Comparative Transcriptomics Reveal Key Sheep (Ovis aries) Hypothalamus LncRNAs that Affect Reproduction
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Animal Processing
2.2. Tissues Acquirement and Sequencing
2.3. Transcriptome Assembly
2.4. LncRNA Identification and Differential Expression Analysis
2.5. Target Gene Prediction of lncRNAs and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Analyses
2.6. Construction of Integral LncRNA–mRNA Interaction Networks
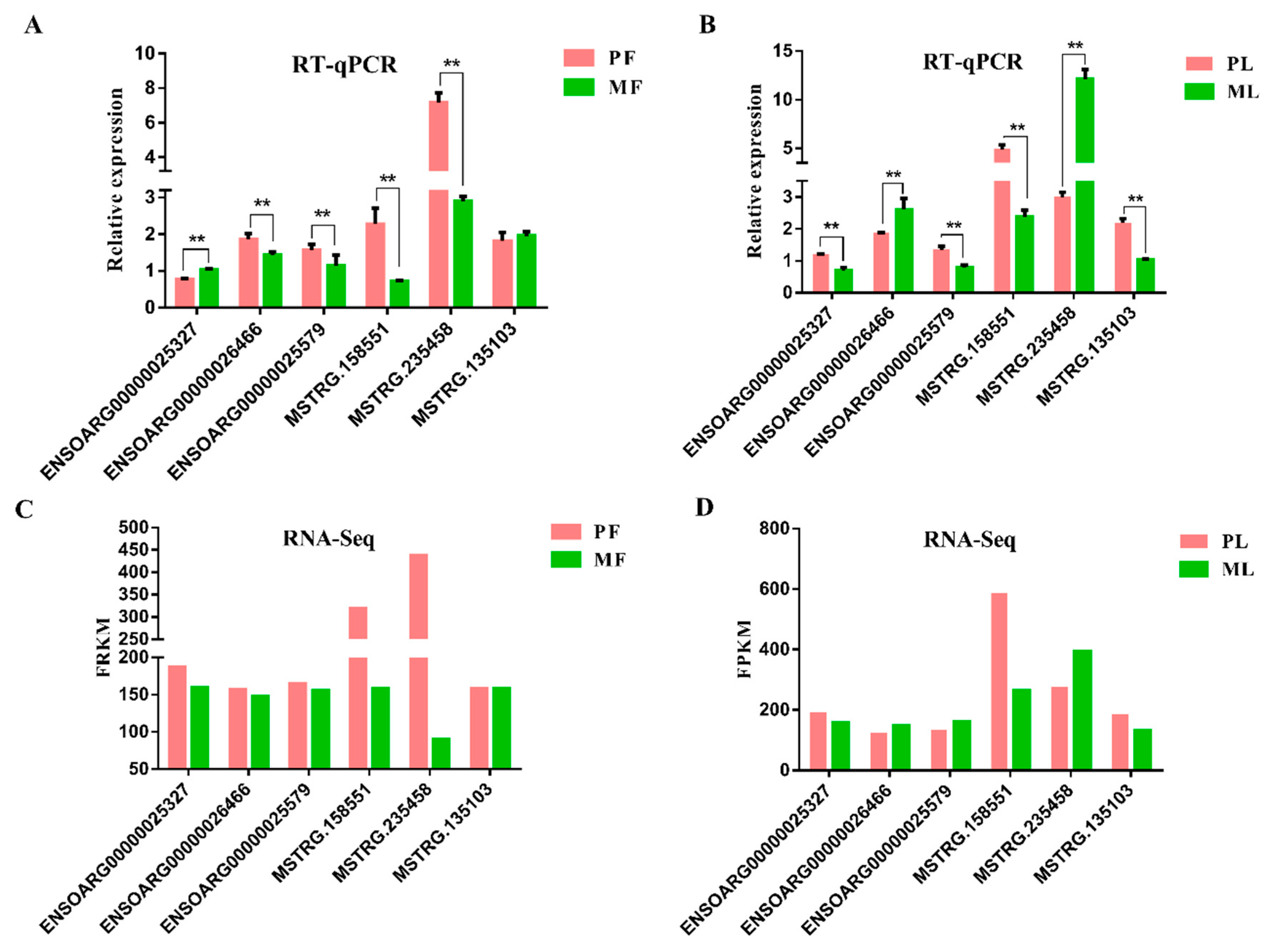
2.7. Data Validation
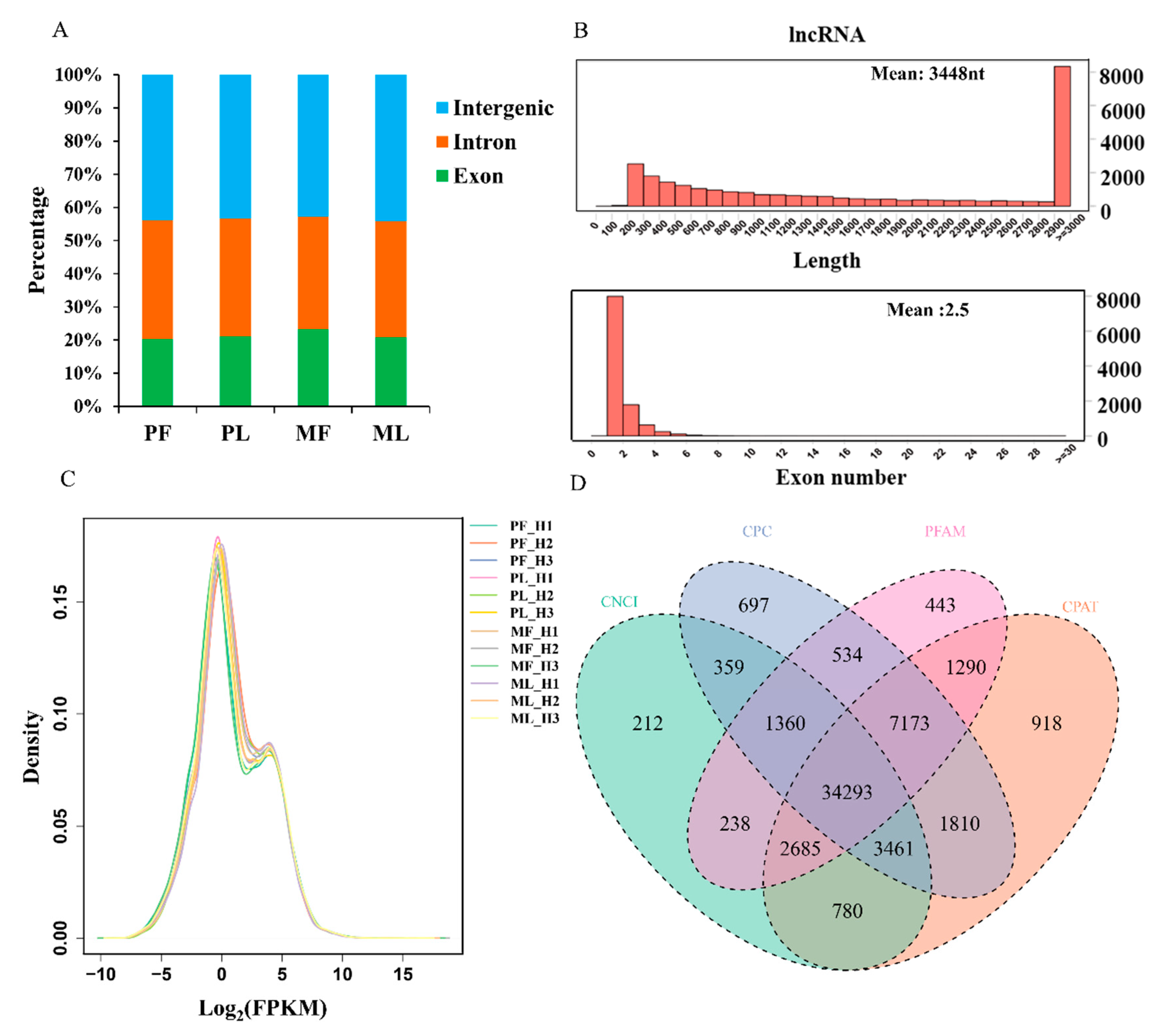
