Detection of Bovine TMEM95 p.Cys161X Mutation in 13 Chinese Indigenous Cattle Breeds
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Collection and Genomic DNA Isolation
2.2. Primer Design and Genotyping by T-ARMS-PCR and Forced PCR-RFLP Methods
2.3. Genomic DNA Pool Construction and DNA Sequencing
3. Results
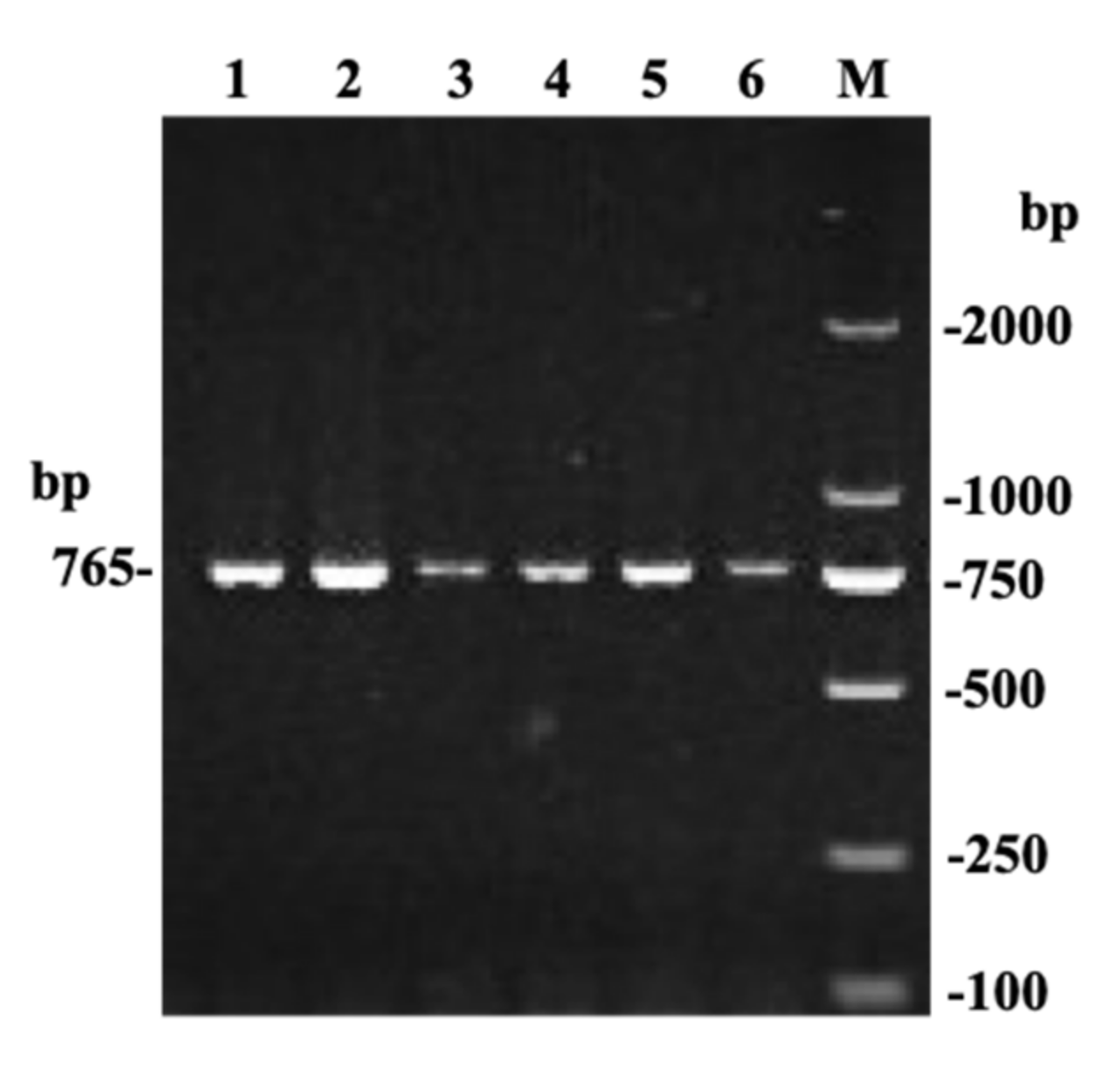
3.1. SNP Identification and Genotyping of the Bovine TMEM95 Gene by T-ARMS-PCR and Forced PCR-RFLP
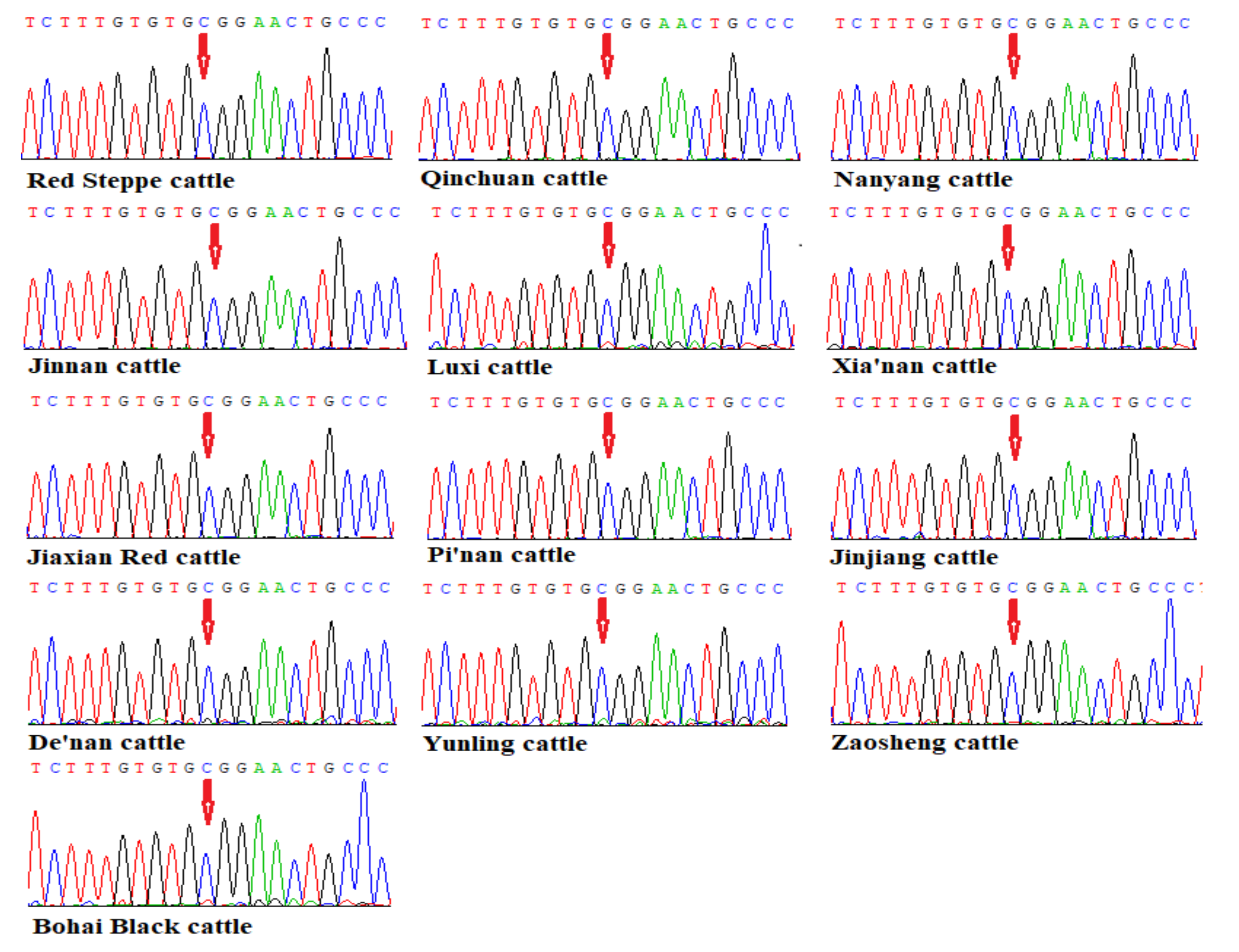
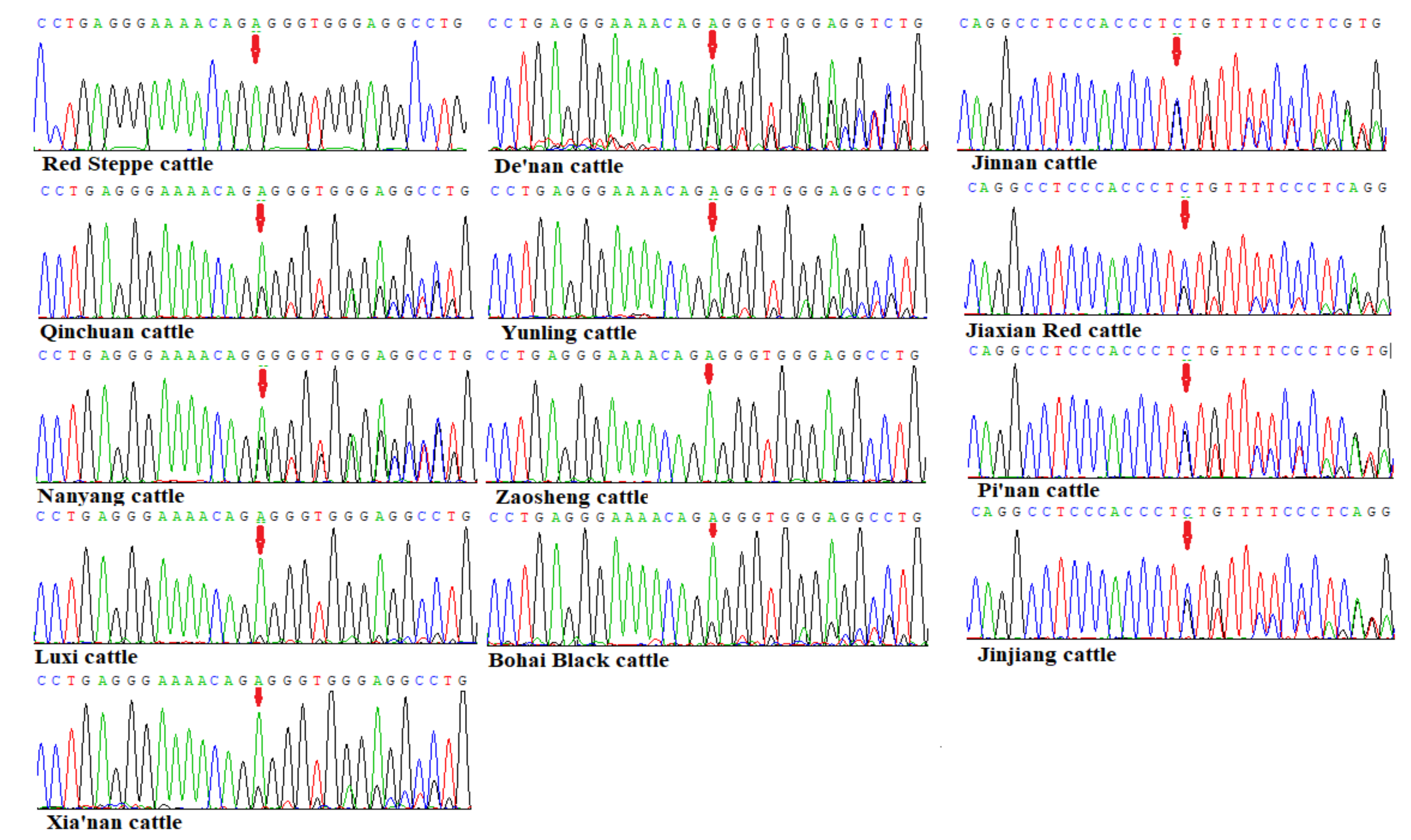
3.2. Sequencing Results of the TMEM95 Gene in 13 Chinese Indigenous Cattle Breeds
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.R.; Franke, D.E.; Saxton, A.M.; Turner, J.W. Two-, three- and four-breed rotational crossbreeding of beef cattle: Reproductive traits. J. Anim. Sci. 1990, 68, 1536–1546. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.; Van Arendonk, J.A. Potential improvements in rate of genetic gain from marker-assisted selection in dairy cattle breeding schemes. J. Dairy Sci. 1992, 75, 1651–1659. [Google Scholar] [CrossRef]
- Donaldson, L.E. Artificial insemination of beef cattle. Aust. Vet. J. 1976, 52, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Thundathil, J.C.; Dance, A.L.; Kastelic, J.P. Fertility management of bulls to improve beef cattle productivity. Theriogenology 2016, 86, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Kastelic, J.P.; Thundathil, J.C. Breeding soundness evaluation and semen analysis for predicting bull fertility. Reprod. Domest. Anim. 2008, 43, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Pausch, H.; Kölle, S.; Wurmser, C.; Schwarzenbacher, H.; Emmerling, R.; Jansen, S.; Trottmann, M.; Fuerst, C.; Götz, K.U.; Fries, R. A nonsense mutation in TMEM95 encoding a nondescript transmembrane protein causes idiopathic male subfertility in cattle. PLoS Genet. 2014, 10, e1004044. [Google Scholar] [CrossRef]
