Gut Microbiota and Mucosal Immunity in the Neonate
Abstract
:1. Introduction
- DEFINITIONS
- Microbiota: the microbial community in a specific niche/environment.
- Microbiome: the total genomic repertoire of a microbial community (microbiota).
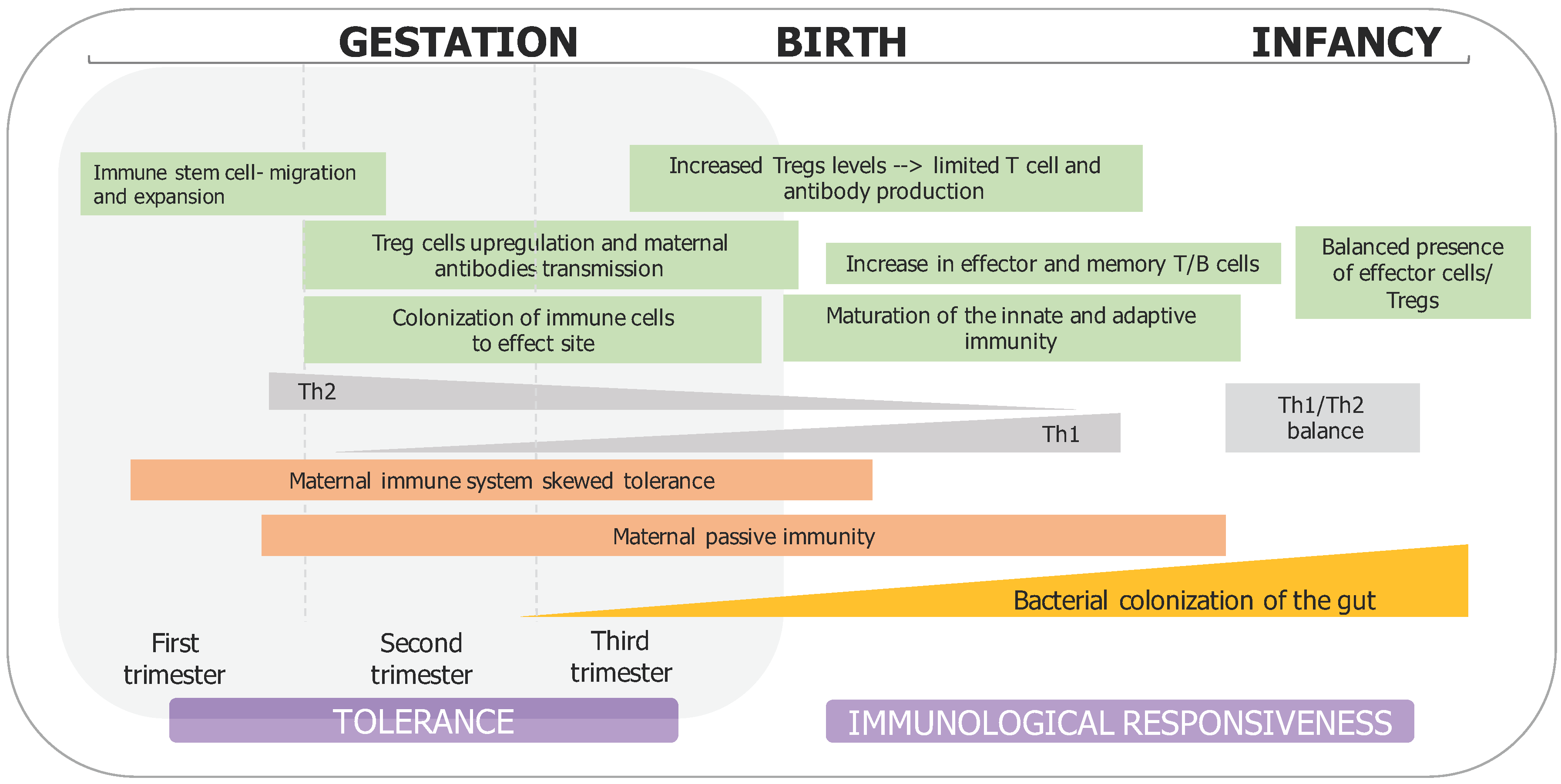
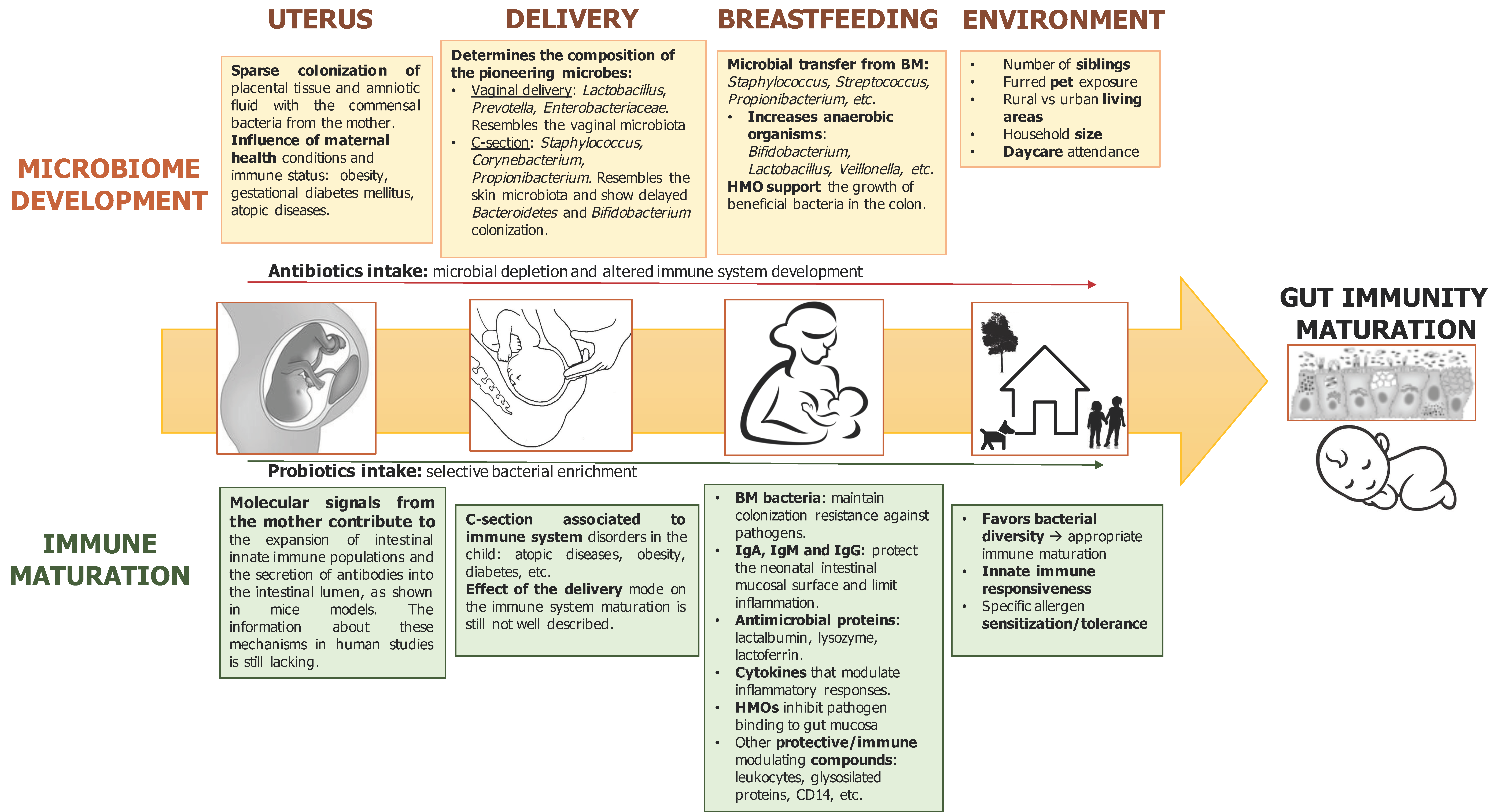
2. Prenatal Microbial Exposure and Mother–Fetus Immune Interaction
3. Perinatal Factors Influencing Infant Microbiota Development and Immune System Maturation
3.1. Environment and Geographical Location
3.2. Mode of Delivery
3.3. Antibiotics Intake during Pregnancy and Early Infancy
4. Postnatal Microbial Establishment and Immune Maturation
Early Colonizers and the Immune System
5. Commensal Recognition and Tolerogenic Features of the Gut Immune System
5.1. Intestinal Epithelial Cells
5.2. Microbial Associated Molecular Patterns and Dendritic Cells
5.3. IgA Antibody–Microbiota Interactions
5.4. Regulatory T Cells
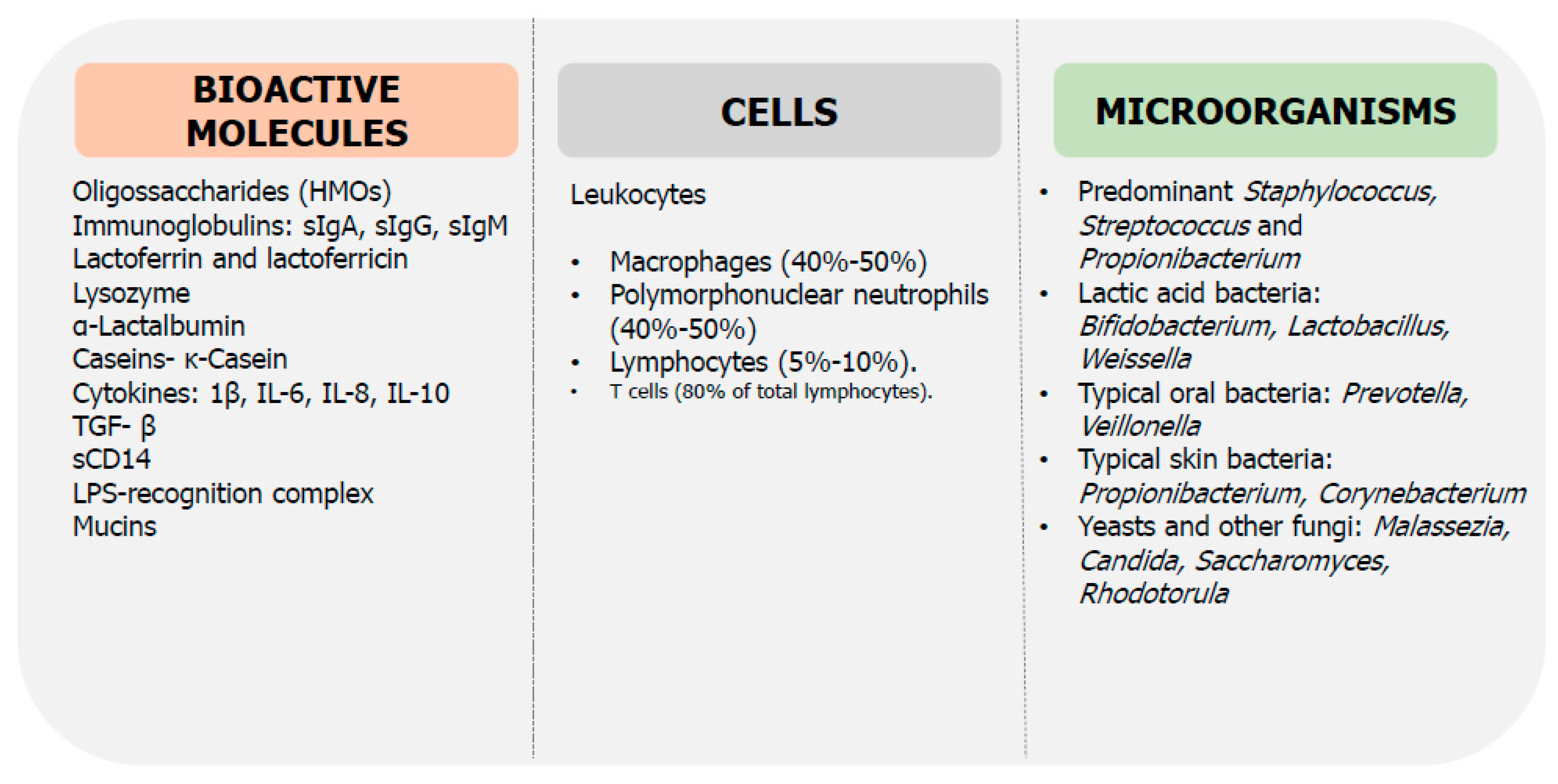
6. Role of Breast Milk on Microbiota and Immunity Maturation
6.1. Breast Milk Microbiota
6.2. Human Milk Oligosaccharides
6.3. Secretory IgA
6.4. Anti-Microbial Bioactive Proteins
6.5. Cytokines
6.6. Cells and Other Immunological Compounds
7. Concluding Remarks
Funding
Conflicts of Interest
References
- Barker, D.J.P. The origins of the developmental origins theory. J. Intern. Med. 2007, 261, 412–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, M.C.; Isolauri, E.; Laitinen, K.; Salminen, S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 2008, 88, 894–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Kling Bäckhed, H.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host Remodeling of the Gut Microbiome and Metabolic Changes during Pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.A.; Wong, R.J.; Shaw, G.; et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, N.T.; Shin, H.; Pizoni, A.; Werlang, I.C.; Matte, U.; Goldani, M.Z.; Goldani, H.A.S.; Dominguez-Bello, M.G. Birth mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Sci. Rep. 2016, 6, 23133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanislawski, M.A.; Dabelea, D.; Wagner, B.D.; Sontag, M.K.; Lozupone, C.A.; Eggesbø, M. Pre-pregnancy weight, gestational weight gain, and the gut microbiota of mothers and their infants. Microbiome 2017, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.; Aho, V.; Pereira, P.; Paulin, L.; Koivusalo, S.B.; Auvinen, P.; Eriksson, J.G. Gut microbiome in gestational diabetes: A cross-sectional study of mothers and offspring 5 years postpartum. Acta Obstet. Gynecol. Scand. 2018, 97, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsson, T.R.; Wu, R.Y.; Jenmalm, M.C. Gut microbiota and allergy: The importance of the pregnancy period. Pediatr. Res. 2015, 77, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Vuillermin, P.J.; Macia, L.; Nanan, R.; Tang, M.L.; Collier, F.; Brix, S. The maternal microbiome during pregnancy and allergic disease in the offspring. Semin. Immunopathol. 2017, 39, 669–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rock, R.; Ben-Amram, H.; Neuman, H.; Hanya, H.; Koren, O.; Youngster, I. The Impact of Antibiotic Use in Breastfeeding Women on the Infant’s Gut Microbiome—A Prospective Cohort Study. Open Forum Infect. Dis. 2017, 4, S232. [Google Scholar] [CrossRef]
- Ma, J.; Prince, A.L.; Bader, D.; Hu, M.; Ganu, R.; Baquero, K.; Blundell, P.; Alan Harris, R.; Frias, A.E.; Grove, K.L.; et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat. Commun. 2014, 5, 3889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, S.; Godfrey, K.M.; McDonald, D.; Treuren, W.V.; Bjørnholt, J.V.; Midtvedt, T.; Moen, B.; Rudi, K.; Knight, R.; Brantsæter, A.L.; et al. Fat and vitamin intakes during pregnancy have stronger relations with a pro-inflammatory maternal microbiota than does carbohydrate intake. Microbiome 2016, 4, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, D.M.; Antony, K.M.; Ma, J.; Prince, A.L.; Showalter, L.; Moller, M.; Aagaard, K.M. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Gomez de Agüero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Funkhouser, L.J.; Bordenstein, S.R. Mom Knows Best: The Universality of Maternal Microbial Transmission. PLoS Biol. 2013, 11, e1001631. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Moles, L.; Gómez, M.; Heilig, H.; Bustos, G.; Fuentes, S.; de Vos, W.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Bacterial Diversity in Meconium of Preterm Neonates and Evolution of Their Fecal Microbiota during the First Month of Life. PLoS ONE 2013, 8, e66986. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef] [PubMed]
