Histological and Comparative Transcriptome Analyses Provide Insights into Small Intestine Health in Diarrheal Piglets after Infection with Clostridium Perfringens Type C
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Bacterium
2.3. Animal Experiments
2.4. Sample Collection
2.5. Clinical Index Records
2.6. RNA Extraction and Analysis
2.7. Library Preparation and Sequencing
2.8. Screening of Differentially Expressed Genes
2.9. GO and KEGG Enrichment Analysis
2.10. RT-qPCR Confirmation
2.11. Statistical Analysis
3. Results
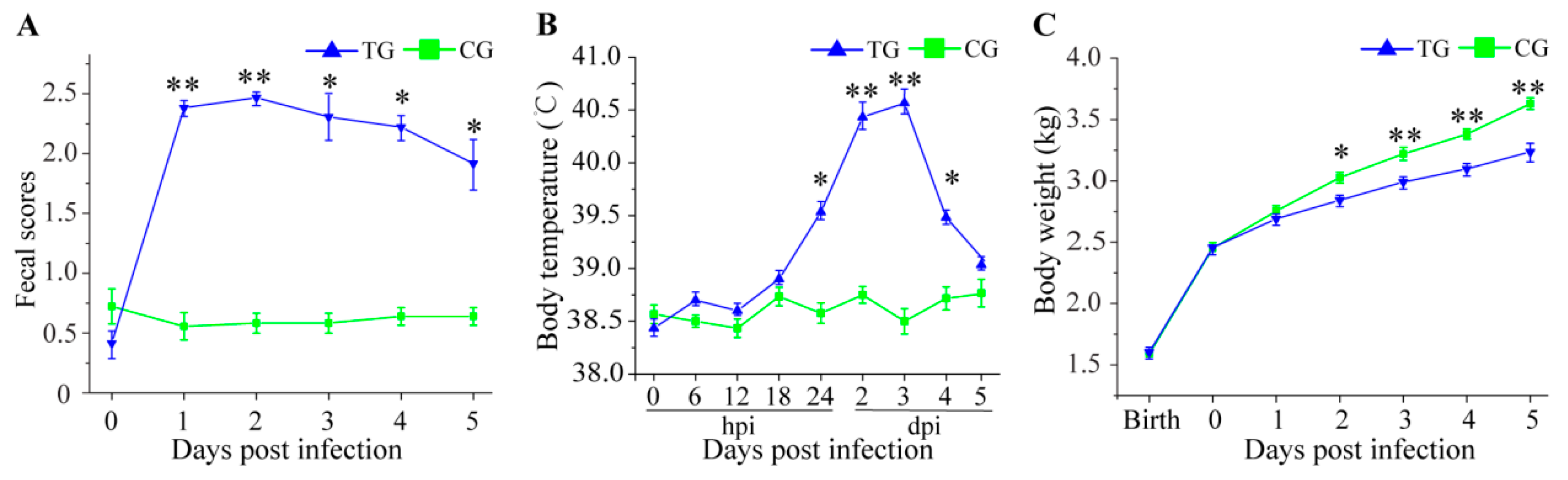
3.1. Physiological Changes in C. perfringens Type C Challenged Piglets
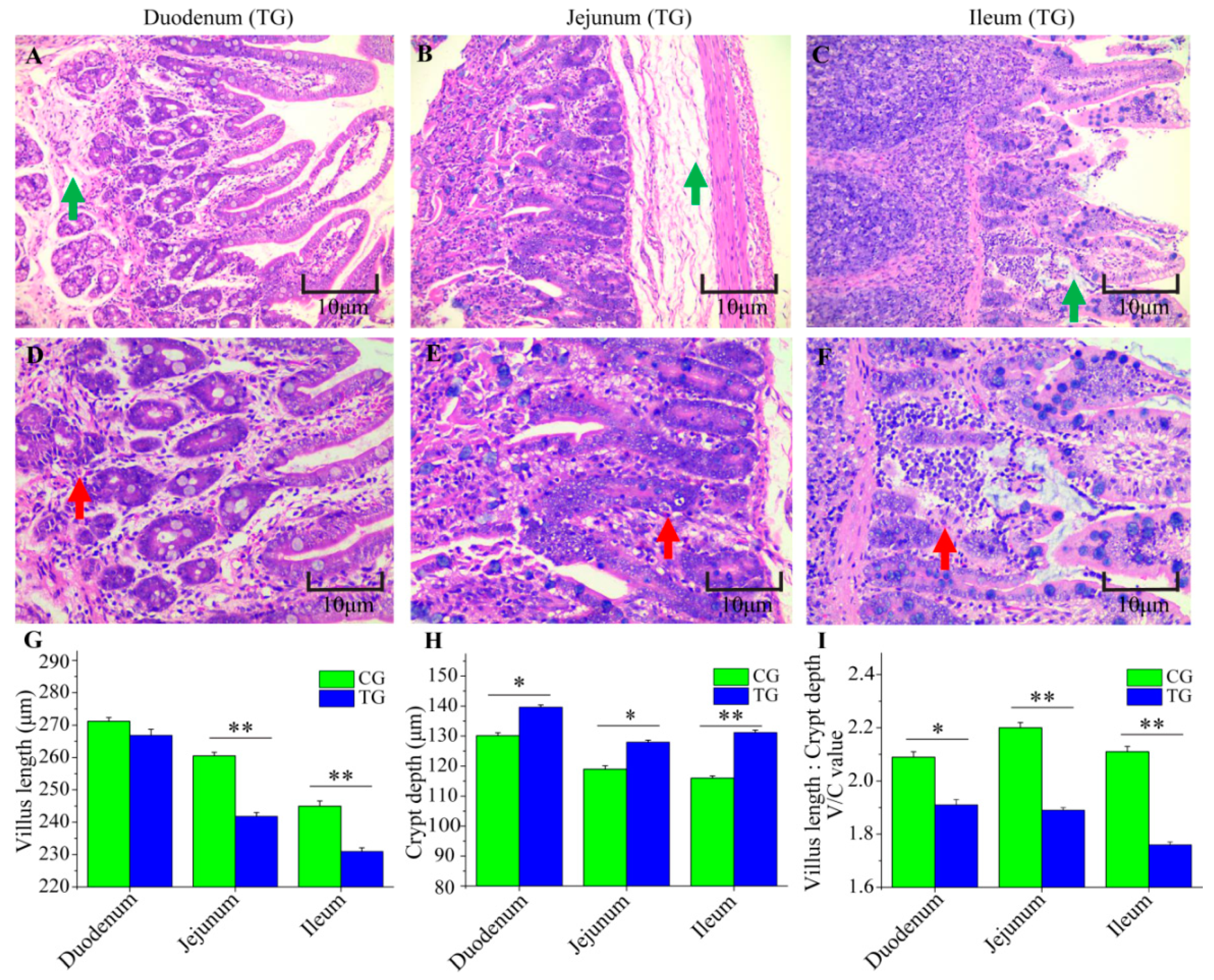
3.2. Changes to the Small Intestine in C. perfringens Type C Challenged Piglets
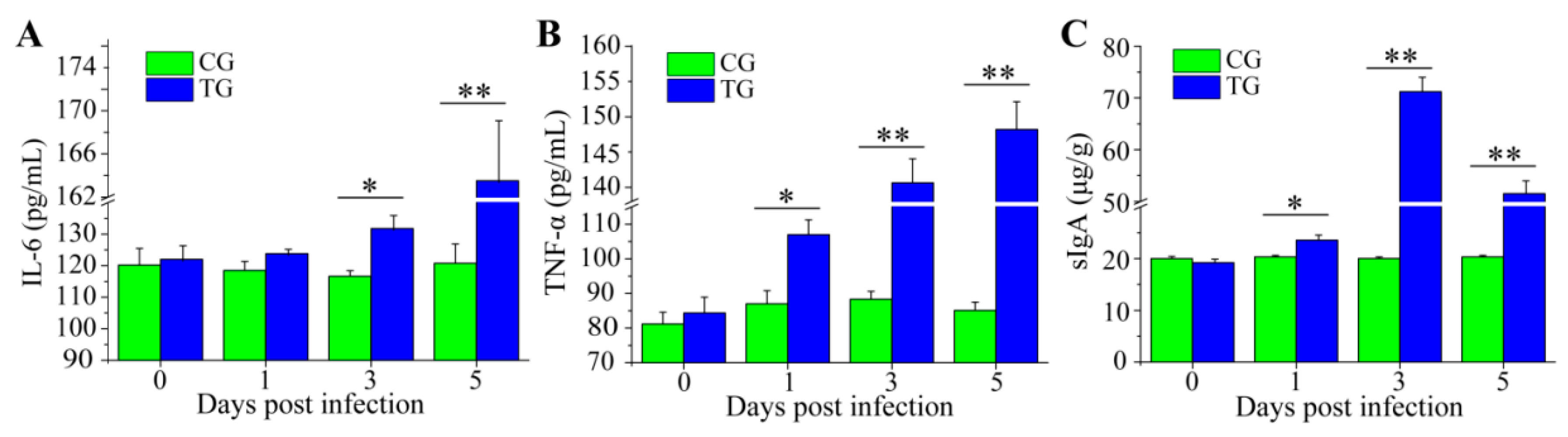
3.3. Dynamic Change of Inflammatory Cytokines and sIgA
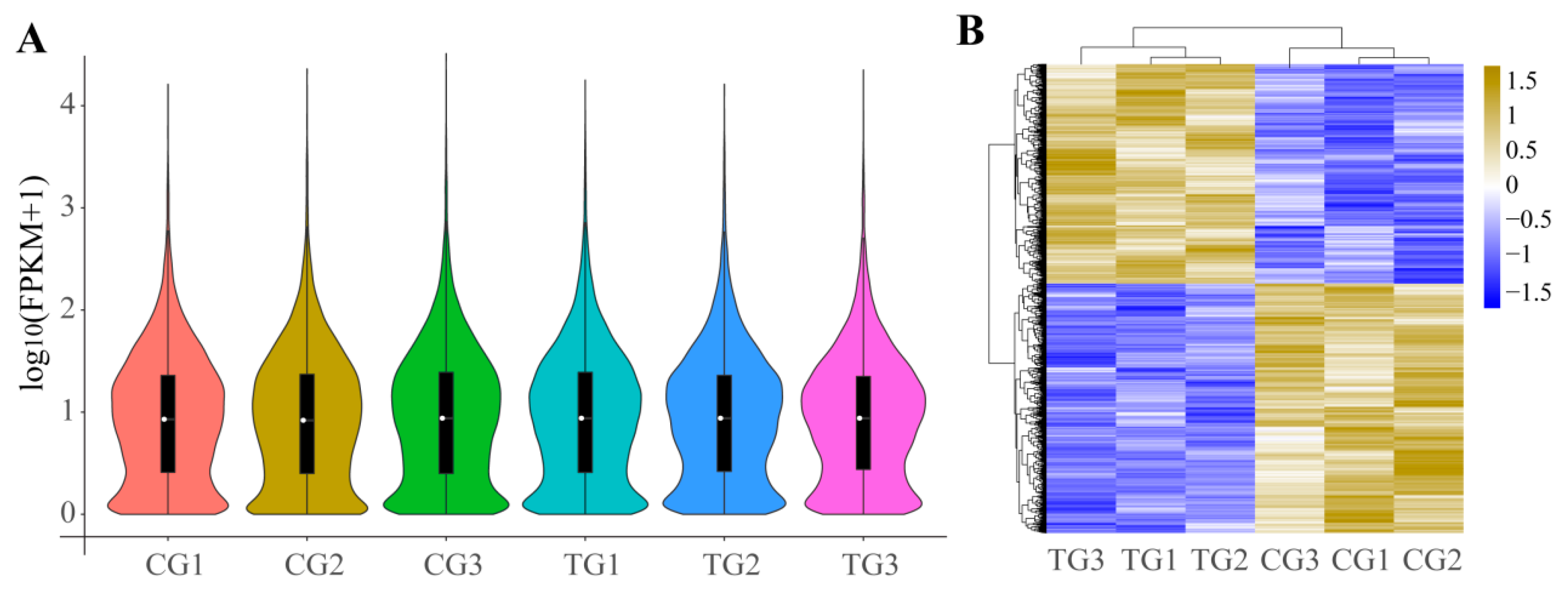
3.4. DEGs of Ileum after Infection
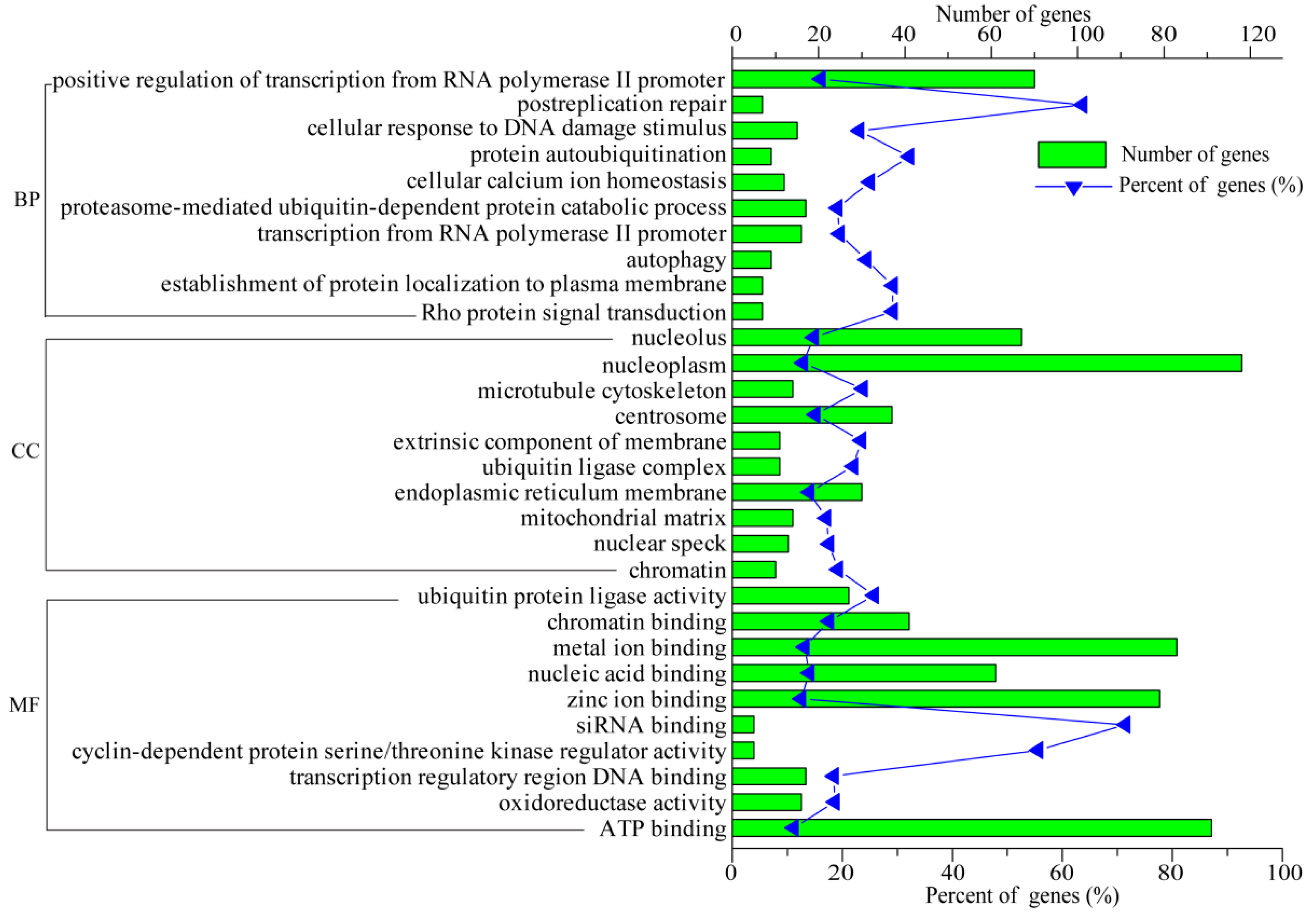
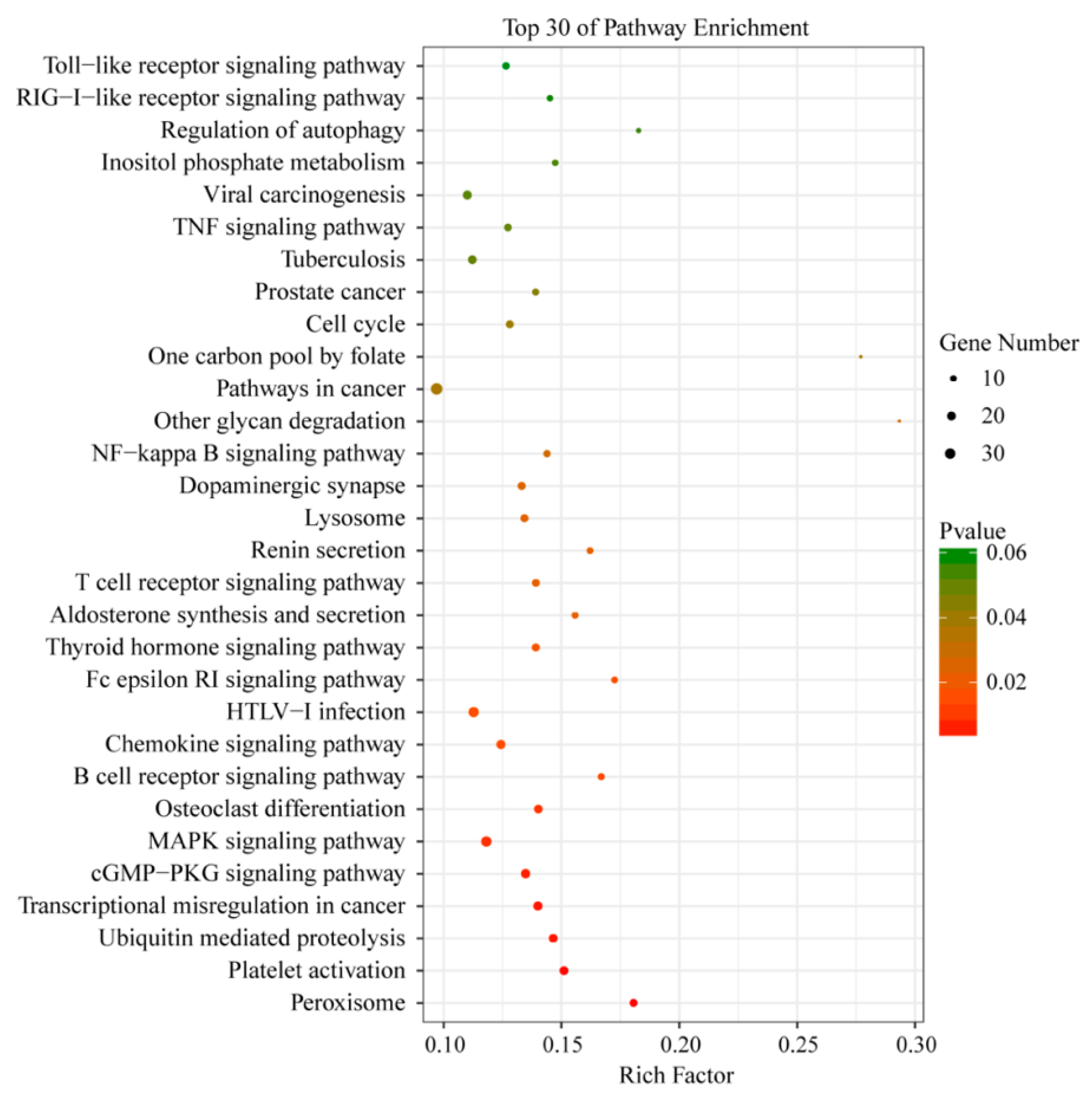
3.5. Functional Analysis of DEGs
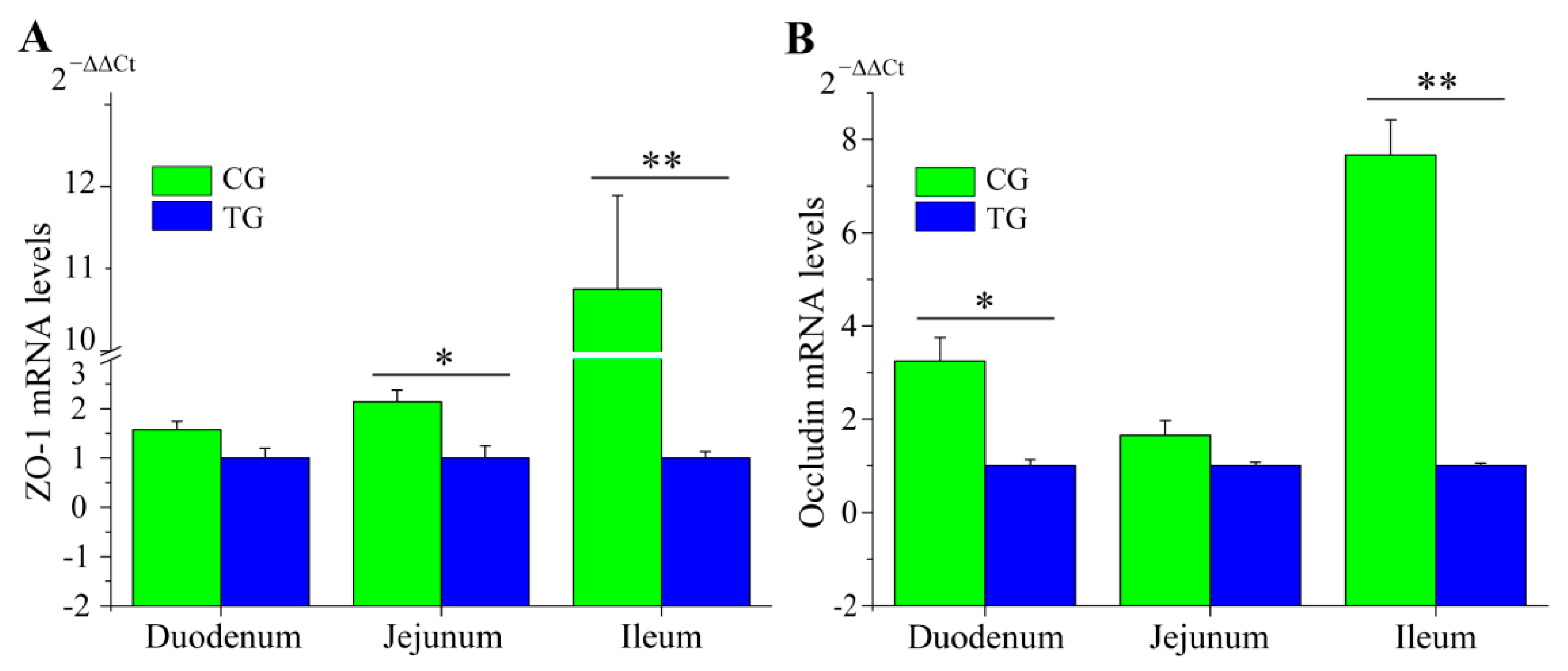
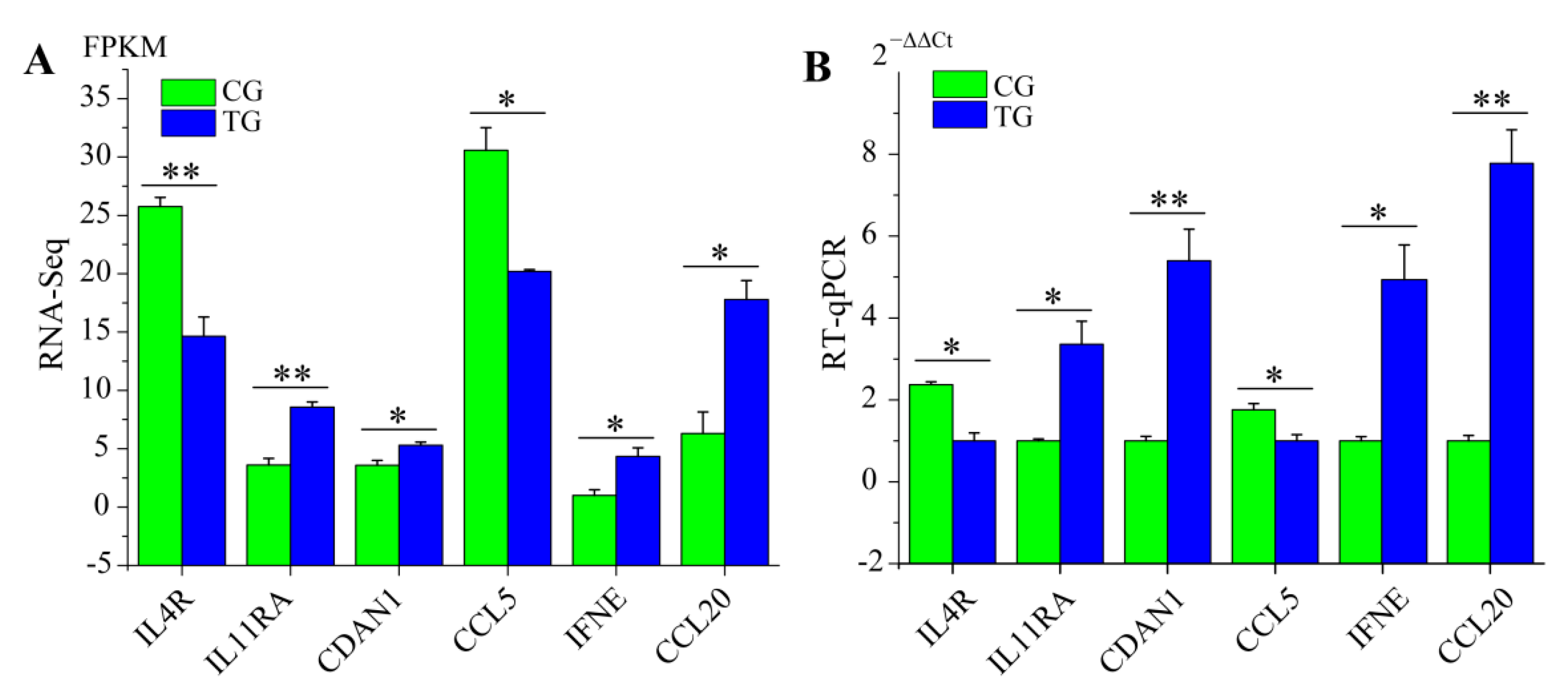
3.6. RT-qPCR Validation of RNA-Seq Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, J.H.; Adams, V.; Bannam, T.L.; Miyamoto, K.; Garcia, J.P.; Uzal, F.A.; Rood, J.I.; McClane, B.A. Toxin plasmids of Clostridium perfringens. Microbiol. Mol. Biol. Rev. 2013, 77, 208–233. [Google Scholar] [CrossRef] [PubMed]
- Songer, J.G.; Uzal, F.A. Clostridial enteric infections in pigs. J. Vet. Diagn. Investig. 2005, 17, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Songer, J.G. Clostridial diseases of small ruminants. Vet. Res. 1998, 29, 219–232. [Google Scholar]
- Uzal, F.A.; McClane, B.A. Recent progress in understanding the pathogenesis of Clostridium perfringens type C infections. Vet. Microbiol. 2011, 153, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Petit, L.; Gibert, M.; Popoff, M.R. Clostridium perfringens: Toxinotype and genotype. Trends Microbiol. 1999, 7, 104–110. [Google Scholar] [CrossRef]
