Lentivirus Susceptibility in Iranian and German Sheep Assessed by Determination of TMEM154 E35K
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Blood Collection
2.2. Ethics Statements
2.3. DNA Extraction
2.4. Quantity and Quality Control of Extracted DNA
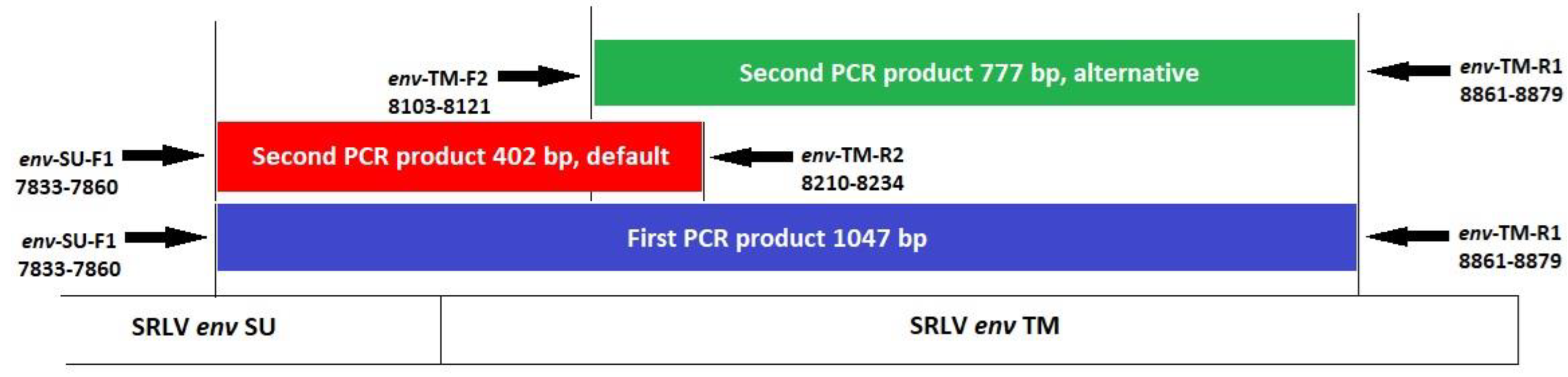
2.5. PCR Amplification for Detection of SRLVs in Iranian Sheep Samples (Set 1)
2.6. Serological Test for Detection of SRLVs in German Sheep Samples (Set 2)
2.7. TMEM154 Genotyping
2.8. Data Analysis
3. Results
3.1. SRLV Infection Status of Iranian and German Sheep Flocks (Sets 1 and 2)
3.2. TMEM154 Genotyping and Association Analyses (Chi-Square and Fisher’s Exact Test)
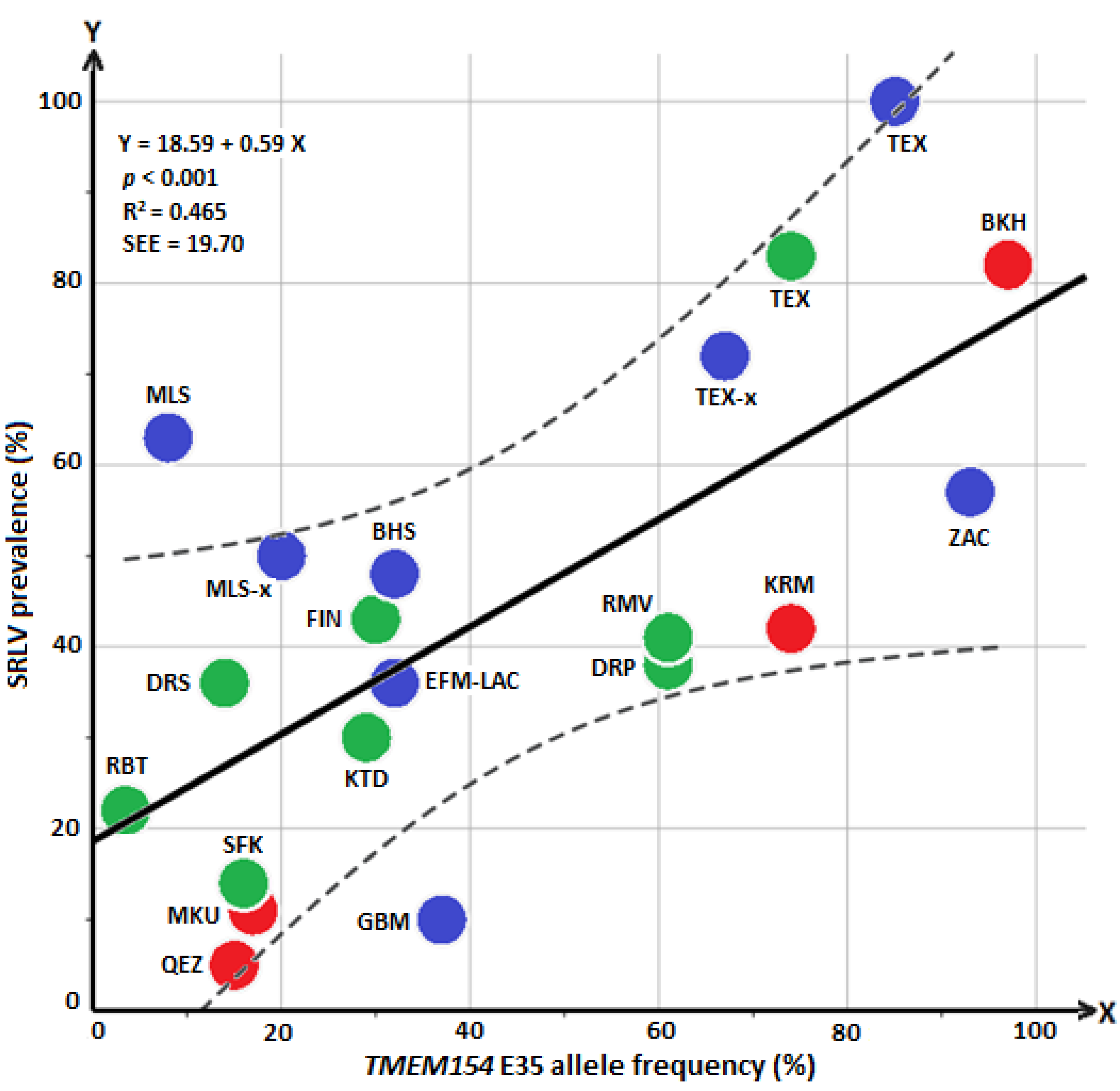
3.3. Regression Analysis Based on a Combination of Previously Published Data and Those of the Present Study
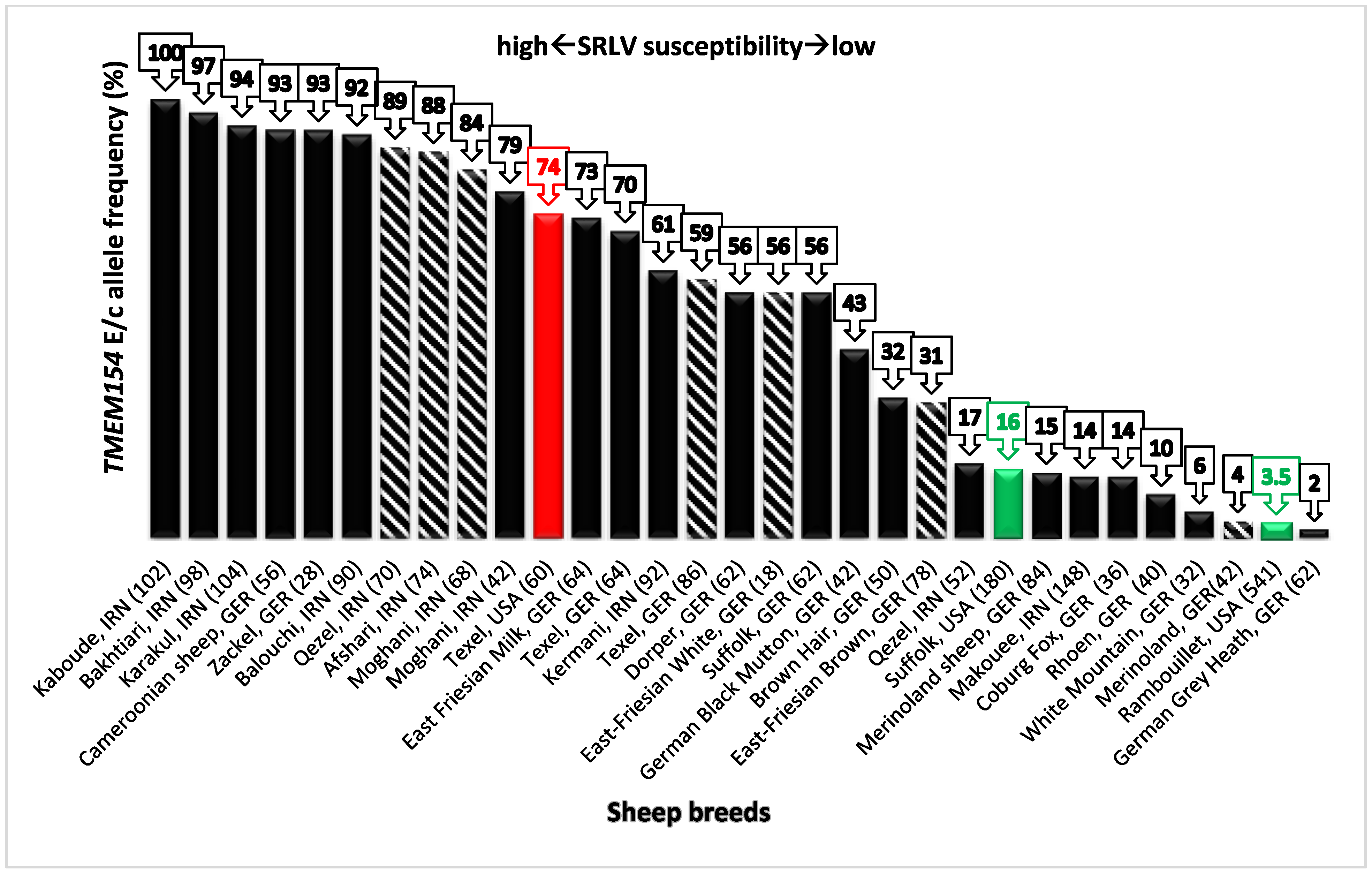
3.4. SRLV Susceptibility in Sheep Breeds of Iran and Germany Based on the Frequency of TMEM154 Alleles E and c
4. Discussion
4.1. Limitation of the Study
5. Conclusions
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
Appendix A
| Breed No. | Breed | Sheep (n) |
|---|---|---|
| 1 | German Grey Heath | 31 |
| 2 | White Moutain | 16 |
| 3 | Rhoen | 20 |
| 4 | Coburg Fox | 18 |
| 5 | Merinoland | 42 |
| 6 | German Black Mutton | 21 |
| 7 | Dorper | 31 |
| 8 | Suffolk | 31 |
| 9 | Texel | 32 |
| 10 | East Friesian Milk | 32 |
| 11 | Cameroonian sheep | 28 |
| Total | 302 |
| Gene | Primer Sequences (5′ → 3′) | Purpose of PCR | Product Size | Reference |
|---|---|---|---|---|
| TMEM154 | CCACAGGAGAGGAGRACACA (forward) GGGCACGTCTCCTGACAGTTT (reverse, FAM-labeled, matching with K allele) GGCACGTCTCCTGACAGTTC (reverse, HEX-labeled, matching with E allele) | genotyping TMEM154 E35K | 40/41 bp | [14] |
| TMEM154 | GCTAGACACTGCCAAGCTTC (forward) TGTCACTGAAACAAGTCATCACT (reverse) | sequencing TMEM154 region around E35K | 788 bp | [14] |