- Gosalbes, M.J.; Llop, S.; Vallès, Y.; Moya, A.; Ballester, F.; Francino, M.P. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 2013, 43, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Jimønez, E.; Fernµndez, L.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Nueno-Palop, C.; Narbad, A.; Olivares, M.; Xaus, J.; Rodríguez, J.M. Isolation of Commensal Bacteria from Umbilical Cord Blood of Healthy Neonates Born by Cesarean Section. Curr. Microbiol. 2005, 51, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Rautava, S.; Collado, M.C.; Salminen, S.; Isolauri, E. Probiotics Modulate Host-Microbe Interaction in the Placenta and Fetal Gut: A. Randomized, Double-Blind, Placebo-Controlled Trial. Neonatology 2012, 102, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Steel, J.H.; Malatos, S.; Kennea, N.; Edwards, A.D.; Miles, L.; Duggan, P.; Reynolds, P.R.; Feldman, R.G.; Sullivan, M.H.F. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr. Res. 2005, 57, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Bearfield, C.; Davenport, E.S.; Sivapathasundaram, V.; Allaker, R.P. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG 2002, 109, 527–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldenberg, R.L.; Hauth, J.C.; Andrews, W.W. Intrauterine Infection and Preterm Delivery. N. Engl. J. Med. 2000, 342, 1500–1507. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, L.F.; Chaiworapongsa, T.; Romero, R. Intrauterine infection and prematurity. Ment. Retard Dev. Disabil. Res. Rev. 2002, 8, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Rodriguez, C.; Holtz, L.R. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 2018, 6, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauder, A.P.; Roche, A.M.; Sherrill-Mix, S.; Bailey, A.; Laughlin, A.L.; Bittinger, K.; Leite, R.; Elovitz, M.A.; Parry, S.; Bushman, F.D. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.N.; Himes, J.E.; Martinez, D.R.; Permar, S.R. The Impact of the Gut Microbiota on Humoral Immunity to Pathogens and Vaccination in Early Infancy. PLOS Pathog. 2016, 12, e1005997. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.-P.; Ricciardi-Castagnoli, P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.F.; Dore, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J.; Donnet-Hughes, A. Bacterial Imprinting of the Neonatal Immune System: Lessons From Maternal Cells? Pediatrics 2007, 119, e724–e732. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Arango, L.F.; Barrett, H.L.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Nitert, M.D. Contributions of the maternal oral and gut microbiome to placental microbial colonization in overweight and obese pregnant women. Sci. Rep. 2017, 7, 2860. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef] [PubMed]
- DiGiulio, D.B. Diversity of microbes in amniotic fluid. Semin. Fetal Neonatal Med. 2012, 17, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.; Saini, S.; Saini, S.R. Periodontitis: A risk for delivery of premature labor and low-birth-weight infants. J. Nat. Sci. Biol. Med. 2010, 1, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Dasanayake, A.P.; Li, Y.; Wiener, H.; Ruby, J.D.; Lee, M.-J. Salivary Actinomyces naeslundii Genospecies 2 and Lactobacillus casei Levels Predict Pregnancy Outcomes. J. Periodontol. 2005, 76, 171–177. [Google Scholar] [CrossRef] [PubMed]
- López, N.J.; Smith, P.C.; Gutierrez, J. Periodontal Therapy May Reduce the Risk of Preterm Low Birth Weight in Women With Peridotal Disease: A randomized Controlled Trial. J. Periodontol. 2002, 73, 911–924. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.M.; Merchant, M.; Kumar, P.N.; Dutta, A.; Mande, S.S. First-trimester vaginal microbiome diversity: A potential indicator of preterm delivery risk. Sci. Rep. 2017, 7, 16145. [Google Scholar] [CrossRef] [PubMed]
- Dortbudak, O.; Eberhardt, R.; Ulm, M.; Persson, G.R. Periodontitis, a marker of risk in pregnancy for preterm birth. J. Clin. Periodontol. 2005, 32, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Stout, M.J.; Lee, I.; Mysorekar, I.U. Placental Microbiome and Its Role in Preterm Birth. Neoreviews 2014, 15, e537–e545. [Google Scholar] [CrossRef] [PubMed]
- Prince, A.L.; Ma, J.; Kannan, P.S.; Alvarez, M.; Gisslen, T.; Harris, R.A.; Sweeney, E.L.; Knox, C.L.; Lambers, D.S.; Jobe, A.H.; et al. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am. J. Obstet. Gynecol. 2016, 214, 627.e1–627.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagpal, R.; Tsuji, H.; Takahashi, T.; Kawashima, K.; Nagata, S.; Nomoto, K.; Yamashiro, Y. Sensitive Quantitative Analysis of the Meconium Bacterial Microbiota in Healthy Term Infants Born Vaginally or by Cesarean Section. Front. Microbiol. 2016, 7, 1997. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Björkstén, B.; Engstrand, L.; Andersson, A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by Caesarean section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Hesla, H.M.; Stenius, F.; Jäderlund, L.; Nelson, R.; Engstrand, L.; Alm, J.; Dicksved, J. Impact of lifestyle on the gut microbiota of healthy infants and their mothers—The ALADDIN birth cohort. FEMS Microbiol. Ecol. 2014, 90, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.J.; Lynch, D.B.; Murphy, K.; Ulaszewska, M.; Jeffery, I.B.; O’Shea, C.A.; Watkins, C.; Dempsey, E.; Mattivi, F.; Tuohy, K.; et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Schuster, M.; Hsiao, W.; Matzinger, P. Uncovering effest of the antibiotics on the host and microbiota using Transkingdom Gene Networks. Gut 2016, 64, 1732–1743. [Google Scholar]