- Songer, J.G. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 1996, 9, 216–234. [Google Scholar] [CrossRef]
- Miclard, J.; Jaggi, M.; Sutter, E.; Wyder, M.; Grabscheid, B.; Posthaus, H. Clostridium perfringens beta-toxin targets endothelial cells in necrotizing enteritis in piglets. Vet. Microbiol. 2009, 137, 320–325. [Google Scholar] [CrossRef]
- Vidal, J.E.; McClane, B.A.; Saputo, J.; Parker, J.; Uzal, F.A. Effects of Clostridium perfringens beta-toxin on the rabbit small intestine and colon. Infect. Immun. 2008, 76, 4396–4404. [Google Scholar] [CrossRef]
- Fisher, D.J.; Fernandez-Miyakawa, M.E.; Sayeed, S.; Poon, R.; Adams, V.; Rood, J.I.; Uzal, F.A.; McClane, B.A. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect. Immun. 2006, 74, 5200–5210. [Google Scholar] [CrossRef]
- Anh Duc, T.; Hong, Y.; Ban, J.; Park, B.; Hoang, T.C.; Hong, Y.H.; Lillehoj, H.S. Analysis of differentially expressed genes in necrotic enteritis-infected fayoumi chickens using RNA sequencing. J. Poult. Sci. 2017, 54, 121–133. [Google Scholar]
- Truong, A.D.; Hong, Y.H.; Lillehoj, H.S. High-throughput sequencing reveals differing immune responses in the intestinal mucosa of two inbred lines afflicted with necrotic enteritis. Vet. Immunol. Immunopathol. 2015, 166, 116–124. [Google Scholar] [CrossRef]
- Yan, Z.; Jiang, T.; Wang, P.; Huang, X.; Yang, Q.; Sun, W.; Gun, S. Circular rna expression profile of spleen in a Clostridium perfringens type C-induced piglet model of necrotizing enteritis. FEBS Open Bio 2018, 8, 1722–1732. [Google Scholar] [CrossRef]
- Kelly, D.; O’Brien, J.J.; McCracken, K.J. Effect of creep feeding on the incidence, duration and severity of post-weaning diarrhoea in pigs. Res. Vet. Sci. 1990, 49, 223–228. [Google Scholar] [CrossRef]