| env Surf-Trans | TAATAARRGTRAGAGCTTACACATATGG (env-SU-F1) CCTGACAGTCCACCCTTTC (env-TM-R1) | diagnosis of SRLV infection, first PCR | 1047 bp | this study |
| env Surf-Trans | TAATAARRGTRAGAGCTTACACATATGG (env-SU-F1) TTACACAGWAGTGTTGATAATGCCA (env-TM-R2) | diagnosis of SRLV infection, second PCR (default) | 402 bp | this study |
| env Surf-Trans | ATGCCATGGTACAGCATGT (env-TM-F2) CCTGACAGTCCACCCTTTC (env-TM-R1) | diagnosis of SRLV infection, second PCR (alternative) | 777 bp | this study |
| AMELX (Capra hircus) | CAGTAGCTCCAGCTCCAGCT (Aml.X-F1) GTGCATCCCTTCATTGGC (Aml.X-R1) | assessment of DNA quality | 300 bp | [24] |
| SRY (Capra hircus) | ATGAATAGAACGGTGCAATCG (Sry-F) GAAGAGGTTTTCCCAAAGGC (Sry-R) | assessment of DNA quality | 116 bp | [24] |
| Breed Subset | MV Status | TMEM154 Genotype Frequencies | ||
|---|---|---|---|---|
| (n Sheep) | (n Sheep) | KK | EK | EE |
| Bakhtiari, IRN (50) | neg (9) | 0.00 (0) | 0.11 (1) | 0.89 (8) |
| pos (41) | 0.00 (0) | 0.05 (2) | 0.95 (39) | |
| Qezel, IRN (20) | neg (19) | 0.74 (14) | 0.21 (4) | 0.05 (1) |
| pos (1) | 1.00 (1) | 0.00 (0) | 0.00 (0) | |
| Texel, GER (39) | neg (0) | 0.00 (0) | 0.00 (0) | 0.00 (0) |
| pos (39) | 0.00 (0) | 0.31 (12) | 0.69 (27) | |
| Zackel, GER (14) | neg (6) | 0.00 (0) | 0.17 (1) | 0.83 (5) |
| pos (8) | 0.00 (0) | 0.12 (1) | 0.88 (7) | |
References
- Peterhans, E.; Greenland, T.; Badiola, J.; Harkiss, G.; Bertoni, G.; Amorena, B.; Eliaszewicz, M.; Juste, R.A.; Kraßnig, R.; Lafont, J.-P.; et al. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet. Res. 2004, 35, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Reina, R.; Berriatua, E.; Luján, L.; Juste, R.; Sánchez, A.; de Andrés, D.; Amorena, B. Prevention strategies against small ruminant lentiviruses: An update. Vet. J. 2009, 182, 31–37. [Google Scholar] [CrossRef] [PubMed]
- White, S.; Knowles, D. Expanding possibilities for intervention against small ruminant lentiviruses through genetic marker-assisted selective breeding. Viruses 2013, 5, 1466–1499. [Google Scholar] [CrossRef] [PubMed]