- Furusawa, Y.; Obata, Y.; Hase, K. Commensal microbiota regulates T cell fate decision in the gut. Semin. Immunopathol. 2015, 37, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T. cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Panea, C.; Nakato, G.; Cebula, A.; Lee, C.; Diez, M.G.; Laufer, T.M.; Ignatowicz, L.; Ivanov, I.I. Segmented Filamentous Bacteria Antigens Presented by Intestinal Dendritic Cells Drive Mucosal Th17 Cell Differentiation. Immunity 2014, 40, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Sprockett, D.; Fukami, T.; Relman, D.A. Role of priority effects in the early-life assembly of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2013, 2, e00458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laursen, M.F.; Zachariassen, G.; Bahl, M.I.; Bergström, A.; Høst, A.; Michaelsen, K.F.; Licht, T.R. Having older siblings is associated with gut microbiota development during early childhood. BMC Microbiol. 2015, 15, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virtanen, S.M.; Takkinen, H.-M.; Nwaru, B.I.; Kaila, M.; Ahonen, S.; Nevalainen, J.; Niinistö, S.; Siljander, H.; Simell, O.; Ilonen, J.; et al. Microbial Exposure in Infancy and Subsequent Appearance of Type 1 Diabetes Mellitus–Associated Autoantibodies. JAMA Pediatr. 2014, 168, 755–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azad, M.B.; Konya, T.; Maughan, H.; Guttman, D.S.; Field, C.J.; Sears, M.R.; Becker, A.B.; Scott, J.A.; Kozyrskyj, A.L. Infant gut microbiota and the hygiene hypothesis of allergic disease: Impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin. Immunol. 2013, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Jatzlauk, G.; Bartel, S.; Heine, H.; Schloter, M.; Krauss-Etschmann, S. Influences of environmental bacteria and their metabolites on allergies, asthma, and host microbiota. Allergy 2017, 72, 1859–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Normand, A.-C.; Sudre, B.; Vacheyrou, M.; Depner, M.; Wouters, I.M.; Noss, I.; Heederik, D.; Hyvarinen, A.; Genuneit, J.; Braun-Fahrlander, C.; et al. Airborne cultivable microflora and microbial transfer in farm buildings and rural dwellings. Occup. Environ. Med. 2011, 68, 849–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steiman, C.A.; Fadrosh, D.; Evans, M.D.; Vrtis, R.F.; Olson, B.; Barnes, K.L.; Seroogy, C.M.; Bendixsen, C.G.; Lynch, S.V.; Gern, J.E. Farm Exposure Influences Skin Microbiota In Infancy. J. Allergy Clin. Immunol. 2018, 141, AB86. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, C.H.F.; Andersen, L.S.F.; Krych, L.; Metzdorff, S.B.; Hasselby, J.P.; Skov, S.; Nielsen, D.S.; Buschard, K.; Hansen, L.H.; Hansen, A.K. Mode of delivery shapes gut colonization pattern and modulates regulatory immunity in mice. J. Immunol. 2014, 193, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S.; Sakwinska, O.; Soh, S.-E.; Ngom-Bru, C.; Brück, W.M.; Berger, B.; Brüssow, H.; Lee, Y.S.; Yap, F.; Chong, Y.-S.; et al. Dynamics of Infant Gut Microbiota Are Influenced by Delivery Mode and Gestational Duration and Are Associated with Subsequent Adiposity. mBio 2015, 6, e02419-14. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.D.; Lieber, A.; Wu, F.; Perez-Perez, G.I.; Chen, Y.; et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra82. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Chen, Q.; Chen, Y.; Bao, Y.; Wu, M.; Zhang, J. Cesarean section without medical indication and risk of childhood asthma, and attenuation by breastfeeding. PLoS ONE 2017, 12, e0184920. [Google Scholar] [CrossRef] [PubMed]
- Gerlich, J.; Benecke, N.; Peters-Weist, A.S.; Heinrich, S.; Roller, D.; Genuneit, J.; Weinmayr, G.; Windstetter, D.; Dressel, H.; Range, U.; et al. Pregnancy and perinatal conditions and atopic disease prevalence in childhood and adulthood. Allergy 2018, 73, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Sevelsted, A.; Stokholm, J.; Bonnelykke, K.; Bisgaard, H. Cesarean Section and Chronic Immune Disorders. Pediatrics 2015, 135, e92–e98. [Google Scholar] [CrossRef] [PubMed]
