4.1. Linking Ecological Outcomes with Soil Health Metrics
This study evaluated the potential of EOV to be used as a proxy assessment of soil health by examining the relationships between EOV indicators and laboratory-based soil health metrics. Our analyses revealed that a small subset of variables can capture a significant portion of variance in ecological and soil health data, demonstrating the potential for EOV to serve as an effective tool for monitoring soil health.
EOV indicators such as community dynamics, vegetation richness, number of functional groups, and herbage mass captured the majority of the total variance associated with our dataset. This indicates that ecological indicators related to plant community diversity play a prominent role in maintaining the ecosystem’s structural and functional integrity, as plants serve as the primary source of OM and energy for sustaining many soil ecosystem functions [
33,
34]. The EHI, which is the sum of 15 ecological indicators assessed in the STM protocol, was also a good indicator to explain the variance of the dataset. Xu et al., 2019 [
24] reported a positive correlation between the EHI and vegetation richness, which aligns with the results of this study. For the HSHT subset, carbon-related parameters such as WEOC and soil respiration were crucial for explaining changes in soil health. Recent studies have highlighted the importance of soil carbon and its link with other soil functions, such as nutrient and water cycling and greenhouse gas emissions, especially in grazing lands [
35,
36,
37,
38]. Additionally, soil carbon responds to management practices, an essential criterion for evaluating soil health [
38]. The results from the PLFA subset highlighted the importance of microbial community structure and its function as major components in soil health assessments. Microbial biomass and specific communities such as fungi, bacteria, and AMF were the most important parameters, which aligns with Yang et al., 2022 [
39].
The interconnectedness between the three subsets is particularly evident in Spearman’s correlation matrix (
Table 6). Notably, EOV parameters had the strongest positive relationships across subsets, particularly with PLFA parameters (e.g., microbial biomass). Total bacteria correlated positively with all EOV indicators, while AMF showed strong correlations with the WCI. This suggests that microbial diversity and activity, often promoted by greater SOM and biodiversity, play a central role in enhancing ecological processes such as nutrient and water cycling [
10]. Total microbial biomass and the functional groups were closely related to vegetation parameters (e.g., vegetation richness) and soil nutrients (e.g., WEOC), which is consistent with previous findings [
39,
40]. Microbial communities play a fundamental role in the biogeochemical cycling of essential nutrients, including carbon, nitrogen, phosphorus, and sulfur, contributing to overall ecosystem productivity [
41]. Furthermore, microbial decomposition of plant and animal residues is essential for nutrient recycling and maintaining soil organic matter [
42].
van Es and Karlen, 2019 [