- Pépin, M.; Vitu, C.; Russo, P.; Mornex, J.-F.; Peterhans, E. Maedi-visna virus infection in sheep: A review. Vet. Res. 1998, 29, 341–367. [Google Scholar] [PubMed]
- Ramírez, H.; Reina, R.; Amorena, B.; de Andrés, D.; Martínez, H.A. Small ruminant lentiviruses: Genetic variability, tropism and diagnosis. Viruses 2013, 5, 1175–1207. [Google Scholar] [CrossRef] [PubMed]
- Herrmann-Hoesing, L.M. Diagnostic assays used to control small ruminant lentiviruses. J. Vet. Diagn. Investig. 2010, 22, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Rosati, S.; Profiti, M.; Lorenzetti, R.; Bandecchi, P.; Mannelli, A.; Ortoffi, M.; Tolari, F.; Ciabatti, I.M. Development of recombinant capsid antigen/transmembrane epitope fusion proteins for serological diagnosis of animal lentivirus infections. J. Virol. Methods 2004, 121, 73–78. [Google Scholar] [CrossRef]
- Herrmann-Hoesing, L.M.; White, S.N.; Lewis, G.S.; Mousel, M.R.; Knowles, D.P. Development and validation of an ovine progressive pneumonia virus quantitative PCR. Clin. Vaccine Immunol. 2007, 14, 1274–1278. [Google Scholar] [CrossRef]
- Keen, I.E.; Hungerford, L.L.; Wittum, T.E.; Kwang, J.; Littledike, E.T. Rick factors for seroprevalence of ovine lentivirus in breeding ewe flocks in Nebraska, USA. Prev. Vet. Med. 1997, 30, 81–94. [Google Scholar] [CrossRef]
- Gates, N.L.; Winward, L.D.; Gorham, J.R.; Shen, D.T. Serologic survey of prevalence of ovine progressive pneumonia in Idaho range sheep. J. Am. Vet. Med. Assoc. 1978, 173, 1575–1577. [Google Scholar]
- Larruskain, A.; Jugo, B. Retroviral infections in sheep and goats: Small ruminant lentiviruses and host interaction. Viruses 2013, 5, 2043–2061. [Google Scholar] [CrossRef] [PubMed]
- Heaton, M.P.; Clawson, M.L.; Chitko-Mckown, C.G.; Leymaster, K.A.; Smith, T.P.L.; Harhay, G.P.; White, S.N.; Herrmann-Hoesing, L.M.; Mousel, M.R.; Lewis, G.S.; et al. Reduced lentivirus susceptibility in sheep with TMEM154 mutations. PLoS Genet. 2012, 8, e1002467. [Google Scholar] [CrossRef] [PubMed]