- Yang, Q.L.; Kong, J.J.; Wang, D.W.; Zhao, S.G.; Gun, S.B. Swine leukocyte antigen-DQA gene variation and its association with piglet diarrhea in Large White, Landrace and Duroc. Asian-Australas. J. Anim. Sci. 2013, 26, 1065–1071. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. Tophat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C.; et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010, 28, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with Tophat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tossou, M.C.B.; Liu, H.; Bai, M.; Chen, S.; Cai, Y.; Duraipandiyan, V.; Liu, H.; Adebowale, T.O.; Al-Dhabi, N.A.; Long, L.; et al. Effect of high dietary tryptophan on intestinal morphology and tight junction protein of weaned pig. BioMed Res. Int. 2016, 2016, 2912418. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Li, X.; Lu, D.; Liu, S.; Suo, X.; Li, Q.; Li, N. Lysozyme improves gut performance and protects against enterotoxigenic Escherichia coli infection in neonatal piglets. Vet. Res. 2018, 49. [Google Scholar] [CrossRef] [PubMed]
- Higgins, S.E.; Wolfenden, A.D.; Tellez, G.; Hargis, B.M.; Porter, T.E. Transcriptional profiling of cecal gene expression in probiotic- and Salmonella-challenged neonatal chicks. Poult. Sci. 2011, 90, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Lillehoj, H.S.; Jang, S.I.; Lee, S.H.; Hong, Y.H.; Cheng, H.H. Transcriptional profiles of host-pathogen responses to necrotic enteritis and differential regulation of immune genes in two inbreed chicken lines showing disparate disease susceptibility. PLoS ONE 2014, 9, e114960. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-E.; Park, H.-T.; Jung, Y.H.; Yoo, H.S. Gene expression profiles of immune-regulatory genes in whole blood of cattle with a subclinical infection of Mycobacterium avium subsp paratuberculosis. PLoS ONE 2018, 13, e0196502. [Google Scholar] [CrossRef]
- Park, S.K.; Jeong, J.Y.; Cho, E.S.; Jeong, Y.D.; Park, C.S. RNA-seq reveals differentially expressed genes of pig vaccinated with modified live attenuated porcine epidemic diarrhea. Pak. J. Zool. 2017, 49, 1107–1110. [Google Scholar] [CrossRef]