- Gurunath, S.; Pandian, Z.; Anderson, R.A.; Bhattacharya, S. Defining infertility–a systematic review of prevalence studies. Hum. Reprod. Update 2011, 17, 575–588. [Google Scholar] [CrossRef]
- Fernandez-Fuertes, B.; Kölle, S.; Lonergan, P. Subfertility in bulls carrying a nonsense mutation in TMEM95 is due to failure to penetrate the zona pellucida. Reprod. Fertil. Dev. 2016, 29, 109–110. [Google Scholar] [CrossRef]
- Fernandez-Fuertes, B.; Laguna-Barraza, R.; Fernandez-Gonzalez, R.; Gutierrez-Adan, A.; Blanco-Fernandez, A.; O’Doherty, A.M.; Di Fenza, M.; Kelly, A.K.; Kölle, S.; Lonergan, P. Subfertility in bulls carrying a nonsense mutation in transmembrane protein 95 is due to failure to interact with the oocyte vestments. Biol. Reprod. 2017, 97, 50–60. [Google Scholar] [CrossRef]
- Fernandez-Fuertes, B.; Lonergan, P. Sperm Transmembrane Protein 95 (TMEM95) is required for sperm–oocyte interaction and successful fertilization but not mucus penetration in cattle. Reprod. Fertil. Dev. 2018, 30, 209–210. [Google Scholar] [CrossRef]
- Yang, Q.X; Zhang, S.H.; Cao, X.K.; Liu, L.L.; Lei, C.Z.; Qi, X.L.; Lin, F.P.; Qu, W.D.; Qi, X.S.; Chen, H.; et al. Application of mathematical expectation (ME) strategy on detecting the low frequency mutation: An example for evaluating 14 bp InDel of the PRNP gene 3′UTR in four Chinese indigenous cattle breeds. Prion 2016, 10, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Yang, Q.; Wang, K.; Yan, H.L.; Pan, C.Y.; Chen, H.; Liu, J.W.; Zhu, H.J.; Qu, L.; Lan, X.Y. Two strongly linked single nucleotide polymorphisms (Q320P and V397I) in GDF9 gene are associated with litter size in cashmere goats. Theriogenology 2019, 125, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Dhillon, S.; Ke, X.Y.; Collins, A.R.; Day, I.N. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001, 29, e88. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.Y.; Collins, A.R.; Ye, S. PCR designer for restriction analysis of various types of sequence mutation. Bioinformatics 2002, 18, 1688–1689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Dang, Y.; Zhang, Q.; Qin, Q.; Lei, C.; Chen, H.; Lan, X. Tetra-primer amplification refractory mutation system PCR (T-ARMS-PCR) rapidly identified a critical missense mutation (P236T) of bovine ACADVL gene affecting growth traits. Gene 2015, 559, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Yang, Q.; Wang, K.; Zhang, S.H.; Pan, C.Y.; Chen, H.; Qu, L.; Yan, H.L.; Lan, X.Y. A novel 12-bp InDel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats. Anim. Genet. 2017, 48, 735–736. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yan, H.L.; Li, J.; Xu, H.; Wang, K.; Zhu, H.J.; Chen, H.; Qu, L.; Lan, X.Y. A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size. Anim. Genet. 2017, 48, 499–500. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, S.L.; Li, J.; Wang, X.; Peng, K.; Lan, X.Y.; Pan, C.Y. Development of a touchdown multiplex PCR method for simultaneously rapidly detecting three novel insertion/deletions (InDels) within one gene: An example for goat GHR gene. Anim. Biotechnol. 2018. [Google Scholar] [CrossRef]
- Cui, Y.; Yan, H.L.; Wang, K.; Xu, H.; Zhang, X.L.; Zhu, H.J.; Liu, J.W.; Qu, L.; Lan, X.Y.; Pan, C.Y. Insertion/deletion within the KDM6A gene is significantly associated with litter size in goat. Front. Genet. 2018, 9, 91. [Google Scholar] [CrossRef]