- Leymaster, K.A.; Chitko-McKown, C.G.; Clawson, M.L.; Harhay, G.P.; Heaton, M.P. Effects of TMEM154 haplotypes 1 and 3 on susceptibility to ovine progressive pneumonia virus following natural exposure in sheep. J. Anim. Sci. 2013, 91, 5114–5121. [Google Scholar] [CrossRef] [PubMed]
- Molaee, V.; Eltanany, M.; Lühken, G. First survey on association of TMEM154 and CCR5 variants with serological maedi-visna status of sheep in German flocks. Vet. Res. 2018, 49, 36. [Google Scholar] [CrossRef] [PubMed]
- Yaman, Y.; Keleş, M.; Aymaz, R.; Sevim, S.; Sezenler, T.; Önaldı, A.T.; Kaptan, C.; Başkurt, A.; Koncagül, S.; Öner, Y.; et al. Association of TMEM154 variants with visna/maedi virus infection in Turkish sheep. Small Rumin. Res. 2019, 177, 61–67. [Google Scholar] [CrossRef]
- Heaton, M.P.; Kalbfleisch, T.S.; Petrik, D.T.; Simpson, B.; Kijas, J.W.; Clawson, M.L.; Chitko-McKown, C.G.; Harhay, G.P.; Leymaster, K.A. Genetic testing for TMEM154 mutations associated with lentivirus susceptibility in sheep. PLoS ONE 2013, 8, 55490. [Google Scholar] [CrossRef]
- Tavakolian, J. An introduction to genetic resources of native farm animals in Iran; Animal Science Genetic Research Institute Press: Tehran, Iran, 2000. (In Persian) [Google Scholar]
- Sayari, M.; Soltanian, S. Serological and pathological study of Maedi-like diseases in mammary glands of sheep of Ahwaz region. Pajouhesh Sazandegi 2001, 2–7. (In Persian) [Google Scholar]
- Azizi, S.; Tajbakhsh, E.; Fathi, F.; Oryan, A.; Momtaz, H.; Goodarzi, M. Maedi in slaughtered sheep: A pathology and polymerase chain reaction study in southwestern Iran. Trop. Anim. Health Prod. 2012, 44, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Norouzi, B.; Taghavi-Razavizadeh, A.; Azizzadeh, M.; Mayameei, A.; Najar-Nezhad-Mashhadi, V. Serological study of small ruminant lentiviruses in sheep population of Khorasan-e-Razavi province in Iran. Vet. Res. Forum 2015, 6, 245–249. [Google Scholar]
