to life history traits. Biogeosci. Discuss. 2018, 16, 847–862. [Google Scholar] [CrossRef]
- Momo, S.T.; Ploton, P.; Martin-Ducup, O.; Lehnebach, R.; Fortunel, C.; Sagang, L.B.T.; Boyemba, F.; Couteron, P.; Fayolle, A.; Libalah, M.; et al. Leveraging Signatures of Plant Functional Strategies in Wood Density Profiles of African Trees to Correct Mass Estimations from Terrestrial Laser Data. Sci. Rep. 2020, 10, 2001. [Google Scholar] [CrossRef]
- Lohbeck, M.; Lebrija-Trejos, E.; Martínez-Ramos, M.; Meave, J.A.; Poorter, L.; Bongers, F. Functional Trait Strategies of Trees in Dry and Wet Tropical Forests Are Similar but Differ in Their Consequences for Succession. PLoS ONE 2015, 10, e0123741. [Google Scholar] [CrossRef]
- Poorter, L.; Rozendaal, D.M.A.; Bongers, F.; de Almeida-Cortez, J.S.; Almeyda Zambrano, A.M.; Álvarez, F.S.; Andrade, J.L.; Villa, L.F.A.; Balvanera, P.; Becknell, J.M.; et al. Wet and dry tropical forests show opposite successional pathways in wood density but converge over time. Nat. Ecol. Evol. 2019, 3, 928–934. [Google Scholar] [CrossRef]
- Lombardo, R.; Beh, E.J.; Lombardo, M.R. Package ‘CAvariants’. Medicine 2017, 57, 947–961. [Google Scholar]
- Lombardo, R.; Beh, E.J.; D’Ambra, L. Non-symmetric correspondence analysis with ordinal variables using orthogonal polynomials. Comput. Stat. Data Anal. 2007, 52, 566–577. [Google Scholar] [CrossRef]
- Molto, Q.; Hérault, B.; Boreux, J.J.; Daullet, M.; Rousteau, A.; Rossi, V. Predicting tree heights for biomass estimates in tropical forests -A test from French Guiana. Biogeosciences 2014, 11, 315021–315030. [Google Scholar] [CrossRef]
- Loubota Panzou, G.J.; Fayolle, A.; Jucker, T.; Phillips, O.L.; Bohlman, S.; Banin, L.F.; Lewis, S.L.; Affum-Baffoe, K.; Alves, L.F.; Antin, C.; et al. Pantropical variability in tree crown allometry. Glob. Ecol. Biogeogr. 2020, 30, 459–475. [Google Scholar] [CrossRef]
- Feldpausch, T.R.; Banin, L.; Phillips, O.L.; Baker, T.R.; Lewis, S.L.; Quesada, C.A.; Affum-Baffoe, K.; Arets, E.J.M.M.; Berry, N.J.; Bird, M.; et al. Height-diameter allometry of tropical forest trees. Biogeosciences 2011, 8, 1081–1106. [Google Scholar] [CrossRef]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.C.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef]
- Fayolle, A.; Doucet, J.L.; Gillet, J.F.; Bourland, N.; Lejeune, P. Tree allometry in Central Africa: Testing the validity of pantropical multi-species allometric equations for estimating biomass and carbon stocks. For. Ecol. Manag. 2013, 305, 29–37. [Google Scholar] [CrossRef]
- Chidumayo, E.N. Estimating tree biomass and changes in root biomass following clear-cutting of Brachystegia-Julbernardia (miombo) woodland in central Zambia. Environ. Conserv. 2013, 41, 54–63. [Google Scholar] [CrossRef]
- Mugasha, W.A.; Mwakalukwa, E.E.; Luoga, E.; Malimbwi, R.E.; Zahabu, E.; Silayo, D.S.; Sola, G.; Crete, P.; Henry, M.; Kashindye, A. Allometric models for estimating tree volume and aboveground biomass in lowland forests of Tanzania. Int. J. For. Res. 2016, 2016, 8076271. [Google Scholar] [CrossRef]
- Mugasha, W.A.; Eid, T.; Bollandsås, O.M.; Malimbwi, R.E.; Chamshama, S.A.O.; Zahabu, E.; Katani, J.Z. Allometric models for prediction of above and belowground biomass of trees in the miombo woodlands of Tanzania. For. Ecol. Manag. 2013, 310, 87–101. [Google Scholar] [CrossRef]
- Liu, B.; Bu, W.; Zang, R. Improved allometric models to estimate the aboveground biomass of younger secondary tropical forests. Glob. Ecol. Conserv. 2023, 41, e02359. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; Available online: http://www.R-project.org/ (accessed on 10 June 2025).