- Lotz, M.; Gütle, D.; Walther, S.; Ménard, S.; Bogdan, C.; Hornef, M.W. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 2006, 203, 973–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiemsma, L.T.; Turvey, S.E. Asthma and the microbiome: Defining the critical window in early life. Allergy Asthma Clin. Immunol. 2017, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Stokholm, J.; Blaser, M.J.; Thorsen, J.; Rasmussen, M.A.; Waage, J.; Vinding, R.K.; Schoos, A.-M.M.; Kunøe, A.; Fink, N.R.; Chawes, B.L.; et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Oh, S.F.; Olszak, T.; Neves, J.F.; Avci, F.Y.; Erturk-Hasdemir, D.; Lu, X.; Zeissig, S.; Blumberg, R.S.; Kasper, D.L. Sphingolipids from a Symbiotic Microbe Regulate Homeostasis of Host Intestinal Natural Killer, T. Cells. Cell 2014, 156, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Seedat, F.; Stinton, C.; Patterson, J.; Geppert, J.; Tan, B.; Robinson, E.R.; McCarthy, N.D.; Uthman, O.A.; Freeman, K.; Johnson, S.A.; et al. Adverse events in women and children who have received intrapartum antibiotic prophylaxis treatment: A. systematic review. BMC Pregnancy Childbirth 2017, 17, 247. [Google Scholar] [CrossRef] [PubMed]
- Stearns, J.C.; Simioni, J.; Gunn, E.; McDonald, H.; Holloway, A.C.; Thabane, L.; Mousseau, A.; Schertzer, J.D.; Ratcliffe, E.M.; Rossi, L.; et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci. Rep. 2017, 7, 16527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokholm, J.; Schjørring, S.; Eskildsen, C.E.; Pedersen, L.; Bischoff, A.L.; Følsgaard, N.; Carson, C.G.; Chawes, B.L.K.; Bønnelykke, K.; Mølgaard, A.; et al. Antibiotic use during pregnancy alters the commensal vaginal microbiota. Clin. Microbiol. Infect. 2014, 20, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Tekieli, L.-S.; Radens, C.M.; Williams, B.L.; N Gabriela Gonzalez-Perez, E.S.; Hicks, A.L.; Gonzalez-Perez, G.; Tekieli, T.M.; N Lamousé-Smith, E.S. Microbiome and Antiviral Immunity Development of the Neonatal Intestinal Maternal Antibiotic Treatment Impacts Maternal Antibiotic Treatment Impacts Development of the Neonatal Intestinal Microbiome and Antiviral Immunity. J. Immunol. 2018, 196, 3768–3779. [Google Scholar]
- Mor, A.; Antonsen, S.; Kahlert, J.; Holsteen, V.; Jørgensen, S.; Holm-Pedersen, J.; Sørensen, H.T.; Pedersen, O.; Ehrenstein, V. Prenatal exposure to systemic antibacterials and overweight and obesity in Danish schoolchildren: A. prevalence study. Int. J. Obes. 2015, 39, 1450–1455. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Zijlmans, M.A.C.; Kuitunen, M.; Kukkonen, K.; Savilahti, E.; Salonen, A.; de Weerth, C.; de Vos, W.M. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome 2017, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Esposito, S. Antibiotic administration and the development of obesity in children. Int. J. Antimicrob. Agents 2016, 47, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Mulder, B.; Pouwels, K.B.; Schuiling-Veninga, C.C.M.; Bos, H.J.; de Vries, T.W.; Jick, S.S.; Hak, E. Antibiotic use during pregnancy and asthma in preschool children: The influence of confounding. Clin. Exp. Allergy 2016, 46, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Salonen, A.; Virta, L.J.; Kekkonen, R.A.; Forslund, K.; Bork, P.; De Vos, W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016, 7, 10410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timm, S.; Schlünssen, V.; Olsen, J.; Ramlau-Hansen, C.H. Prenatal antibiotics and atopic dermatitis among 18-month-old children in the Danish National Birth Cohort. Clin. Exp. Allergy 2017, 47, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.D.; Bergholt, T.; Bouaziz, O.; Arpi, M.; Eriksson, F.; Rasmussen, S.; Keiding, N.; Løkkegaard, E.C. Broad-spectrum antibiotic treatment and subsequent childhood type 1 diabetes: A nationwide Danish cohort study. PLoS ONE 2016, 11, e0161654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candon, S.; Perez-Arroyo, A.; Marquet, C.; Valette, F.; Foray, A.P.; Pelletier, B.; Milani, C.; Ventura, M.; Bach, J.F.; Chatenoud, L. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS ONE 2015, 10, e0125448. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; Godovannyi, A.; Ma, C.; Zhang, Y.; Ahmadi-Vand, Z.; Dai, C.; Gorzelak, M.A.; Chan, Y.; Chan, J.M.; Lochner, A.; et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016, 10, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Slykerman, R.F.; Thompson, J.; Waldie, K.E.; Murphy, R.; Wall, C.; Mitchell, E.A. Antibiotics in the first year of life and subsequent neurocognitive outcomes. Acta Paediatr. Int. J. Paediatr. 2017, 106, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The Key Role of Segmented Filamentous Bacteria in the Coordinated Maturation of Gut Helper, T. Cell Responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Nishimura, J.; Shima, T.; Umesaki, Y.; Yamamoto, M.; Onoue, M.; Yagita, H.; Ishii, N.; Evans, R.; Honda, K.; et al. ATP drives lamina propria TH17 cell differentiation. Nature 2008, 455, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Grasa, L.; Abecia, L.; Forcén, R.; Castro, M.; de Jalón, J.A.G.; Latorre, E.; Alcalde, A.I.; Murillo, M.D. Antibiotic-Induced Depletion of Murine Microbiota Induces Mild Inflammation and Changes in Toll-Like Receptor Patterns and Intestinal Motility. Microb. Ecol. 2015, 70, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Tourneur, E.; Chassin, C. Neonatal immune adaptation of the gut and its role during infections. Clin. Dev. Immunol. 2013, 2013, 270301. [Google Scholar] [CrossRef] [PubMed]