- Stephenson, W.; Donlin, L.T.; Butler, A.; Rozo, C.; Bracken, B.; Rashidfarrokhi, A.; Goodman, S.M.; Ivashkiv, L.B.; Bykerk, V.P.; Orange, D.E.; et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun. 2018, 9, 791. [Google Scholar] [CrossRef]
- Yu, F.; Duan, C.; Zhang, X.; Yao, D.; Si, G.; Gao, Y.; Gao, Z.; Umer, F.; Guo, X. RNA-seq analysis reveals different gene ontologies and pathways in rheumatoid arthritis and kashin-beck disease. Int. J. Rheum. Dis. 2018, 21, 1686–1694. [Google Scholar] [CrossRef]
- Damiani, C.; Maspero, D.; Di Filippo, M.; Colombo, R.; Pescini, D.; Graudenzi, A.; Westerhoff, H.V.; Alberghina, L.; Vanoni, M.; Mauri, G. Integration of single-cell RNA-seq data into population models to characterize cancer metabolism. PLoS Comput. Biol. 2019, 15, e1006733. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Qin, X.L.; Yang, J.Y.; Liao, Y.W.; Wu, X.Z.; Zheng, H.P. Identification and expression analysis of ceftriaxone resistance-related genes in neisseria gonorrhoeae integrating RNA-seq data and qRT-PCR validation. J. Glob. Antimicrob. Resist. 2019, 16, 202–209. [Google Scholar] [CrossRef]
- Sargeant, H.R.; McDowall, K.J.; Miller, H.M.; Shaw, M.-A. Dietary zinc oxide affects the expression of genes associated with inflammation: Transcriptome analysis in piglets challenged with ETEC k88. Vet. Immunol. Immunopathol. 2010, 137, 120–129. [Google Scholar] [CrossRef]
- Trevisi, P.; Latorre, R.; Priori, D.; Luise, D.; Archetti, I.; Mazzoni, M.; D’Inca, R.; Bosi, P. Effect of feed supplementation with live yeast on the intestinal transcriptome profile of weaning pigs orally challenged with Escherichia coli F4. Animal 2017, 11, 33–44. [Google Scholar] [CrossRef]
- Gaur, U.; Xiong, Y.Y.; Luo, Q.P.; Yuan, F.Y.; Wu, H.Y.; Qiao, M.; Wimmers, K.; Li, K.; Mei, S.Q.; Liu, G.S. Breed-specific transcriptome response of spleen from six to eight week old piglet after infection with Streptococcus suis type 2. Mol. Biol. Rep. 2014, 41, 7865–7873. [Google Scholar] [CrossRef]