- Réjou-Méchain, M.; Tanguy, A.; Piponiot, C.; Chave, J.; Hérault, B. Biomass: An R Package for Estimating Above-Ground Biomass and Its Uncertainty in Tropical Forests. Methods Ecol. Evol. 2017, 8, 1163–1167. [Google Scholar] [CrossRef]
- Vaissie, A.P.; Monge, A.; Husson, F.; Husson, M.F. Factoshiny. Available online: http://factominer.free.fr/graphs/factoshiny.html (accessed on 20 September 2021).
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. R Package Version 1.3-5. 2021. Available online: https://cran.r-project.org/web/packages/agricolae/index.html (accessed on 10 June 2025).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Szoecs, E. Vegan: Community Ecology Package [Internet]. The R Foundation. (CRAN: Contributed Packages). 2001. Available online: https://CRAN.R-project.org/package=vegan (accessed on 10 June 2025).
- Pélissier, R.; Couteron, P.; Dray, S.; Sabatier, D. Consistency between ordination techniques and diversity measurements: Two strategies for species occurrence data. Ecology 2003, 84, 242–251. [Google Scholar] [CrossRef]
- Chidumayo, E.N.; Gumbo, D.J. The Dry Forests and Woodlands of Africa: Managing for Products and Services; Routledge: London, UK, 2010. [Google Scholar] [CrossRef]
- Schmitz, A. La Végétation de la Plaine de Lubumbashi: Région d‘Elisabethville (Haut-Katanga); Publication INEAC: Bruxelles, Belgium, 1971; 388p. [Google Scholar]
- Paul, D. La végétation du Katanga et de ses sols métallifères. Bull. Société R. Bot. Belg. 1958, 90, 127–283. [Google Scholar]
- Mugasha, W.A.; Bollandsås, O.M.; Eid, T. Relationships between diameter and height of trees in natural tropical forest in Tanzania. South. For. 2013, 75, 221–237. [Google Scholar] [CrossRef]
- Chidumayo, E.N. Species structure in Zambian miombo woodland. J. Trop. Ecol. 1987, 3, 109–118. [Google Scholar] [CrossRef]
- Backéus, I.; Pettersson, B.; Strömquist, L.; Ruffo, C. Tree communities and structural dynamics in miombo (Brachystegia-Julbernardia) woodland, Tanzania. For. Ecol. Manag. 2006, 230, 171–178. [Google Scholar] [CrossRef]
- Ryan, C.M.; Williams, M.; Grace, J. Above- and belowground carbon stocks in a miombo woodland landscape of mozambique. Biotropica 2011, 43, 423–432. [Google Scholar] [CrossRef]
- Gao, Y.; Xia, S.; Wang, P.; Xi, X.; Nie, S.; Wang, C. Lidar remote sensing meets weak supervision: Concepts, methods, and perspectives. arXiv 2025, arXiv:2503.18384. [Google Scholar] [CrossRef]
- Brede, B.; Terryn, L.; Barbier, N.; Bartholomeus, H.M.; Calders, K.; Krishna, S.M.; Lau, A.; Levick, S.R.; Raumonen, P.; Verbeeck, H.; et al. Remote Sensing of Environment Non-destructive estimation of individual tree biomass: Allometric models, terrestrial and UAV laser scanning. Remote Sens. Environ. 2022, 280, 113180. [Google Scholar] [CrossRef]
- Lewis, S.L.; Sonké, B.; Sunderland, T.; Begne, S.K.; Lopez-Gonzalez, G.; Van Der Heijden, G.M.; Phillips, O.L.; Affum-Baffoe, K.; Baker, T.R.; Banin, L.; et al. Above-ground biomass and structure of 260 African tropical forests. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120295. [Google Scholar] [CrossRef] [PubMed]