- Noverr, M.C.; Falkowski, N.R.; McDonald, R.A.; McKenzie, A.N.; Huffnagle, G.B. Development of Allergic Airway Disease in Mice following Antibiotic Therapy and Fungal Microbiota Increase: Role of Host Genetics, Antigen, and Interleukin-13. Infect. Immun. 2005, 73, 30–38. [Google Scholar] [CrossRef] [PubMed]
- West, C.E.; Jenmalm, M.C.; Prescott, S.L. The gut microbiota and its role in the development of allergic disease: A. wider perspective. Clin. Exp. Allergy 2015, 45, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Wopereis, H.; Oozeer, R.; Knipping, K.; Belzer, C.; Knol, J. The first thousand days—Intestinal microbiology of early life: Establishing a symbiosis. Pediatr. Allergy Immunol. 2014, 25, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Chassard, C. New Insights in Gut Microbiota Establishment in Healthy Breast Fed Neonates. PLoS ONE 2012, 7, e44595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houghteling, P.D.; Walker, W.A. Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J. Pediatr. Gastroenterol. Nutr. 2015, 60, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Weng, M.; Walker, W.A. The role of gut microbiota in programming the immune phenotype. J. Dev. Orig. Health Dis. 2013, 4, 203–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PrabhuDas, M.; Adkins, B.; Gans, H.; King, C.; Levy, O.; Ramilo, O.; Siegrist, C.-A. Challenges in infant immunity: Implications for responses to infection and vaccines. Nat. Immunol. 2011, 12, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Olszak, T.; An, D.; Zeissig, S.; Vera, M.P.; Richter, J.; Franke, A.; Glickman, J.N.; Siebert, R.; Baron, R.M.; Kasper, D.L.; et al. Microbial Exposure During Early Life Has Persistent Effects on Natural Killer, T. Cell Function. Science 2012, 336, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.M.K.; Nair, L.; Alegre, M.-L. The interplay between the intestinal microbiota and the immune system. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 9–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kollmann, T.R.; Levy, O.; Montgomery, R.R.; Goriely, S. Innate Immune Function by Toll-like Receptors: Distinct Responses in Newborns and the Elderly. Immunity 2012, 37, 771–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeuthen, L.H.; Fink, L.N.; Frokiaer, H. Epithelial cells prime the immune response to an array of gut-derived commensals towards a tolerogenic phenotype through distinct actions of thymic stromal lymphopoietin and transforming growth factor-β? Immunology 2008, 123, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Smits, H.H.; Engering, A.; van der Kleij, D.; de Jong, E.C.; Schipper, K.; van Capel, T.M.M.; Zaat, B.A.J.; Yazdanbakhsh, M.; Wierenga, E.A.; van Kooyk, Y.; et al. Selective probiotic bacteria induce IL-10–producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell–specific intercellular adhesion molecule 3–grabbing nonintegrin. J. Allergy Clin. Immunol. 2005, 115, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Figdor, C.G.; van Kooyk, Y.; Adema, G.J. C-type lectin receptors on dendritic cells and langerhans cells. Nat. Rev. Immunol. 2002, 2, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Swiatczak, B.; Rescigno, M. How the interplay between antigen presenting cells and microbiota tunes host immune responses in the gut. Semin. Immunol. 2012, 24, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Konieczna, P.; Groeger, D.; Ziegler, M.; Frei, R.; Ferstl, R.; Shanahan, F.; Quigley, E.M.M.; Kiely, B.; Akdis, C.A.; O’Mahony, L. Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: Potential role for myeloid and plasmacytoid dendritic cells. Gut 2012, 61, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Granucci, F.; Zanoni, I. Role of Toll like receptor-activated dendritic cells in the development of autoimmunity. Front. Biosci. 2008, 13, 4817–4826. [Google Scholar] [CrossRef] [PubMed]
- Rutella, S.; Locatelli, F. Intestinal dendritic cells in the pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 2011, 17, 3761–3775. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macpherson, A.J.; Yilmaz, B.; Limenitakis, J.P.; Ganal-Vonarburg, S.C. IgA Function in Relation to the Intestinal Microbiota. Annu. Rev. Immunol. 2018, 36, 359–381. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, A.; Chen, K.; Chorny, A. Immunoglobulin responses at the mucosal interface. Annu. Rev. Immunol. 2011, 29, 273–293. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Gatto, D.; Sainsbury, E.; Harriman, G.R.; Hengartner, H.; Zinkernagel, R.M. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000, 288, 2222–2226. [Google Scholar] [CrossRef] [PubMed]