- Splichalova, A.; Jenistova, V.; Splichalova, Z.; Splichal, I. Colonization of preterm gnotobiotic piglets with probiotic lactobacillus rhamnosus gg and its interference with Salmonella typhimurium. Clin. Exp. Immunol. 2019, 195, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, Y.; Zhang, Z.; You, J.; Yang, Y.; Li, X. Protective immunity of a multivalent vaccine candidate against piglet diarrhea caused by enterotoxigenic Escherichia coli (ETEC) in a pig model. Vaccine 2018, 36, 723–728. [Google Scholar] [CrossRef]
- Sayeed, S.; Uzal, F.A.; Fisher, D.J.; Saputo, J.; Vidal, J.E.; Chen, Y.; Gupta, P.; Rood, J.I.; McClane, B.A. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol. Microbiol. 2008, 67, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Lentle, R.G.; de Loubens, C. A review of mixing and propulsion of chyme in the small intestine: Fresh insights from new methods. J. Comp. Physiol. B 2015, 185, 369–387. [Google Scholar] [CrossRef]
- Al Masri, S.; Hunigen, H.; Al Aiyan, A.; Rieger, J.; Zentek, J.; Richardson, K.; Plendl, J. Influence of age at weaning and feeding regimes on the postnatal morphology of the porcine small intestine. J. Swine Health Prod. 2015, 23, 186–203. [Google Scholar]
- Adhikari, P.; Cosby, D.E.; Cox, N.A.; Franca, M.S.; Williams, S.M.; Gogal, R.M.; Ritz, C.W.; Kim, W.K. Effect of dietary fructooligosaccharide supplementation on internal organs salmonella colonization, immune response, ileal morphology, and ileal immunohistochemistry in laying hens challenged with salmonella enteritidis. Poult. Sci. 2018, 97, 2525–2533. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.L.; Liu, Z.Z.; Jiang, J.C.; Yu, Y.; Zhang, Q. Differential gene expression profiling of porcine epithelial cells infected with three enterotoxigenic Escherichia coli strains. BMC Genom. 2012, 13, 330. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Akhtar, S.; Choudhry, M.A. Alteration in intestine tight junction protein phosphorylation and apoptosis is associated with increase in IL-18 levels following alcohol intoxication and burn injury. Biochim. Biophys. Acta-Mol. Basis Dis. 2012, 1822, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 2018, 16, 457–470. [Google Scholar] [CrossRef]