- Sakhaee, E.; Khalili, M. Serological study of Maedi-Visna virus among sheep flocks in Kerman province of Iran. Iran. J. Virol. 2010, 4, 29–33. [Google Scholar] [CrossRef]
- Sasani, F.; Javanbakht, J.; Hemmatzadeh, F.; Moghadam, M.R.; Hassan, M.A.M. Evaluation of histopathological on maedi disease with serological confirmation in North-East of Iran. Res. J. Infect. Dis. 2013, 1, 5. [Google Scholar] [CrossRef]
- Montgomery, G.W.; Sise, J.A. Extraction of DNA from sheep white blood cells. New Zeal. J. Agric. Res. 1990, 33, 437–441. [Google Scholar] [CrossRef]
- Phua, A.C.Y.; Abdullah, R.B.; Mohamed, Z. A PCR-based sex determination method for possible application in caprine gender selection by simultaneous amplification of the Sry and Aml-X genes. J. Reprod. Dev. 2003, 49, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G. Practical Statistics for Medical Research; Chapman and Hall/CRC: London, UK, 1990; ISBN 9780412276309. [Google Scholar]
- Clarke, G.M.; Anderson, C.A.; Pettersson, F.H.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Basic statistical analysis in genetic case-control studies. Nat. Protoc. 2011, 6, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Sider, L.H.; Heaton, M.P.; Chitko-McKown, C.G.; Harhay, G.P.; Smith, T.P.; Leymaster, K.A.; Laegreid, W.W.; Clawson, M.L. Small ruminant lentivirus genetic subgroups associate with sheep TMEM154 genotypes. Vet. Res. 2013, 44, 64. [Google Scholar] [CrossRef] [PubMed]
- Straub, O.C. Maedi–Visna virus infection in sheep. History and present knowledge. Comp. Immunol. Microbiol. Infect. Dis. 2004, 27, 1–5. [Google Scholar] [CrossRef]
- Muz, D.; Oǧuzoǧlu, T.Ç.; Rosati, S.; Reina, R.; Bertolotti, L.; Burgu, I. First molecular characterization of visna/maedi viruses from naturally infected sheep in Turkey. Arch. Virol. 2013, 158, 559–570. [Google Scholar] [CrossRef] [PubMed]