- Lindner, C.; Wahl, B.; Föhse, L.; Suerbaum, S.; Macpherson, A.J.; Prinz, I.; Pabst, O. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J. Exp. Med. 2012, 209, 365–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renz, H.; Brandtzaeg, P.; Hornef, M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2012, 12, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004, 303, 1662–1665. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M. Intestinal microbiota and its effects on the immune system. Cell. Microbiol. 2014, 16, 1004–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, D.A.; Siracusa, M.C.; Abt, M.C.; Kim, B.S.; Kobuley, D.; Kubo, M.; Kambayashi, T.; LaRosa, D.F.; Renner, E.D.; Orange, J.S.; et al. Commensal bacteria–derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 2012, 18, 538–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmore, J.R.; Gaudette, B.T.; Gomez Atria, D.; Hashemi, T.; Jones, D.D.; Gardner, C.A.; Cole, S.D.; Misic, A.M.; Beiting, D.P.; Allman, D. Commensal Microbes Induce Serum IgA Responses that Protect against Polymicrobial Sepsis. Cell Host Microbe 2018, 23, 302.e3–311.e3. [Google Scholar] [CrossRef] [PubMed]
- Peterson, D.A.; McNulty, N.P.; Guruge, J.L.; Gordon, J.I. IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis. Cell Host Microbe 2007, 2, 328–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunker, J.J.; Erickson, S.A.; Flynn, T.M.; Henry, C.; Koval, J.C.; Meisel, M.; Jabri, B.; Antonopoulos, D.A.; Wilson, P.C.; Bendelac, A. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017, 358, eaan6619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dzidic, M.; Abrahamsson, T.R.; Artacho, A.; Björkstén, B.; Collado, M.C.; Mira, A.; Jenmalm, M.C. Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. J. Allergy Clin. Immunol. 2017, 139, 1017.e14–1025.e14. [Google Scholar] [CrossRef] [PubMed]
- Weiner, H.L.; da Cunha, A.P.; Quintana, F.; Wu, H. Oral tolerance. Immunol. Rev. 2011, 241, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.A.E.; Ng, D.C.K.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal Bacterial Colonization Induces Mutualistic Regulatory, T. Cell Responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Ohue, R.; Hashimoto, K.; Nakamoto, M.; Furukawa, Y.; Masuda, T.; Kitabatake, N.; Tani, F. Bacterial Heat Shock Protein 60, GroEL, Can Induce the Conversion of Naïve T Cells into a CD4+ CD25+ Foxp3-Expressing Phenotype. J. Innate Immun. 2011, 3, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.; Prioult, G.; Hacini-Rachinel, F.; Moine, D.; Bruttin, A.; Ngom-Bru, C.; Labellie, C.; Nicolis, I.; Berger, B.; Mercenier, A.; et al. Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T.-cell response. FEMS Microbiol. Ecol. 2012, 79, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y.; Itoh, K.; Honda, K. The induction of Treg cells by gut-indigenous Clostridium. Curr. Opin. Immunol. 2012, 24, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Finamore, A.; Roselli, M.; Britti, M.S.; Merendino, N.; Mengheri, E. Lactobacillus rhamnosus GG and Bifidobacterium animalis MB5 Induce Intestinal but Not Systemic Antigen-Specific Hyporesponsiveness in Ovalbumin-Immunized Rats. J. Nutr. 2012, 142, 375–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-Like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maizels, R.M.; McSorley, H.J. Regulation of the host immune system by helminth parasites. J. Allergy Clin. Immunol. 2016, 138, 666–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.; McCoy, K.D.; Macpherson, A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007, 19, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Wong, M.H.; Thelin, A.; Hansson, L.; Falk, P.G.; Gordon, J.I. Molecular Analysis of Commensal Host-Microbial Relationships in the Intestine. Science 2001, 291, 881–884. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Vaughan, E.E. Probiotic and Gut Lactobacilli and Bifidobacteria: Molecular Approaches to Study Diversity and Activity. Annu. Rev. Microbiol. 2009, 63, 269–290. [Google Scholar] [CrossRef] [PubMed]
- Stappenbeck, T.S.; Hooper, L.V.; Gordon, J.I. Nonlinear partial differential equations and applications: Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 2002, 99, 15451–15455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deplancke, B.; Gaskins, H.R. Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001, 73, 1131S–1141S. [Google Scholar] [CrossRef] [PubMed]
- Walker, A. Breast milk as the gold standard for protective nutrients. J. Pediatr. 2010, 156, S3–S7. [Google Scholar] [CrossRef] [PubMed]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Langa, S.; Martín, V.; Maldonado, A.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Howie, P.W.; Forsyth, J.S.; Ogston, S.A.; Clark, A.; Florey, C.D. Protective effect of breast feeding against infection. BMJ 1990, 300, 11–16. [Google Scholar] [CrossRef] [PubMed]
- César, J.A.; Victora, C.G.; Barros, F.C.; Santos, I.S.; Flores, J.A. Impact of breast feeding on admission for pneumonia during postneonatal period in Brazil: Nested case-control study. BMJ 1999, 318, 1316–1320. [Google Scholar] [CrossRef] [PubMed]