43] assessed soil health parameters from three long-term trials and found different correlations among variables from individual sites, suggesting that results can be impacted by soil properties (e.g., soil types) and management practices. According to Stanley et al., 2024 [
44], climatic, edaphic, and plant ecophysiology factors, along with optimal grazing management, also play a crucial role in soil organic carbon (SOC) sequestration. It is important to note that our study comprised a diverse range of grazing management practices, with diverse land uses, soil types, and a wide range of physical, chemical, and biological soil properties, which allowed for a comprehensive assessment of soil health. Such diversity ensures that the findings are not limited to a single management approach, enhancing their applicability across a wide range of landscapes.
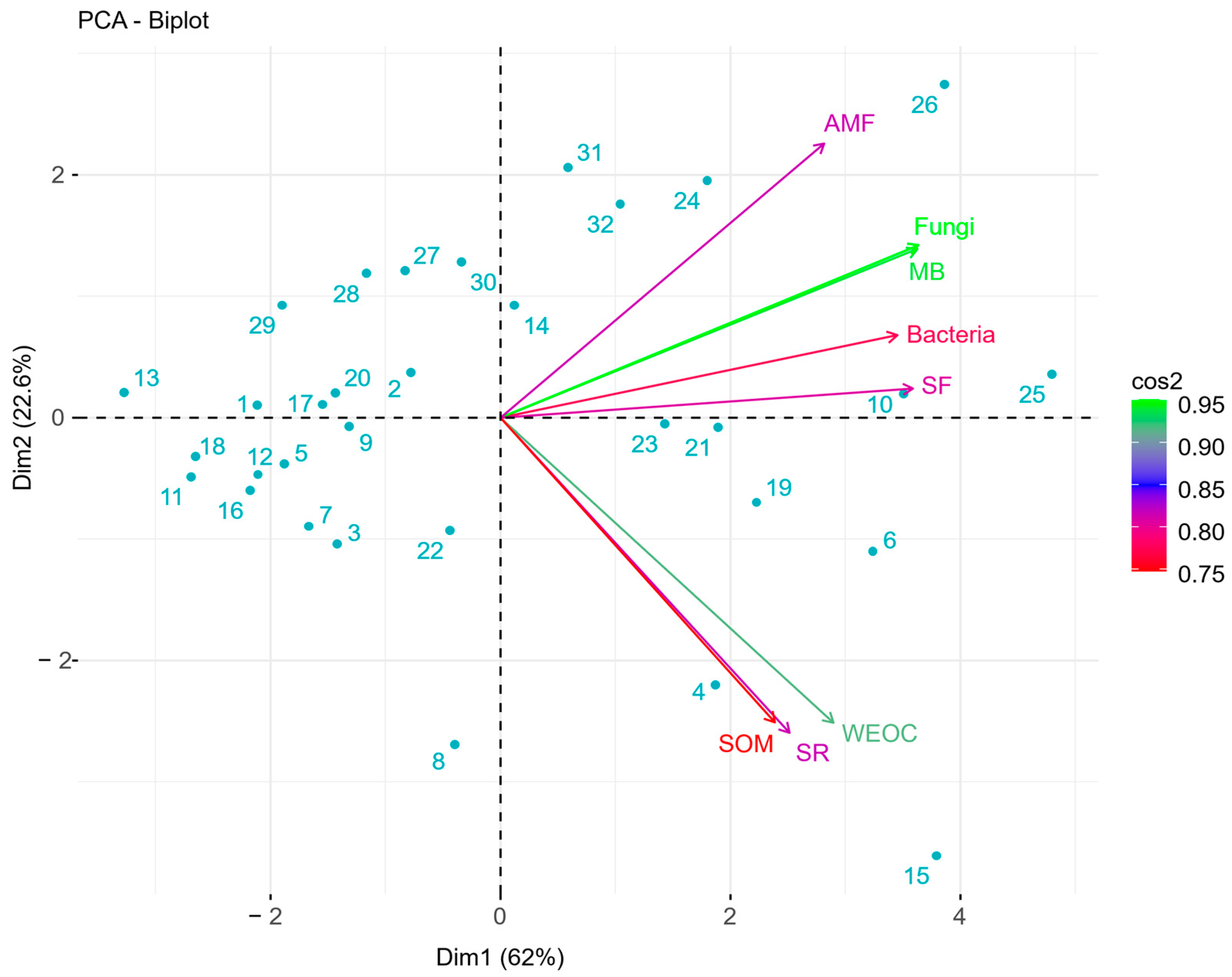
The PCA biplot (
Figure 2) revealed distinct groups of sampling sites across four quadrants. Variables grouped in two clusters, one for HSHT parameters and the other for PLFA parameters, suggesting that sites are differentiated based on shared characteristics related to soil health metrics [
45]. The right-side quadrants were characterized by greater values for both HSHT and PLFA parameters, representing the healthiest soils. The increase in WEOC, SOM, and total microbial biomass in those quadrants compared to the left-side quadrants were 38, 62, and 186%, respectively. The structured behavior of variables suggests that soils with enriched microbial diversity and greater carbon and SOM support a healthier ecological state [
46], as corroborated by the greater EOV values in those quadrants. These sites demonstrated active nutrient cycling and carbon availability, which fostered microbial growth and diversity, thereby supporting more sustainable and resilient ecosystems. Ecological outcomes verification could be an option for identifying efficient nutrient cycling, carbon and SOM availability, and robust microbial communities, which are essential for maintaining ecosystem functionality. Previous studies described the potential of microbial diversity to alter terrestrial ecosystem processes, soil functional stability [
47,
48], and the amount of carbon and SOM. Highlighting the relationships between physical, chemical, and biological soil parameters underscores the importance of a holistic approach towards soil health management.
4.2. Strengthening the Link Between EOV and Soil Test Metrics—Modeling HSHT and PLFA Parameters
The sequential approach, allowing only one group of variables as covariates in the first step of development (e.g., EOV to predict PLFA parameters) and then providing two groups at a second level (e.g., EOV + HSHT to predict PLFA parameters), provided more accurate models. Overall, more complex models consistently enhanced predictive ability compared to simpler models [
49]. However, although prediction performance is likely to be improved at the expense of model complexity [
50], the trade-off between the on-farm availability of variable inputs and prediction accuracy must be carefully considered [
51]. Models with increasing complexity may include covariates that are costly and infeasible to obtain [
52].
