1. Introduction
The chicken karyotype consists of 39 chromosome pairs, including one sex chromosome pair. With the development of sequencing technology, the chicken W chromosome was sequenced recently [
1], but some genomic regions remain poorly annotated due to their complex sequence composition. These include microchromosome 16 (GGA16) and 19 (GGA19). Microchromosomes are denser in genetic material compared with macrochromosomes. The size of GGA16 is estimated at 9–11 Mb, but only 0.5 Mb have been assembled with known sequences [
2,
3]. GGA16 is of great interest because many genes on this chromosome are closely related to infectious diseases responses. The nucleolus organizing region (NOR), which is a cluster of genes that encode ribosomal RNAs 28S, 18S and 5.8S, is located on GGA16. The NOR copy number and expression level affect cell growth, cell differentiation and protein synthesis capacity [
4,
5]. The chicken major histocompatibility complex (MHC) has also been mapped to GGA16 [
6]. Proteins encoded by the MHC are widely involved in viral and bacterial disease resistance and immune response traits of chickens [
7]. Class I and II MHCs include the
BF and
BL genes which determine resistance and susceptibility to various diseases [
8].The class IV MHC encodes erythrocyte antigens, which are used for MHC haplotype classification. CD1 genes constitute a family of conserved cell-surface proteins with crucial roles in the immune system related to antigen presentation to T cells encoded by MHC [
5,
9].
There are three types of immunoglobulins, including IgM, IgA and IgY, which have been identified in chickens. IgY is the major antibody in chickens [
10]. IgY accumulates in egg yolk and provides the offspring with powerful immunity against avian pathogens [
10]. A genome-wide association study (GWAS) was performed for IgY levels and antibody responses to sheep red-blood-cells (SRBC) using the chicken 60k high-density single nucleotide polymorphism (SNP) array [
11]. There were five SNPs found to be significantly associated with the serum IgY levels. These SNPs were found on chicken chromosome 16: 5,661,311 to 4,412,041 and chromosome 11: 4,412,041 to 5,661,311 [
11]. There were five SNPs in chromosome 19 (spanning 8,974,480 to 9,730,805) and three SNPs in chromosome 11 (spanning 13,531,246 to 14,463,528) which were shown to be associated with antibody responses to sheep red-blood-cells (SRBC).
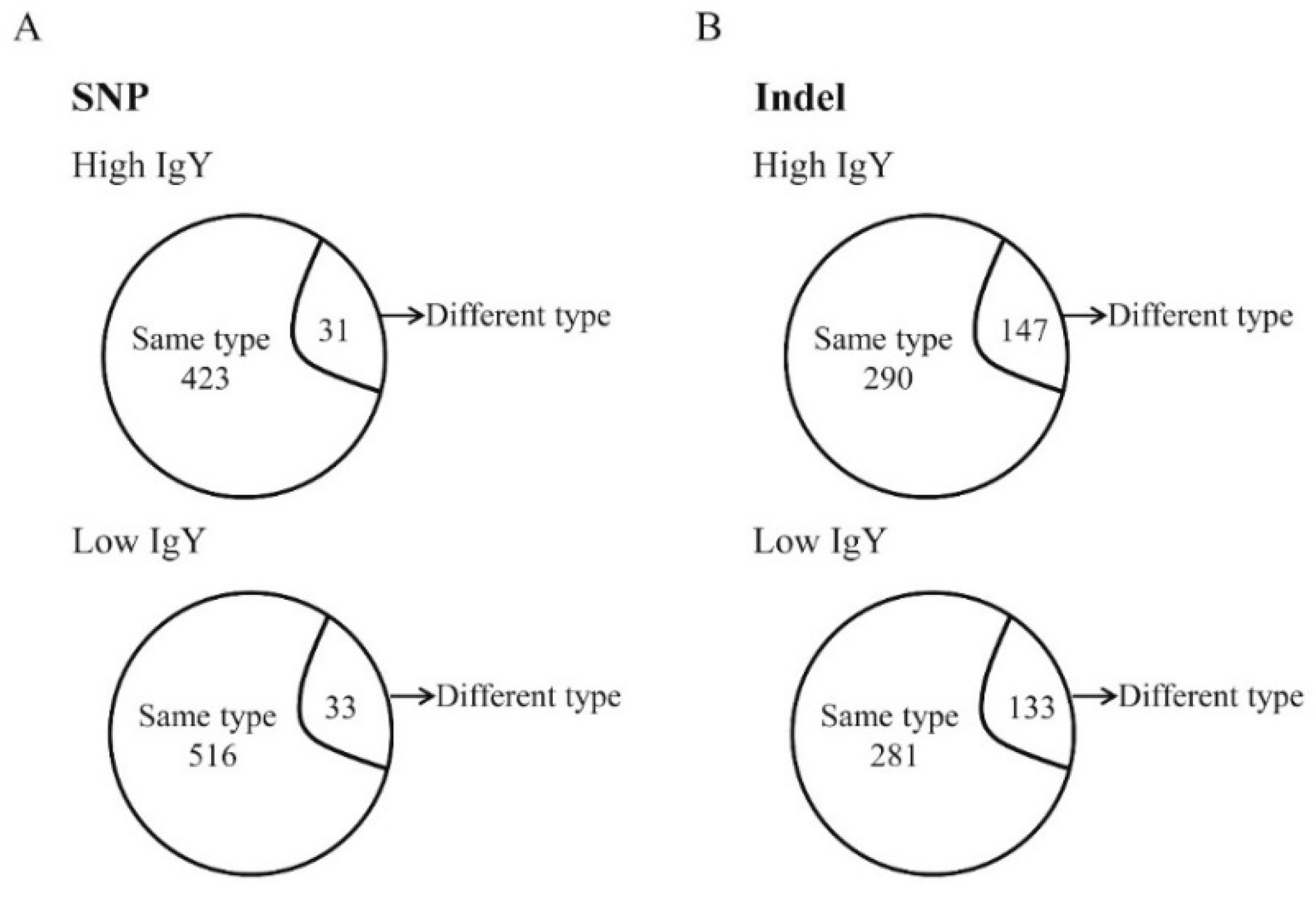
In the present study, SNPs were screened in the above-mentioned genomic regions between high and low IgY chickens in the White Leghorn and Beijing-You lines. The capture chip was designed based on all known sequences of chromosome 16 and the genomic regions covering SNPs that have been associated with IgY levels and antibody responses to SRBC in other chromosomes. The captured DNA was then sequenced using high-throughput sequencing technology. Thousands of genetic variations were identified between the high and low IgY chickens, indicating substantial genetic differences between the lines.
2. Materials and Methods
2.1. Animals and Sample Collection