| Model | Performance Criteria | Coefficient Estimates (±SE) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | RSE | AIC | BIC | S | B | a | b | c | d | |
| (m1) log (AGB) ~ a + b × log (DBH) | 0.91 | 0.37 | 193.90 | 203.99 | 16.34 | 2.78 | −2.6230 (±0.1792) | 2.6338 (±0.0584) | - | - |
| (m2) log (AGB) ~ a + b × log (DBH2 × H) | 0.90 | 0.38 | 198.19 | 208.28 | 15.67 | −0.27 | −2.9297 (±0.1881) | 0.9920 (±0.0222) | - | - |
| (m3) log (AGB) ~ a + b × log (DBH2 × H × WDi) | 0.92 | 0.34 | 154.62 | 164.70 | 12.65 | −0.71 | −2.4368 (±0.1585) | 0.9790 (±0.0196) | - | - |
| (m4) log (AGB) ~ a + b × log (DBH) + c × log (H) + d × log (WDi) | 0.93 | 0.32 | 137.15 | 153.96 | 11.73 | 0.53 | −2.2499 (±0.1974) | 1.0871 (±0.0378) | 0.6485 (±0.0853) | 1.2101 (±0.2124) |
| Models | 10-Fold CV | Spatial CV | ||||
|---|---|---|---|---|---|---|
| R2 | S | B | R2 | S | B | |
| (m1) AGB_kg = 0.0718 × DBH2.6338 | 0.91 | 16.41 | 2.92 | 0.91 | 19.45 | 3.52 |
| (m2) AGB_kg = 0.0534 × (DBH2 × H)0.992 | 0.87 | 15.75 | −0.21 | 0.87 | 16.11 | 2 |
| (m3) AGB_kg = 0.0927 × (D2 × H × WDi)0.979 | 0.90 | 12.72 | −0.64 | 0.90 | 12.86 | 0.22 |
| (m4) AGB_kg = 0.1054× DBH1.0871 × H0.6485 × WDi1.210 | 0.91 | 14.70 | 3.22 | 0.90 | 17.13 | 3.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muledi, J.I.; Momo, S.T.; Ploton, P.; Kamukenge, A.L.; Ibey, W.K.; Pamavesi, B.M.; Mushabaa, B.A.; Shutcha, M.N.; Mwenze, D.N.; Sonké, B.; et al. Allometric Equations for Aboveground Biomass Estimation in Wet Miombo Forests of the Democratic Republic of the Congo Using Terrestrial LiDAR. Environments 2025, 12, 260. https://doi.org/10.3390/environments12080260
Muledi JI, Momo ST, Ploton P, Kamukenge AL, Ibey WK, Pamavesi BM, Mushabaa BA, Shutcha MN, Mwenze DN, Sonké B, et al. Allometric Equations for Aboveground Biomass Estimation in Wet Miombo Forests of the Democratic Republic of the Congo Using Terrestrial LiDAR. Environments. 2025; 12(8):260. https://doi.org/10.3390/environments12080260
Chicago/Turabian StyleMuledi, Jonathan Ilunga, Stéphane Takoudjou Momo, Pierre Ploton, Augustin Lamulamu Kamukenge, Wilfred Kombe Ibey, Blaise Mupari Pamavesi, Benoît Amisi Mushabaa, Mylor Ngoy Shutcha, David Nkulu Mwenze, Bonaventure Sonké, and et al. 2025. "Allometric Equations for Aboveground Biomass Estimation in Wet Miombo Forests of the Democratic Republic of the Congo Using Terrestrial LiDAR" Environments 12, no. 8: 260. https://doi.org/10.3390/environments12080260
APA StyleMuledi, J. I., Momo, S. T., Ploton, P., Kamukenge, A. L., Ibey, W. K., Pamavesi, B. M., Mushabaa, B. A., Shutcha, M. N., Mwenze, D. N., Sonké, B., Tshanika, U. M., Bamuninga, B. T., Ndikumagenge, C., & Barbier, N. (2025). Allometric Equations for Aboveground Biomass Estimation in Wet Miombo Forests of the Democratic Republic of the Congo Using Terrestrial LiDAR. Environments, 12(8), 260. https://doi.org/10.3390/environments12080260






