The Blood and Muscle Expression Pattern of the Equine TCAP Gene during the Race Track Training of Arabian Horses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
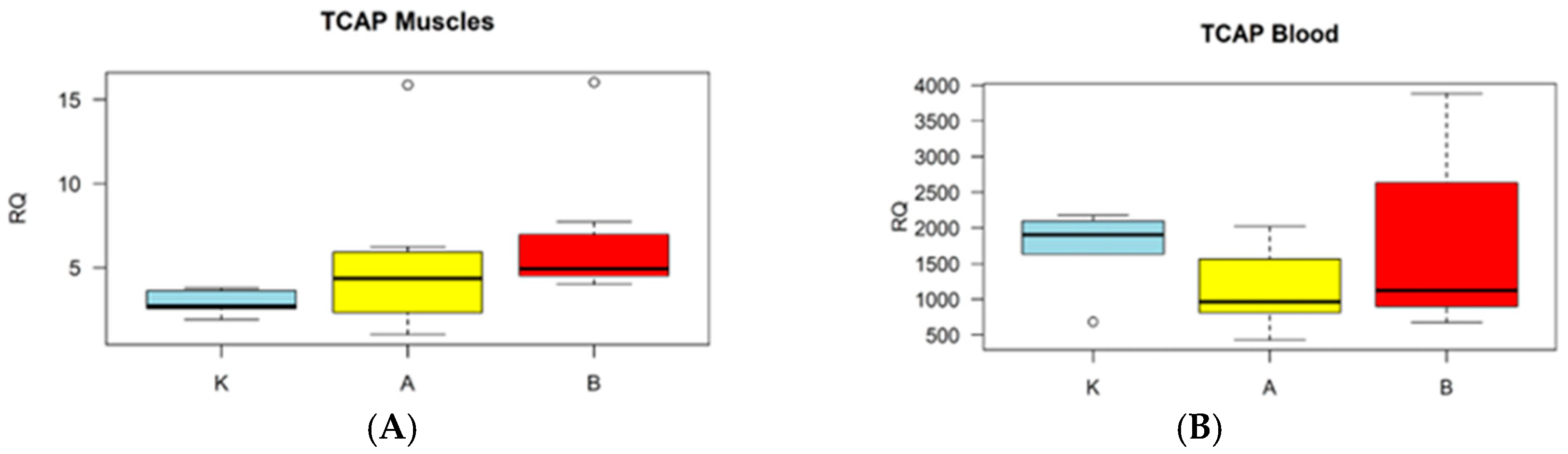
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Stefaniuk, M.; Ropka-Molik, K. RNA sequencing as a powerful tool in searching for genes influencing health and performance traits of horses. J. Appl. Genet. 2016, 57, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Hill, E.W.; Gu, J.; Eivers, S.S.; Fonseca, R.G.; McGivney, B.A.; Govindarajan, P.; Orr, N.; Katz, L.M.; MacHugh, D.E. A sequence polymorphism in MSTN predicts sprinting ability and racing stamina in thoroughbred horses. PLoS ONE 2010, 5, e8645. [Google Scholar] [CrossRef]
- Andersson, L.S.; Larhammar, M.; Memic, F.; Wootz, H.; Schwochow, D.; Rubin, C.J.; Patra, K.; Arnason, T.; Wellbring, L.; Hjälm, G.; et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 2012, 488, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Żukowski, K.; Piórkowska, K.; Gurgul, A.; Bugno-Poniewierska, M. Transcriptome profiling of Arabian horse blood during training regimens. BMC Genet. 2017, 18, 31. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Żukowski, K.; Piórkowska, K.; Bugno-Poniewierska, M. Exercise-induced modification of the skeletal muscle transcriptome in Arabian horses. Physiol. Genom. 2017, 149, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Piórkowska, K.; Żukowski, K.; Bugno-Poniewierska, M. Molecular characterization of the apoptosis-related SH3RF1 and SH3RF2 genes and their association with exercise performance in Arabian horses. BMC Vet. Res. 2018, 14, 237. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Musiał, A.D.; Piórkowska, K.; Szmatoła, T. Sequence analysis and expression profiling of the equine ACTN3 gene during exercise in Arabian horses. Gene 2019, 685, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Szmatoła, T.; Piórkowska, K.; Bugno-Poniewierska, M. The use of the SLC16A1 gene as a potential marker to predict race performance in Arabian horses. BMC Genet. 2019, in press. [Google Scholar]
- Gurgul, A.; Jasielczuk, I.; Semik-Gurgul, E.; Pawlina-Tyszko, K.; Stefaniuk-Szmukier, M.; Szmatoła, T.; Polak, G.; Tomczyk-Wrona, I.; Bugno-Poniewierska, M. A genome-wide scan for diversifying selection signatures in selected horse breeds. PLoS ONE 2019, 14, e0210751. [Google Scholar] [CrossRef]
- Gunn, H.M. Muscle, bone and fat proportions and the muscle distribution of Thoroughbreds and other horses. In Equine Exercise Physiology 2; ICEEP Publications: Davis, CA, USA, 1987; pp. 253–264. [Google Scholar]