- Lepine, A.F.P.; de Wit, N.; Oosterink, E.; Wichers, H.; Mes, J.; de Vos, P. Lactobacillus acidophilus attenuates Salmonella-induced stress of epithelial cells by modulating tight-junction genes and cytokine responses. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ji, H.; Wang, S.; Liu, H.; Zhang, W.; Zhang, D.; Wang, Y. Probiotic lactobacillus plantarum promotes intestinal barrier function by strengthening the epithelium and modulating gut microbiota. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Hendrikx, T.; Schnabl, B. Antimicrobial proteins: Intestinal guards to protect against liver disease. J. Gastroenterol. 2019, 54, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.A.; Uppada, S.; Achkar, I.W.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight junction proteins and signaling pathways in cancer and inflammation: A functional crosstalk. Front. Physiol. 2019, 9. [Google Scholar] [CrossRef]
- He, F.; Wu, C.; Li, P.; Li, N.; Zhang, D.; Zhu, Q.; Ren, W.; Peng, Y. Functions and signaling pathways of amino acids in intestinal inflammation. BioMed Res. Int. 2018, 2018, 9171905. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Hunter, C.A.; Jones, S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015, 16, 448–457. [Google Scholar] [CrossRef]
- Mihara, M.; Hashizume, M.; Yoshida, H.; Suzuki, M.; Shiina, M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin. Sci. 2012, 122, 143–159. [Google Scholar] [CrossRef]