| Flock No. | Country | Province | Breed | Sampled Sheep (n) | Positive 1 | |
|---|---|---|---|---|---|---|
| 1 | Iran | Western Azarbaijan | Makouee | 9 | 1 (11%) | |
| 2 | Iran | Western Azarbaijan | Makouee | 7 | 0 (0%) | |
| 3 | Iran | Western Azarbaijan | Makouee | 5 | 0 (0%) | |
| 4 | Iran | Western Azarbaijan | Makouee | 7 | 1 (14%) | |
| 5 | Iran | Western Azarbaijan | Makouee | 9 | 1 (11%) | |
| 6 | Iran | Western Azarbaijan | Makouee | 10 | 0 (0%) | |
| 7 | Iran | Western Azarbaijan | Makouee | 10 | 1 (10%) | |
| 8 | Iran | Western Azarbaijan | Makouee | 4 | 0 (0%) | |
| 9 | Iran | Western Azarbaijan | Makouee | 13 | 0 (0%) | |
| 10 | Iran | Western Azarbaijan | Qezel | 6 | 0 (0%) | |
| 11 | Iran | Western Azarbaijan | Qezel | 20 | 1 (5%) | |
| 12 | Iran | Ardebil | Moghani | 21 | 0 (0%) | |
| 13 | Iran | Chaharmahal-Va-Bakhtiari | Bakhtiari | 18 | 16 (89%) | |
| 14 | Iran | Chaharmahal-Va-Bakhtiari | Bakhtiari | 15 | 10 (67%) | |
| 15 | Iran | Chaharmahal-Va-Bakhtiari | Bakhtiari | 17 | 15 (88%) | |
| 16 | Iran | Fars | Kaboudeh | 10 | 0 (0%) | |
| 17 | Iran | Fars | Kaboudeh | 21 | 0 (0%) | |
| 18 | Iran | Fars | Kaboudeh | 12 | 0 (0%) | |
| 19 | Iran | Fars | Kaboudeh | 8 | 0 (0%) | |
| 20 | Iran | Kerman | Kermani | 14 | 3 (21%) | |
| 21 | Iran | Kerman | Kermani | 22 | 12 (55%) | |
| 22 | Iran | Kerman | Kermani | 10 | 0 (0%) | |
| 23 | Iran | Khorasan Razavi | Balouchi | 10 | 0 (0%) | |
| 24 | Iran | Khorasan Razavi | Balouchi | 2 | 0 (0%) | |
| 25 | Iran | Khorasan Razavi | Balouchi | 10 | 0 (0%) | |
| 26 | Iran | Khorasan Razavi | Balouchi | 23 | 0 (0%) | |
| 27 | Iran | Khorasan Razavi | Karakul | 12 | 0 (0%) | |
| 28 | Iran | Khorasan Razavi | Karakul | 11 | 0 (0%) | |
| 29 | Iran | Khorasan Razavi | Karakul | 9 | 0 (0%) | |
| 30 | Iran | Khorasan Razavi | Karakul | 20 | 0 (0%) | |
| 31 | Germany | Schleswig-Holstein | Texel | 39 | 39 (100%) | |