- Munblit, D.; Peroni, D.; Boix-Amorós, A.; Hsu, P.; Land, B.; Gay, M.; Kolotilina, A.; Skevaki, C.; Boyle, R.; Collado, M.; et al. Human Milk and Allergic Diseases: An Unsolved Puzzle. Nutrients 2017, 9, 894. [Google Scholar] [CrossRef] [PubMed]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.S. The immune system of human milk: Antimicrobial, antiinflammatory and immunomodulating properties. Pediatr. Infect. Dis. J. 1993, 12, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C.; et al. Distinct Patterns in Human Milk Microbiota and Fatty Acid Profiles Across Specific Geographic Locations. Front. Microbiol. 2016, 7, 1619. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, M.; Wu, S.; Lebrilla, C.B.; Chapkin, R.S.; Ivanov, I.; Donovan, S.M. Fecal Microbiota Composition of Breast-Fed Infants is Correlated With Human Milk Oligosaccharides Consumed. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Fitzstevens, J.L.; Smith, K.C.; Hagadorn, J.I.; Caimano, M.J.; Matson, A.P.; Brownell, E.A. Systematic Review of the Human Milk Microbiota. Nutr. Clin. Pract. 2017, 32, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Chichlowski, M.; De Lartigue, G.; German, J.B.; Raybould, H.E.; Mills, D.A. Bifidobacteria Isolated from Infants and Cultured on Human Milk Oligosaccharides Affect Intestinal Epithelial Function. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.A.; Mills, D.A. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010, 18, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Ropero, M.P.; Martín, R.; Sierra, S.; Lara-Villoslada, F.; Rodríguez, J.M.; Xaus, J.; Olivares, M. Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J. Appl. Microbiol. 2007, 102, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Diaz-Ropero, M.P.; Martin, R.; Rodriguez, J.M.; Xaus, J. Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J. Appl. Microbiol. 2006, 101, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boix-Amorós, A.; Collado, M.C.; Mira, A. Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front. Microbiol. 2016, 7, 492. [Google Scholar] [CrossRef] [PubMed]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the Diversity and Temporal Stability of Bacterial Communities in Human Milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 2016, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.A.; Iyengar, R.S. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr. Res. 2015, 77, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Boix-Amorós, A.; Martinez-Costa, C.; Querol, A.; Collado, M.C.; Mira, A. Multiple Approaches Detect the Presence of Fungi in Human Breastmilk Samples from Healthy Mothers. Sci. Rep. 2017, 7, 13016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshioka, H.; Iseki, K.; Fujita, K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics 1983, 72, 317–321. [Google Scholar] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van De Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smilowitz, J.; Lebrilla, C.; Mills, D.; German, J.; Freeman, S. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 2014, 34, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Hanson, L.A. Comparative immunological studies of the immune globulins of human milk and of blood serum. Int. Arch. Allergy Appl. Immunol. 1961, 18, 241–267. [Google Scholar] [CrossRef] [PubMed]
- Telemo, E.; Hanson, L.A. Antibodies in milk. J. Mammary Gland Biol. Neoplasia 1996, 1, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.A. Role of nutrients and bacterial colonization in the development of intestinal host defense. J. Pediatr. Gastroenterol. Nutr. 2000, 30, S2–S7. [Google Scholar] [CrossRef] [PubMed]
- Newburg, D.S.; Walker, W.A. Protection of the Neonate by the Innate Immune System of Developing Gut and of Human Milk. Pediatr. Res. 2007, 61, 2–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayashree, S.; Bhan, M.K.; Kumar, R.; Bhandari, N.; Sazawal, S. Protection against neonatal rotavirus infection by breast milk antibodies and trypsin inhibitors. J. Med. Virol. 1988, 26, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Carrero, J.C.; Cervantes-Rebolledo, C.; Aguilar-Díaz, H.; Díaz-Gallardo, M.Y.; Laclette, J.P.; Morales-Montor, J. The role of the secretory immune response in the infection by Entamoeba histolytica. Parasite Immunol. 2007, 29, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.J.; Buckley, R.H.; Wakil, S.S.; McAllister, D.C.; David, R.J.; Faix, R.G. Elevated IgA concentration in milk produced by mothers delivered of preterm infants. J. Pediatr. 1981, 99, 389–393. [Google Scholar] [CrossRef]
- Jahani, S.; Shakiba, A.; Jahani, L. The Antimicrobial Effect of Lactoferrin on Gram-Negative and Gram-Positive Bacteria. Int. J. Infect. 2015, 2, e27954. [Google Scholar] [CrossRef]