The animal experiments were carried out in accordance with the Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China) and the study was approved by the Animal Management Committee of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (IAS-CAAS, Beijing, China). Ethical approval regarding animal survival was given by the animal ethics committee of IAS-CAAS (approval number: IASCAAS-AE20140615).
Chickens used in the present study were of the White Leghorn and Beijing-You lines. Beijing-You chickens, who were not subjected to artificial selection, were chosen for this study according to their IgY content. White Leghorn chickens were the result of 7 generations of artificial selection based on anti-SRBC titers. The IgY levels of 527 White Leghorn and 726 Beijing-You chickens were measured by enzyme-linked immune sorbent assay (ELISA). Chickens were divided into high and low IgY groups based on the IgY levels. There was a 5.1-fold difference in the IgY levels between high and low IgY White Leghorn chickens, and a 6.2-fold difference between high-and low-IgY Beijing-You chickens (
Table 1).
Forty animals of both varieties with the highest and lowest IgY levels divided into four groups. Each group was randomly and equally divided into four repeats, each containing 10 individuals. Blood was collected from each animal and genomic DNA was isolated using phenol–chloroform extraction. DNA from the 10 individuals of each repeat was pooled together and sent for high-throughput sequencing by HiSeq2500 for SNPs and Indels.
Genomic DNA was obtained from 34 and 37 animals from lines that were selected for susceptibility and resistance to Marek’s disease respectively, from researchers at Aarhus University (Aarhus, Denmark). This DNA was utilized for sequence analysis of BF1.
2.2. ELISA
Serum IgY levels of chickens from both lines were determined using an IgY Chicken ELISA Kit (Abcam) according to the manufacturer’s instructions.
2.3. Capture Sequencing, Sequence Alignment and Data Analysis
The sequence capture chip was designed by Roche Nimblegen to cover a 3.268 Mb region accounting for 97.73% of the 3.371 Mb region associated with the specific phenotypes of interest. Sequences generated by high-throughput sequencing were aligned to the chicken reference genome (galGal4) by SOAP aligner. High confidence SNPs and Indels were identified by SAM tools with the criteria of coverage >4 and FDR < 0.01. Sequences generated by high-throughput sequencing were aligned to the chicken reference genome (galGal4) to identify SNPs and Indels.
2.4. Validation of SNPs and Indels
A total of 24 SNPs covering the second-generation sequencing region were randomly selected for validation. A mutation common to both Beijing-You and White Leghorns, plus a unique Indel, with high-IgY were validated. Validation populations were selected from the high-throughput sequence samples. In particular, DNA was pooled from 10 individuals in each group to verify SNPs. Regarding the Indel validation, all sequencing samples were used. Primers were designed around the SNPs and Indels, and PCR amplifications of multiple samples were carried out using High Fidelity PCR Super Mix (Transgen, Biotech). PCR products were sequenced using an ABI PRISM® 3100 Genetic Analyzer.