- Kang, Z.; Jiang, E.; Wang, K.; Pan, C.; Chen, H.; Yan, H.; Zhu, H.; Liu, J.; Qu, L.; Lan, X. Goat membrane associated ring-CH-type finger 1 (MARCH1) mRNA expression and association with litter size. Theriogenology 2019, 128, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cai, H.; Yang, Q.; Shi, T.; Pan, C.; Lei, C.; Dang, R.; Chen, H.; Lan, X. Identification of novel alternative splicing transcript and expression analysis of bovine TMEM95 gene. Gene 2016, 575, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, O.; Convertini, P.; Zhang, Z.Y.; Wen, Y.; Shen, M.L.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–30. [Google Scholar] [CrossRef] [PubMed]
| Methods | Primers (5′-3′) | a Regions | Genotype Pattern (bp) |
|---|---|---|---|
| T-ARMS-PCR | F(outer): CCTCACCCCCACCCAGATCTCTGAGCTC (1731-1758) | Intron 5 to Exon 6 | Product of outer primer = 312 CC = 312 + 196; CA = 312 + 196 + 168; AA = 312 + 168. |
| R(outer): ACCTGAGGGAAAACAGAGGGTGGGAGGC (2015-2042) | |||
| F(inner A): CTCGGATCCTGCTCCTCTTTGTGCGC (1847-1872) | |||
| R(inner C): GGGACACCCAGGAGCAGGGCAGTTTCT (1872-1898) | |||
| Forced PCR-RFLP | F: AAGCTCGGATCCTGCTCCTCTTTGTGCG (1844-1869) | Exon 6 | Hha I (GCG↓C) AA = 253; AC = 253 + 228 + 25; CC = 228 + 25. |
| R: GGCTAGGCTCTGTCCTCGTTT (2076-2096) | |||
| Sequencing | F: GTGAGTAAGAAAGGGAAGGGGTCG (1498-1519) | Exon 4 to Exon 6 | 765 |
| R: ACCATCTGACACTGGGACTA (2243-2262) |
| Breeds | Total (n) | Allelic Frequency (%) | Genotypic Frequency (%) | |||
|---|---|---|---|---|---|---|
| Cys | X | Cys/Cys | Cys/X | X/X | ||
| Red Steppe cattle | 135 | 270 (100) | 0 (0) | 135 (100) | 0 (0) | 0 (0) |