This study has identified key covariates to predict both HSHT and PLFA parameters. Water infiltration time was identified as the most important covariate to predict HSHT variables. There were significant negative relationships between water infiltration time, soil respiration, WEOC, and SOM in both categories of model complexity (
Table 7 and
Table 8). Conversely, the time that it takes for water to infiltrate (i.e., 1/WI) is positively related to soil respiration, WEOC, and SOM, aligning with previous studies [
43,
53]. The broad range of soil types assessed in this study confirms the impact of inherent soil properties, such as structure and texture, on water infiltration [
53]. As emphasized by Bagnall et al., 2022 [
53], identifying soil parameters, including soil hydraulic function, is critical for understanding the impact of management practices. Given the significance of water infiltration, it is essential to highlight the simplicity and practicality of this measurement for producers, as it provides a valuable tool for predicting other soil health variables.
At the first level of complexity, rainfall was identified as having a positive relationship with SOM (
Table 7), whereas at the second level, several covariates were selected. Mean annual precipitation was also identified as a significant predictor of SOC elsewhere [
54,
55]. At the second level of complexity, the MCI, water infiltration, Gram-negative bacteria, and the F:B ratio were negatively related to SOM, while the ECI, bare ground, litter, trees, and saprophytic fungi presented significant positive relationships with SOM (
Table 8). The ecological indicators related to the MCI are live organisms (i.e., macrofauna), litter incorporation and decomposition, and bare soil, whereas ECI indicators are limited to live canopy abundance and bare ground (
Table S3). Previous studies have identified vegetation type as a key factor influencing SOC stocks, with effects attributed to the length and area of the root system and the amount and chemical composition of deposited litter [
54,
56].
Fanin et al., 2019 [
57] reported that Gram-negative bacteria are more dependent on simple carbon compounds derived from plants (e.g., alkyl and N-alkyl compounds), whereas the Gram-positive bacteria use more SOM-derived carbon sources, which are more recalcitrant (e.g., carbonyl, aryl, and ketone compounds) and stable. A negative relationship between the F:B ratio and SOC is also reported in the literature, where less fungal biomass has been associated with less soil carbon sequestration [
58]. Six et al., 2006 [
59] explained that shifts toward fungal dominance may increase SOC and reduce its turnover rate due to enhanced fungal-mediated soil aggregation and/or shifts in the microbial biomass physiology [
58]. Still, saprophytic fungi were identified as a key covariate predicting SOC (
Table 8), which agrees with Spearman’s correlation matrix (
Table 6). In our study, saprophytic fungi exhibited a positive correlation with WEOC. This relationship can be attributed to the findings of Zhao et al., 2024 [
60], who demonstrated that hyphae of saprophytic fungi secrete extracellular biopolymers that enhance soil aggregate stability, which is correlated with SOC.
Rainfall was negatively related to total microbial biomass, total fungi, and AMF (
Table 9). Soil moisture, which is affected by rainfall gradients, is known to affect microbial community composition [
61]. Microorganisms need water to maintain their physiological status, and soil moisture also affects the availability of both substrate and oxygen for microbes’ growth [
62]. Hawkes et al., 2011 [
61] reported that fungi respond directly to rainfall levels, with more abundant, diverse, and consistent communities under drought conditions and less abundant, less diverse, and more variable communities during wetter periods. AMF requires oxygen for their metabolic processes, and their functionality may be compromised when the soil becomes waterlogged and anaerobic. Further studies have corroborated the strong effect of rainfall on the composition and function of soil fungal communities, especially AMF, whereas saprophytic fungi were not affected [
61,
63].
The main EOV-specific vegetation parameters used to predict PLFA parameters were the proportion of shrubs and trees and vegetation richness (
Table 9). Plant diversity influences soil microbial biodiversity via two main pathways [
64]: first, by increasing the net primary productivity, and second, by leading towards a greater diversity of litter and root exudates [
65]. Zhao et al., 2014 [
64] mentioned that specific plant species may be more important and sometimes prevail over plant diversity in controlling soil microbial biodiversity. Plant diversity explained one-third of the variance in total plant biomass, whereas specific plant species accounted for about two-thirds [
66].
The WEON was selected as a covariate as it was positively related to predicting all HSHT parameters at the second level of complexity (
Table 10). This positive relationship between nitrogen and total microbial biomass and bacteria was also reported in Australia [
67]. Total nitrogen was also found to be the most important variable correlated with PLFA parameters, including total microbial biomass, total bacteria, Gram-negative bacteria, total fungi, and other diversity indexes [
68]. Soil inorganic nitrogen, however, seemed to be more important than WEON in explaining changes in bacterial and fungal communities in forest ecosystems in China [
62]. Another parameter that is frequently reported as an important modulator of soil microbes’ growth is soil pH [
67]. However, our results showed that pH was not selected to predict any PLFA parameters. Soil calcium concentration, which is related to pH, was positively correlated to saprophytic fungi.