- Grijalva, C.G.; Chen, L.; Delzell, E.; Baddley, J.W.; Beukelman, T.; Winthrop, K.L.; Griffin, M.R.; Herrinton, L.J.; Liu, L.; Ouellet-Hellstrom, R.; et al. Initiation of tumor necrosis factor-alpha antagonists and the risk of hospitalization for infection in patients with autoimmune diseases. Jama 2011, 306, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, T.; Mitoma, H.; Harashima, S.-I.; Tsukamoto, H.; Shimoda, T. Transmembrane TNF-alpha: Structure, function and interaction with anti-TNF agents. Rheumatology 2010, 49, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Pabst, O. New concepts in the generation and functions of IgA. Nat. Rev. Immunol. 2012, 12, 821–832. [Google Scholar] [CrossRef]
- Mantis, N.J.; Rol, N.; Corthesy, B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Minambres, A.; Eid, S.G.; Mangan, N.E.; Pade, C.; Lim, S.S.; Matthews, A.Y.; de Weerd, N.A.; Hertzog, P.J.; Mak, J. Interferon epsilon promotes hiv restriction at multiple steps of viral replication. Immunol. Cell Biol. 2017, 95, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Bergkamp, J.; Blecha, F. Molecular evolution of the porcine type I interferon family: Subtype-specific expression and antiviral activity. PLoS ONE 2014, 9, e112378. [Google Scholar] [CrossRef]
- Hielpos, M.S.; Ferrero, M.C.; Fernandez, A.G.; Bonetto, J.; Giambartolomei, G.H.; Fossati, C.A.; Baldi, P.C. CCL20 and beta-defensin 2 production by human lung epithelial cells and macrophages in response to brucella abortus infection. PLoS ONE 2015, 10, e0140408. [Google Scholar] [CrossRef] [PubMed]
- Guesdon, W.; Auray, G.; Pezier, T.; Bussiere, F.I.; Drouet, F.; Le Vern, Y.; Marquis, M.; Potiron, L.; Rabot, S.; Bruneau, A.; et al. CCL20 displays antimicrobial activity against cryptosporidium parvum, but its expression is reduced during infection in the intestine of neonatal mice. J. Infect. Dis. 2015, 212, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
| Gene | Accession Number | Sequence (5′-3′) | Product Size (bp) | Reference |
|---|---|---|---|---|
| Occludin | NM_001163647.2 | TCCTGGGTGTGATGGTGTTC | 144 | [21] |
| CGTAGAGTCCAGTCACCGCA | ||||
| ZO-1 | XM_021098827.1 | TGAGTTTGATAGTGGCGTTG | 298 | [22] |
| TGGGAGGATGCTGTTGTC | ||||