| 32 | Germany | Bayern | Merinoland | 49 | 31 (63%) | |
| 33 | Germany | Nordrhein-Westfalen | Brown Hair | 25 | 12 (48%) | |
| 34 | Germany | Baden-Württemberg | Zackel | 14 | 8 (57%) | |
| Breed Subset | MV Status | TMEM154 Genotype Frequencies | p Value | RR | 95% Cl | p Value | ||
|---|---|---|---|---|---|---|---|---|
| (n Sheep) | (n Sheep) | KK | EK | EE | ||||
| Makouee, IRN (35) | neg (31) | 0.71 (22) | 0.29 (9) | 0.00 (0) | 0.118 | 2.18 | 0.35 to 13.53 | 0.402 |
| pos (4) | 0.50 (2) | 0.25 (1) | 0.25 (1) | |||||
| Kermani, IRN (36) | neg (21) | 0.09 (2) | 0.24 (5) | 0.67 (14) | 0.047 | 0.48 | 0.24 to 0.96 | 0.038 |
| pos (15) | 0.33 (5) | 0.00 (0) | 0.67 (10) | |||||
| Merinoland, GER (49) | neg (18) | 1.00 (18) | 0.00 (0) | 0.00 (0) | 0.0197 | 1.782 | 1.359 to 2.337 | <0001 |
| pos (31) | 0.74 (23) | 0.26 (8) | 0.00 (0) | |||||
| Brown Hair, GER (25) | neg (13) | 0.85 (11) | 0.15 (2) | 0.00 (0) | 0.00002 | 20 | 1.313 to 304.494 | 0.0311 |
| pos (12) | 0.00 (0) | 0.83 (10) | 0.17 (2) | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molaee, V.; Otarod, V.; Abdollahi, D.; Lühken, G. Lentivirus Susceptibility in Iranian and German Sheep Assessed by Determination of TMEM154 E35K. Animals 2019, 9, 685. https://doi.org/10.3390/ani9090685
Molaee V, Otarod V, Abdollahi D, Lühken G. Lentivirus Susceptibility in Iranian and German Sheep Assessed by Determination of TMEM154 E35K. Animals. 2019; 9(9):685. https://doi.org/10.3390/ani9090685
Chicago/Turabian StyleMolaee, Vahid, Vahid Otarod, Darab Abdollahi, and Gesine Lühken. 2019. "Lentivirus Susceptibility in Iranian and German Sheep Assessed by Determination of TMEM154 E35K" Animals 9, no. 9: 685. https://doi.org/10.3390/ani9090685
APA StyleMolaee, V., Otarod, V., Abdollahi, D., & Lühken, G. (2019). Lentivirus Susceptibility in Iranian and German Sheep Assessed by Determination of TMEM154 E35K. Animals, 9(9), 685. https://doi.org/10.3390/ani9090685