- Crook, T.C.; Cruickshank, S.E.; McGowan, C.M.; Stubbs, N.; Wakeling, J.M.; Wilson, A.M.; Payne, R.C. Comparative anatomy and muscle architecture of selected hind limb muscles in the Quarter Horse and Arab. J. Anat. 2008, 212, 144–152. [Google Scholar] [CrossRef]
- Kawai, M.; Minami, Y.; Sayama, Y.; Kuwano, A.; Hiraga, A.; Miyata, H. Muscle fiber population and biochemical properties of whole body muscles in Thoroughbred horses. Anat. Rec. 2009, 292, 1663–1669. [Google Scholar] [CrossRef]
- Jürimäe, J.; Abernethy, P.J.; Blake, K.; McEniery, M.T. Changes in the myosin heavy chain isoform profile of the triceps brachii muscle following 12 weeks of resistance training. Eur. J. App. Physiol. Occup. Physiol. 1996, 74, 287–292. [Google Scholar] [CrossRef]
- Rivero, J.L.; Ruz, A.; Martí-Korff, S.; Estepa, J.C.; Aguilera-Tejero, E.; Werkman, J.; Sobotta, M.; Lindner, A. Effects of intensity and duration of exercise on muscular responses to training of thoroughbred racehorses. J. Appl. Physiol. 1985, 102, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.L.; Klueber, K.M. Skeletal muscle following tonic overload: Functional and structural analysis. Med. Sci. Sports Exerc. 1991, 23, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Pinotsis, N.; Lange, S.; Song, Y.H.; Popov, A.; Mavridis, I.; Mayans, O.M.; Gautel, M.; Wilmanns, M. Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature 2006, 439, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Osio, A.; Tan, L.; Chen, S.N.; Lombardi, R.; Nagueh, S.F.; Shete, S.; Roberts, R.; Willerson, J.T.; Marian, A.J. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ. Res. 2007, 100, 766–768. [Google Scholar] [CrossRef] [PubMed]
- Faulkne, R.G.; Pallavicini, A.; Comelli, A.; Salamon, M.; Bortoletto, G.; Ievolella, C.; Trevisan, S.; Kojic, S.; Vecchia, F.D.; Laveder, P.; et al. FATZ, a filamin-, actinin-, and telethonin-binding protein of the Z-disc of skeletal muscle. J. Biol. Chem. 2000, 275, 41234–41242. [Google Scholar] [CrossRef] [PubMed]
- Kojic, S.; Medeot, E.; Guccione, E.; Krmac, H.; Zara, I.; Martinelli, V.; Valle, G.; Faulkner, G. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. J. Mol. Biol. 2004, 339, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Stefaniuk, M.; Ropka-Molik, K.; Piórkowska, K.; Bereta, A.; Szpar, P.; Czerwonka, Z.; Podstawski, Z. Evaluation of minimally invasive muscle biopsy method for genetic analysis in horse. Ann. Anim. Sci. 2015, 15, 621–627. [Google Scholar] [CrossRef]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 1993, 15, 532–537. [Google Scholar] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; Available online: http://www.R-project.org (accessed on 17 March 2019).