4. Discussion
Based on the results of a previous genome-wide association study (GWAS),a capture-sequencing approach was performed to identify variants in defined genomic regions among chickens with high- and low-IgY levels in the White Leghorn and Beijing-You line. In total, 35,154 SNPs and 23,309 Indels were identified. Among these mutations, of particular interest were the genetic mutations in proximal promoters and CDS regions, as such mutations lead to changes in promoter activity or amino acid sequences. Transcription factors can bind to promoter regions and initiate gene transcription [
13]. Genetic mutations in these regions can block the binding of transcription factors and thus suppress gene transcriptional activity. Amino acid changes in critical positions may even cause changes in protein structure. There were three and five genes shown to contain SNPs in proximal promoter regions in the high- and low-IgY chickens, among which the
TAP2 gene was identified in both groups. There were five genes, including
TAP2,
BLEC2,
BF1,
BF2,
TAP1 and
TRIM41, that have common non-synonymous SNPs in CDS regions in the high IgY group in both White Leghorn and Beijing-You chickens.
TAP1,
BF1,
BF2 and
TRIM7.1 contained non-synonymous SNPs in their CDS regions in the low-IgY groups of both lines. Both the high- and low-IgG groups contained SNPs in the CDS regions of
TAP1, BF1 and
BF2.
Many of the genes with mutations in the proximal promoter and CDS regions play crucial roles in host immune function. TAP1 forms a heterodimeric complex with TAP2 that has transporter activity. The TAP complex is involved in transporting antigens to the endoplasmic reticulum to associate with MHC class I and promote immune responses against infection [
14]. ICP47 encoded by the herpes simplex virus (HSV) is known to inhibit peptide binding to TAP and impairs peptide loading into MHC class I molecules [
14,
15]. In the present study, SNPs were found in
TAP1 in the CDS regions of high-IgY chickens and in both the CDS regions and proximal promoter regions of low-IgY chickens. SNPs were also found in
TAP2 in CDS regions and proximal promoter, but only in the high IgY chickens. In addition to the SNPs, a 9 bp deletion was identified in the region overlapping the TAP 3′UTR, and the genomic DNA region of
BF1, indicating the potential impact of
TAP1 and
TAP2 mutations on chicken immune molecules.
The chicken MHC B region encodes class I and II molecules. Different haplotypes determine the resistance or susceptibility to infectious diseases [
8]. BF1 and BF2 are encoded by the chicken MHC class I genes. As the dominant MHC class I gene,
BF2 is transcribed more abundantly than
BF1 [
16]. Both BF1 and BF2 are deeply involved in antigen processing and immune response. In the present study, SNPs located in both the proximal promoter regions and CDS regions of
BF2 were identified in low-IgY chickens as well as in the CDS regions of
BF2 in high-IgY chickens. In addition to the SNPs, a nine base pairs deletion was identified in the intron of
BF1. As
BF1 is located in the chicken MHC which has strong effects on host resistance to infectious pathogens, this deletion was examined in well-known chicken selection lines for Marek’s disease resistance from Aarhus University [
12]. No deletions were found in either the susceptible or the resistant lines, but SNPs in three bases were identified in the resistant chicken line at the same locus as the 9 bp deletion. These results strongly implicated BF1′s role in resistance to Marek’s disease.
The two tripartite (TRIM) motif containing proteins, TRIM41 and TRIM7.1, have been shown to contain SNPs in the CDS. The TRIM protein family includes more than 100 members with a conserved TRIM motif and E3 ubiquitin ligase activity [
17,
18]. Most TRIM proteins are closely associated with the degradation of exogenous and endogenous proteins through the ubiquitin-proteasome pathway. TRIM41 blocks the transcription and replication of hepatitis B virus (HBV) by inhibiting the enhancer II activity of HBV in human hepatoma cells [
19]. Previous studies have shown that TRIM7 can promote lung cancer development and a knockdown of TRIM7 reduced tumor growth [
20]. TRIM7 and TRIM41 both belong to the TRIM-B30.2 protein family and are located in a region surrounding the chicken BF/BL region of the B complex on chromosome 16 [
21]. Non-synonymous SNPs were found in the CDS regions of
TRIM7 and
TRIM41 in low IgY chickens and high IgY chickens respectively, indicating the potential involvement of these genes in the selection for IgY level.