| Qinchuan cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| Nanyang cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| Jinnan cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| Luxi cattle | 30 | 60 (100) | 0 (0) | 30 (100) | 0 (0) | 0 (0) |
| Xia’nan cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| Jiaxian Red cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| Pi’nan cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| Jinjiang cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| De’nan cattle | 30 | 60 (100) | 0 (0) | 30 (100) | 0 (0) | 0 (0) |
| Yunling cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| Zaosheng cattle | 30 | 60 (100) | 0 (0) | 30 (100) | 0 (0) | 0 (0) |
| Bohai Black cattle | 60 | 120 (100) | 0 (0) | 60 (100) | 0 (0) | 0 (0) |
| Total | 765 | 1530 (100) | 0 (0) | 765 (100) | 0 (0) | 0 (0) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Peng, K.; Zhang, G.; Cao, Y.; Zhang, M.; Chen, H.; Lei, C.; Lan, X.; Zhao, Y. Detection of Bovine TMEM95 p.Cys161X Mutation in 13 Chinese Indigenous Cattle Breeds. Animals 2019, 9, 444. https://doi.org/10.3390/ani9070444
Zhang S, Peng K, Zhang G, Cao Y, Zhang M, Chen H, Lei C, Lan X, Zhao Y. Detection of Bovine TMEM95 p.Cys161X Mutation in 13 Chinese Indigenous Cattle Breeds. Animals. 2019; 9(7):444. https://doi.org/10.3390/ani9070444
Chicago/Turabian StyleZhang, Sihuan, Kun Peng, Guoliang Zhang, Yang Cao, Meng Zhang, Hong Chen, Chuzhao Lei, Xianyong Lan, and Yumin Zhao. 2019. "Detection of Bovine TMEM95 p.Cys161X Mutation in 13 Chinese Indigenous Cattle Breeds" Animals 9, no. 7: 444. https://doi.org/10.3390/ani9070444
APA StyleZhang, S., Peng, K., Zhang, G., Cao, Y., Zhang, M., Chen, H., Lei, C., Lan, X., & Zhao, Y. (2019). Detection of Bovine TMEM95 p.Cys161X Mutation in 13 Chinese Indigenous Cattle Breeds. Animals, 9(7), 444. https://doi.org/10.3390/ani9070444