- Huxley, H.; Hanson, J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature 1954, 173, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Herzog, W. The multiple roles of titin in muscle contraction and force production. Biophys. Rev. 2018, 10, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Gregorio, C.C.; Trombitás, K.; Centner, T.; Kolmerer, B.; Stier, G.; Kunke, K.; Suzuki, K.; Obermayr, F.; Herrmann, B.; Granzier, H.; et al. The NH2 terminus of titin spans the Z-disc: Its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J. Cell. Biol. 1998, 143, 1013–1027. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.H.; Gao, M.; Pinotsis, N.; Wilmanns, M.; Schulten, K. Mechanical strength of the titin Z1Z2-telethonin complex. Structure 2006, 3, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Knoll, S.; Fürst, K.; Thomas, S.; Villanueva Baselga, S.; Stoll, A.; Schaefer, S.; Pützer, B.M. Dissection of cell context-dependent interactions between HBx and p53 family members in regulation of apoptosis: A role for HBV-induced HCC. Cell Cycle 2011, 10, 3554–3565. [Google Scholar] [CrossRef] [PubMed]
- Dahlqvist, J.R.; Voss, L.G.; Lauridsen, T.; Krag, T.O.; Vissing, J. A pilot study of muscle plasma protein changes after exercise. Muscle Nerve 2014, 49, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Yang, J.; Zhu, J.; Xu, X. Depletion of zebrafish Tcap leads to muscular dystrophy via disrupting sarcomere-membrane interaction, not sarcomere assembly. Hum. Mol. Genet. 2009, 18, 4130–4140. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, M.; Horita, Z.; Nemoto, M.; Langdon, T.G. Processing of metals by equal-channel angular pressing. J. Mate. Sci. 2001, 36, 2835–2843. [Google Scholar] [CrossRef]
- Nicholas, G.; Thomas, M.; Langley, B.; Somers, W.; Patel, K.; Kemp, C.F.; Sharma, M.; Kambadur, R. Titin-cap associates with, and regulates secretion of, Myostatin. J. Cell Physiol. 2002, 1, 120–131. [Google Scholar] [CrossRef]
- Vega, R.B.; Konhilas, J.P.; Kelly, D.P.; Leinwand, L.A. Molecular Mechanisms Underlying Cardiac Adaptation to Exercise. Cell Metab. 2017, 25, 1012–1026. [Google Scholar] [CrossRef] [PubMed]
- Frey, N.; Olson, E.N. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 2002, 277, 13998–14004. [Google Scholar] [CrossRef]
- Franzini-Armstrong, C.; Protasi, F.; Ramesh, V. Shape, size, and distribution of Ca (2+) release units and couplons in skeletal and cardiac muscles. Biophys. J. 1999, 77, 1528–1539. [Google Scholar] [CrossRef]
- Hayashi, T.; Arimura, T.; Itoh-Satoh, M.; Ueda, K.; Hohda, S.; Inagaki, N.; Takahashi, M.; Hori, H.; Yasunami, M.; Nishi, H.; et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J. Am. Coll. Cardiol. 2004, 44, 2192–2201. [Google Scholar] [CrossRef]
| Ensemble Accession Number | Gene Symbol | Primer Sequence | Amplicon Length |
|---|---|---|---|
| ENSECAG00000011860 | TCAP | TCAPF:GGCTGAATGGAAGGATCTGA TCAPR:TGGTAGGGCAGCTGGTACTC | 192 |
| ENSECAG00000022051 | GAPDH | GAPDHF:GCCGTAACTTCTGTGCTGTG GAPDHR:AATGAAGGGGTCATTGATGG | 156 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefaniuk-Szmukier, M.; Szmatoła, T.; Łątka, J.; Długosz, B.; Ropka-Molik, K. The Blood and Muscle Expression Pattern of the Equine TCAP Gene during the Race Track Training of Arabian Horses. Animals 2019, 9, 574. https://doi.org/10.3390/ani9080574
Stefaniuk-Szmukier M, Szmatoła T, Łątka J, Długosz B, Ropka-Molik K. The Blood and Muscle Expression Pattern of the Equine TCAP Gene during the Race Track Training of Arabian Horses. Animals. 2019; 9(8):574. https://doi.org/10.3390/ani9080574
Chicago/Turabian StyleStefaniuk-Szmukier, Monika, Tomasz Szmatoła, Joanna Łątka, Bogusława Długosz, and Katarzyna Ropka-Molik. 2019. "The Blood and Muscle Expression Pattern of the Equine TCAP Gene during the Race Track Training of Arabian Horses" Animals 9, no. 8: 574. https://doi.org/10.3390/ani9080574
APA StyleStefaniuk-Szmukier, M., Szmatoła, T., Łątka, J., Długosz, B., & Ropka-Molik, K. (2019). The Blood and Muscle Expression Pattern of the Equine TCAP Gene during the Race Track Training of Arabian Horses. Animals, 9(8), 574. https://doi.org/10.3390/ani9080574