5. Conclusions
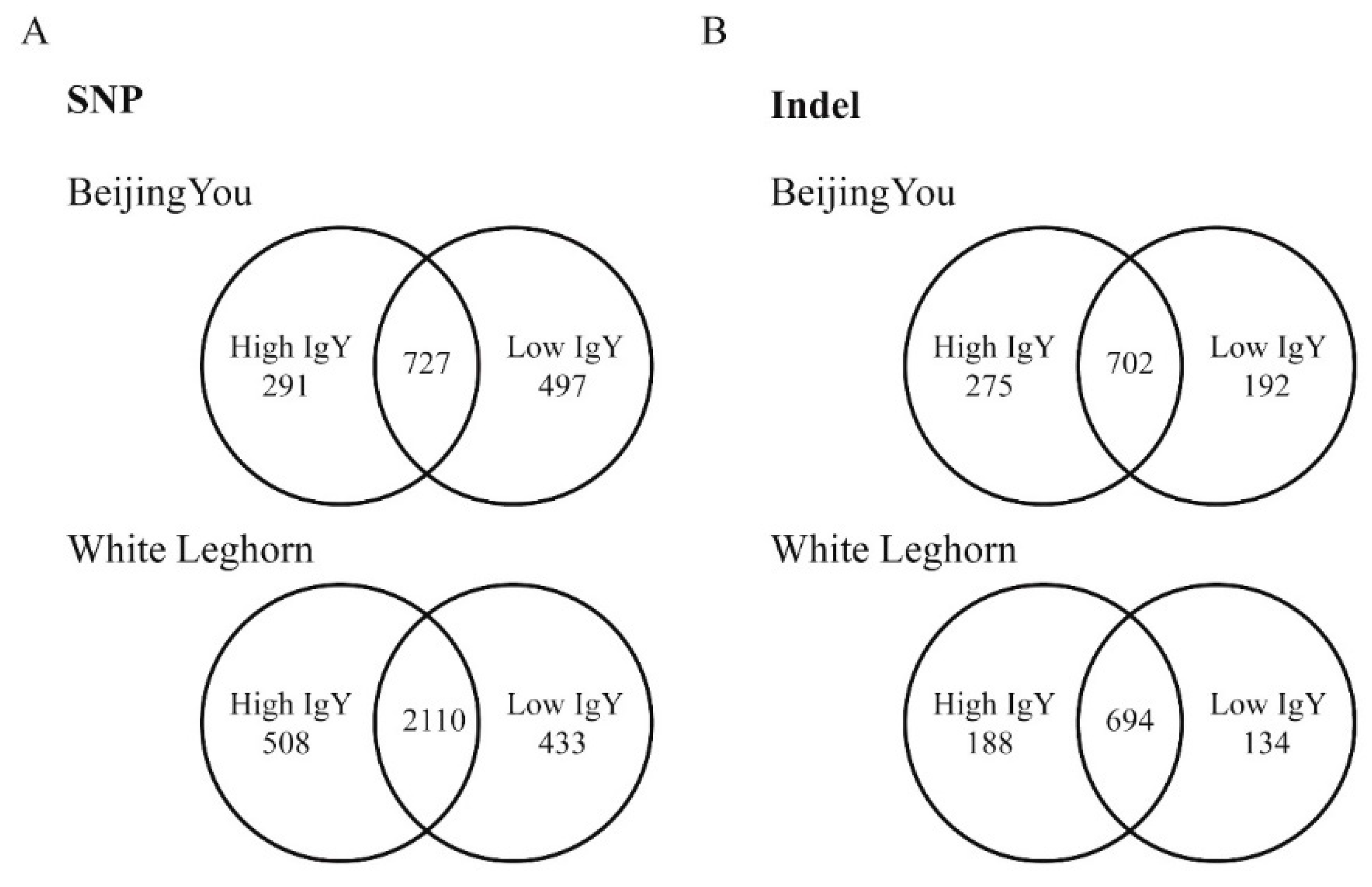
In the present study, SNPs and Indels were identified in genomic regions associated with IgY level in chickens. A large number of genetic mutations were identified in the studied genomic regions, revealing abundant genetic diversity between high and low IgY chickens. The chickens used in this study showed substantial differences in IgY levels. In total, it was found that 35,154 and 829 Indels respectively, in the high and low lines. Among them, 18 in CDS regions and 8 in proximal promoters were shared between White Leghorn and Beijing-You chickens with high IgY level. Among low-IgY White Leghorn and Beijing-You chickens, 14 and 11 shared common SNPs in CDS regions and proximal promoters respectively. In addition to the above SNPs common between the two chicken breeds, other identified SNPs and Indels were specific to White Leghorn or Beijing-You chickens, indicating the fundamental role of genetic mutations in modulating antibody levels. This study contributes valuable insights into the understanding of immune gene enriched microchromosome of chickens.