- Fernandes, K.E.; Carter, D.A. The Antifungal Activity of Lactoferrin and Its Derived Peptides: Mechanisms of Action and Synergy with Drugs against Fungal Pathogens. Front. Microbiol. 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Van der Strate, B.W.; Beljaars, L.; Molema, G.; Harmsen, M.; Meijer, D.K. Antiviral activities of lactoferrin. Antivir. Res. 2001, 52, 225–239. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, G.A. Iron chelators as therapeutic agents against Pneumocystis carinii. Antimicrob. Agents Chemother. 1994, 38, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.F.; Kierszenbaum, F. Lactoferrin effects on phagocytic cell function. I. Increased uptake and killing of an intracellular parasite by murine macrophages and human monocytes. J. Immunol. 1985, 134, 4176–4183. [Google Scholar] [PubMed]
- Håversen, L.; Ohlsson, B.G.; Hahn-Zoric, M.; Hanson, L.A.; Mattsby-Baltzer, I. Lactoferrin down-regulates the LPS-induced cytokine production in monocytic cells via NF-κ B. Cell. Immunol. 2002, 220, 83–95. [Google Scholar] [CrossRef]
- Mastromarino, P.; Capobianco, D.; Campagna, G.; Laforgia, N.; Drimaco, P.; Dileone, A.; Baldassarre, M.E. Correlation between lactoferrin and beneficial microbiota in breast milk and infant’s feces. BioMetals 2014, 27, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Ellison, R.T.; Giehl, T.J.; Giehl, T.J. Killing of gram-negative bacteria by lactoferrin and lysozyme. J. Clin. Investig. 1991, 88, 1080–1091. [Google Scholar] [CrossRef] [PubMed]
- Lönnerdal, B. Bioactive Proteins in Human Milk: Mechanisms of Action. J. Pediatr. 2010, 156, S26–S30. [Google Scholar] [CrossRef] [PubMed]
- Strömqvist, M.; Falk, P.; Bergström, S.; Hansson, L.; Lönnerdal, B.; Normark, S.; Hernell, O. Human milk κ-casein and inhibition of Helicobacter pylori adhesion to human gastric mucosa. J. Pediatr. Gastroenterol. Nutr. 1995, 21, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Brenmoehl, J.; Ohde, D.; Wirthgen, E.; Hoeflich, A. Cytokines in milk and the role of TGF-β. Best Pract. Res. Clin. Endocrinol. Metab. 2018, 32, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Glocker, E.-O.; Kotlarz, D.; Boztug, K.; Gertz, E.M.; Schäffer, A.A.; Noyan, F.; Perro, M.; Diestelhorst, J.; Allroth, A.; Murugan, D.; et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 2009, 361, 2033–2045. [Google Scholar] [CrossRef] [PubMed]
- Berg, D.J.; Davidson, N.; Kühn, R.; Müller, W.; Menon, S.; Holland, G.; Thompson-Snipes, L.; Leach, M.W.; Rennick, D. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4+ TH1-like responses. J. Clin. Investig. 1996, 98, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Oddy, W.H.; Halonen, M.; Martinez, F.D.; Lohman, I.C.; Stern, D.A.; Kurzius-Spencer, M.; Guerra, S.; Wright, A.L. TGFβ in human milk is associated with wheeze in infancy. J. Allergy Clin. Immunol. 2003, 112, 723–728. [Google Scholar] [CrossRef]
- Kalliomäki, M.; Ouwehand, A.; Arvilommi, H.; Kero, P.; Isolauri, E. Transforming growth factor-β in breast milk: A potential regulator of atopic disease at an early age. J. Allergy Clin. Immunol. 1999, 104, 1251–1257. [Google Scholar] [CrossRef]
- Hassiotou, F.; Geddes, D.T.; Hartmann, P.E. Cells in Human Milk. J. Hum. Lact. 2013, 29, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Hepworth, A.R.; Metzger, P.; Tat Lai, C.; Trengove, N.; Hartmann, P.E.; Filgueira, L. Maternal and infant infections stimulate a rapid leukocyte response in breastmilk. Clin. Transl. Immunol. 2013, 2, e3. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Martos, I.; Jiménez, E.; de Andrés, J.; Rodríguez-Alcalá, L.M.; Tavárez, S.; Manzano, S.; Fernández, L.; Alonso, E.; Fontecha, J.; Rodríguez, J.M. Milk and blood biomarkers associated to the clinical efficacy of a probiotic for the treatment of infectious mastitis. Benef. Microbes 2016, 7, 305–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, K.; Labéta, M.O.; Schiffrin, E.J.; Donnet-Hughes, A. Soluble CD14 in human breast milk and its role in innate immune responses. Acta Odontol. Scand. 2001, 59, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Karaca, S.; Civelek, E.; Karaca, M.; Sahiner, U.M.; Ozgul, R.K.; Kocabas, C.N.; Polimanti, R.; Sekerel, B.E. Allergy-specific Phenome-Wide Association Study for Immunogenes in Turkish Children. Sci. Rep. 2016, 6, 33152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yolken, R.H.; Peterson, J.A.; Vonderfecht, S.L.; Fouts, E.T.; Midthun, K.; Newburg, D.S. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J. Clin. Investig. 1992, 90, 1984–1991. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dzidic, M.; Boix-Amorós, A.; Selma-Royo, M.; Mira, A.; Collado, M.C. Gut Microbiota and Mucosal Immunity in the Neonate. Med. Sci. 2018, 6, 56. https://doi.org/10.3390/medsci6030056
Dzidic M, Boix-Amorós A, Selma-Royo M, Mira A, Collado MC. Gut Microbiota and Mucosal Immunity in the Neonate. Medical Sciences. 2018; 6(3):56. https://doi.org/10.3390/medsci6030056
Chicago/Turabian StyleDzidic, Majda, Alba Boix-Amorós, Marta Selma-Royo, Alex Mira, and Maria Carmen Collado. 2018. "Gut Microbiota and Mucosal Immunity in the Neonate" Medical Sciences 6, no. 3: 56. https://doi.org/10.3390/medsci6030056