| IL4R | NM_214340.1 | GTGGCCCATCTGCCTATCC | 161 | |
| CTGAGCCTGCTCTGTTCTCG | ||||
| IL11RA | XM_021064672.1 | CCGCAACAGTGTCGCTAGT | 201 | |
| CCACAGAGACCTTCCCCAAA | ||||
| CDAN1 | XM_021097154.1 | TTTTGAGAAGGGCTTGGGCA | 160 | |
| ATCCGGAGTCTCACCCAAGA | ||||
| CCL5 | NM_001129946.1 | TGCTTCTTGCTCTTGTCCCA | 189 | |
| GTGCCAAGGGTCCAAAGTTC | ||||
| IFNE | NM_001105310.1 | GTGTCTGCCACACCGGAAAA | 160 | |
| GTGGCTTTCCTCCCAACCAT | ||||
| CCL20 | XM_005672261.3 | ATCTGGGTGAAACAAGCCGT | 185 | |
| TGGACAAGTCCAAAGAGGCA | ||||
| β-actin | XM_003124280.5 | AGGCGGACTGTTAGTTGCAT | 187 | [12] |
| TGTCACCTTCACCGTTCCAG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Z.; Cai, L.; Huang, X.; Sun, W.; Li, S.; Wang, P.; Yang, Q.; Jiang, T.; Gun, S. Histological and Comparative Transcriptome Analyses Provide Insights into Small Intestine Health in Diarrheal Piglets after Infection with Clostridium Perfringens Type C. Animals 2019, 9, 269. https://doi.org/10.3390/ani9050269
Yan Z, Cai L, Huang X, Sun W, Li S, Wang P, Yang Q, Jiang T, Gun S. Histological and Comparative Transcriptome Analyses Provide Insights into Small Intestine Health in Diarrheal Piglets after Infection with Clostridium Perfringens Type C. Animals. 2019; 9(5):269. https://doi.org/10.3390/ani9050269
Chicago/Turabian StyleYan, Zunqiang, Lijuan Cai, Xiaoyu Huang, Wenyang Sun, Shouhu Li, Pengfei Wang, Qiaoli Yang, Tiantuan Jiang, and Shuangbao Gun. 2019. "Histological and Comparative Transcriptome Analyses Provide Insights into Small Intestine Health in Diarrheal Piglets after Infection with Clostridium Perfringens Type C" Animals 9, no. 5: 269. https://doi.org/10.3390/ani9050269
APA StyleYan, Z., Cai, L., Huang, X., Sun, W., Li, S., Wang, P., Yang, Q., Jiang, T., & Gun, S. (2019). Histological and Comparative Transcriptome Analyses Provide Insights into Small Intestine Health in Diarrheal Piglets after Infection with Clostridium Perfringens Type C. Animals, 9(5), 269. https://doi.org/10.3390/ani9050269