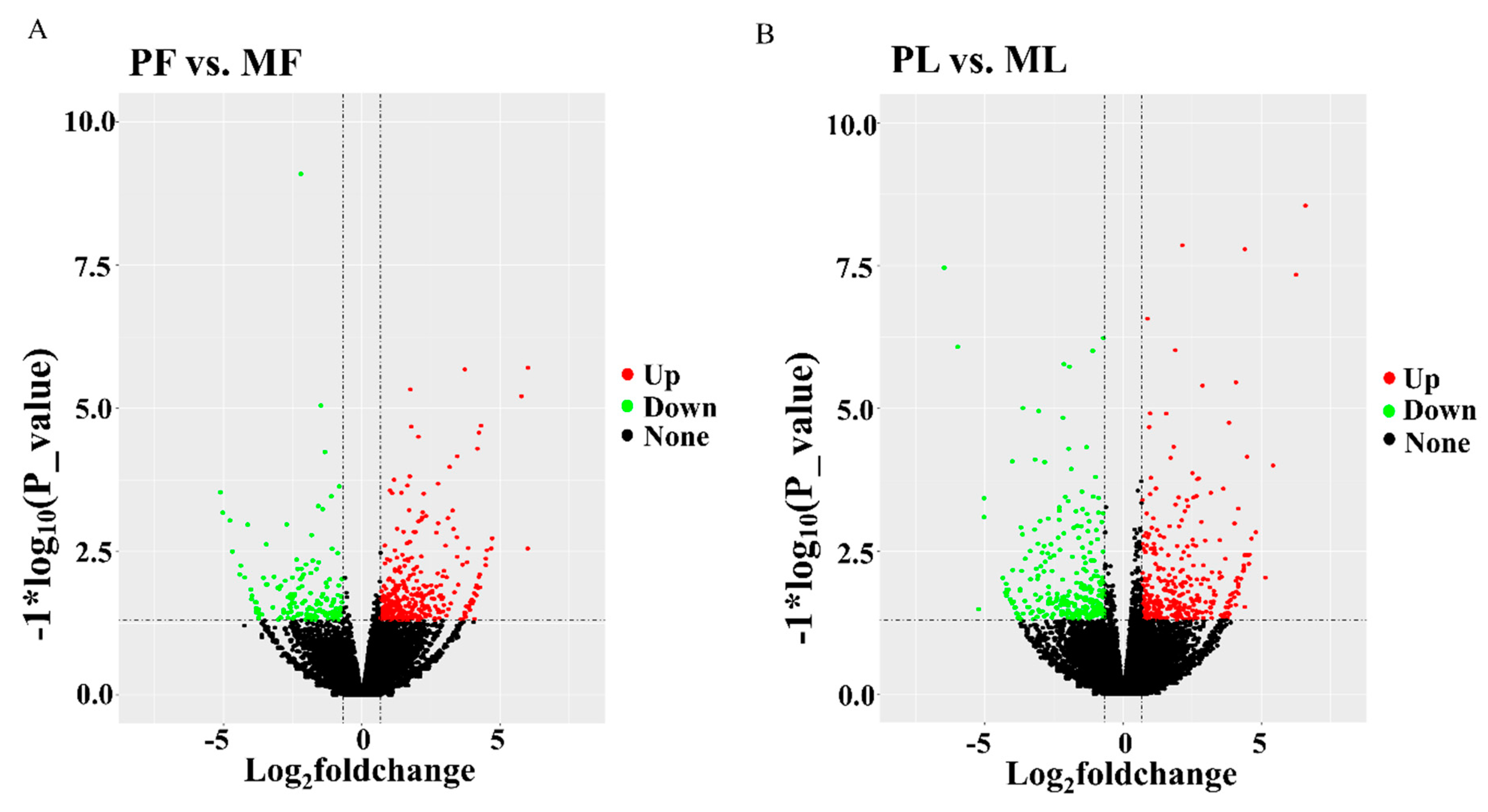
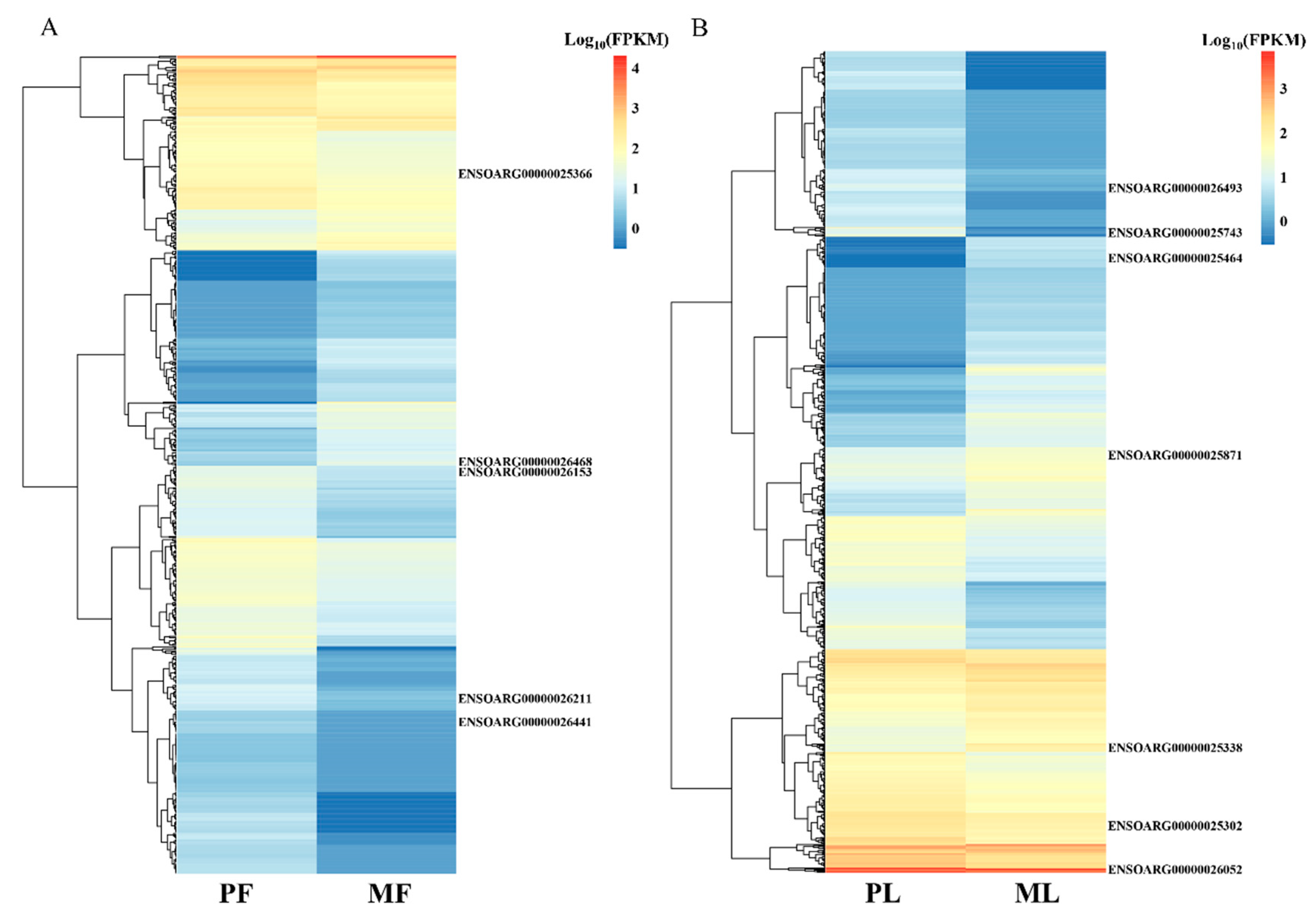
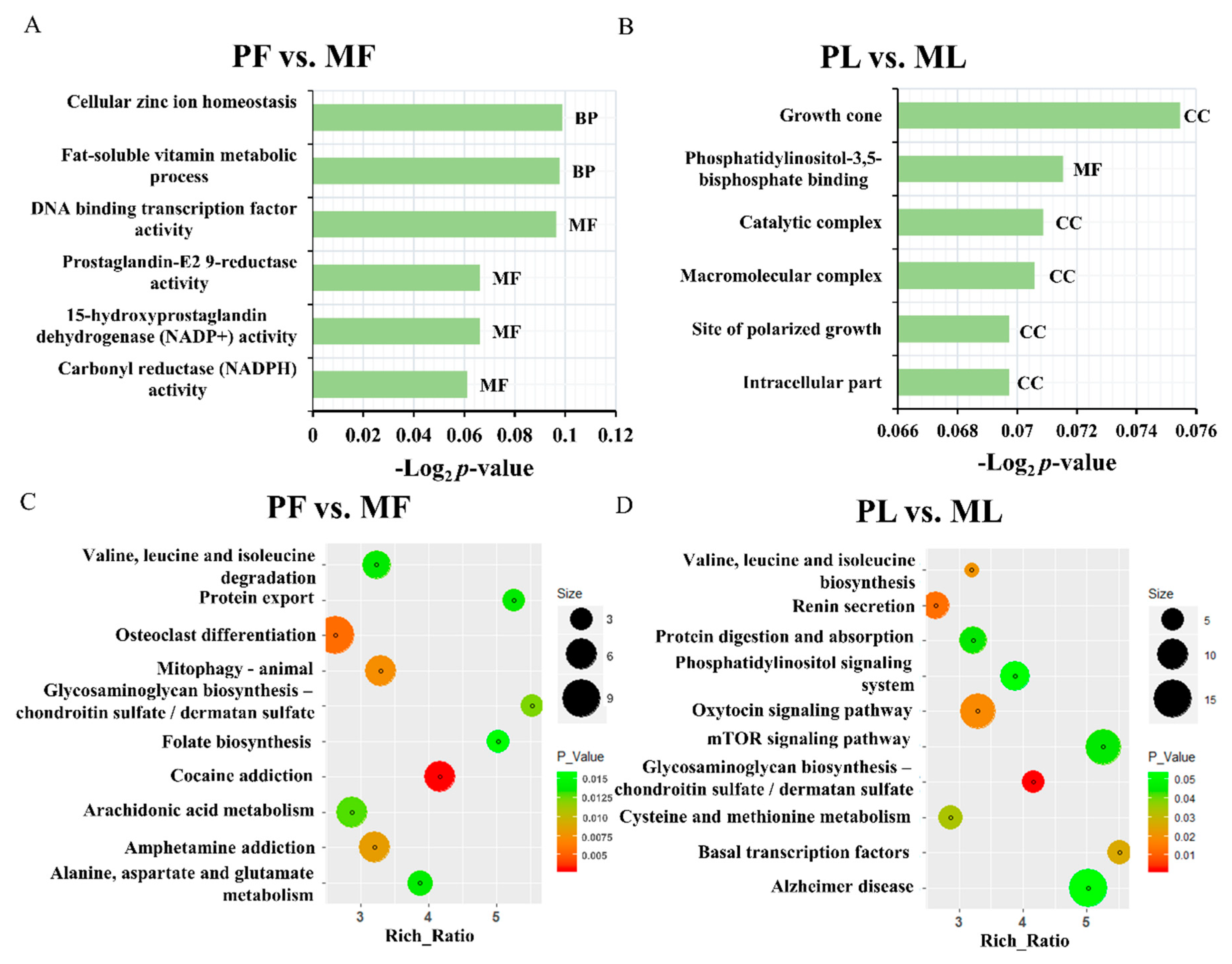
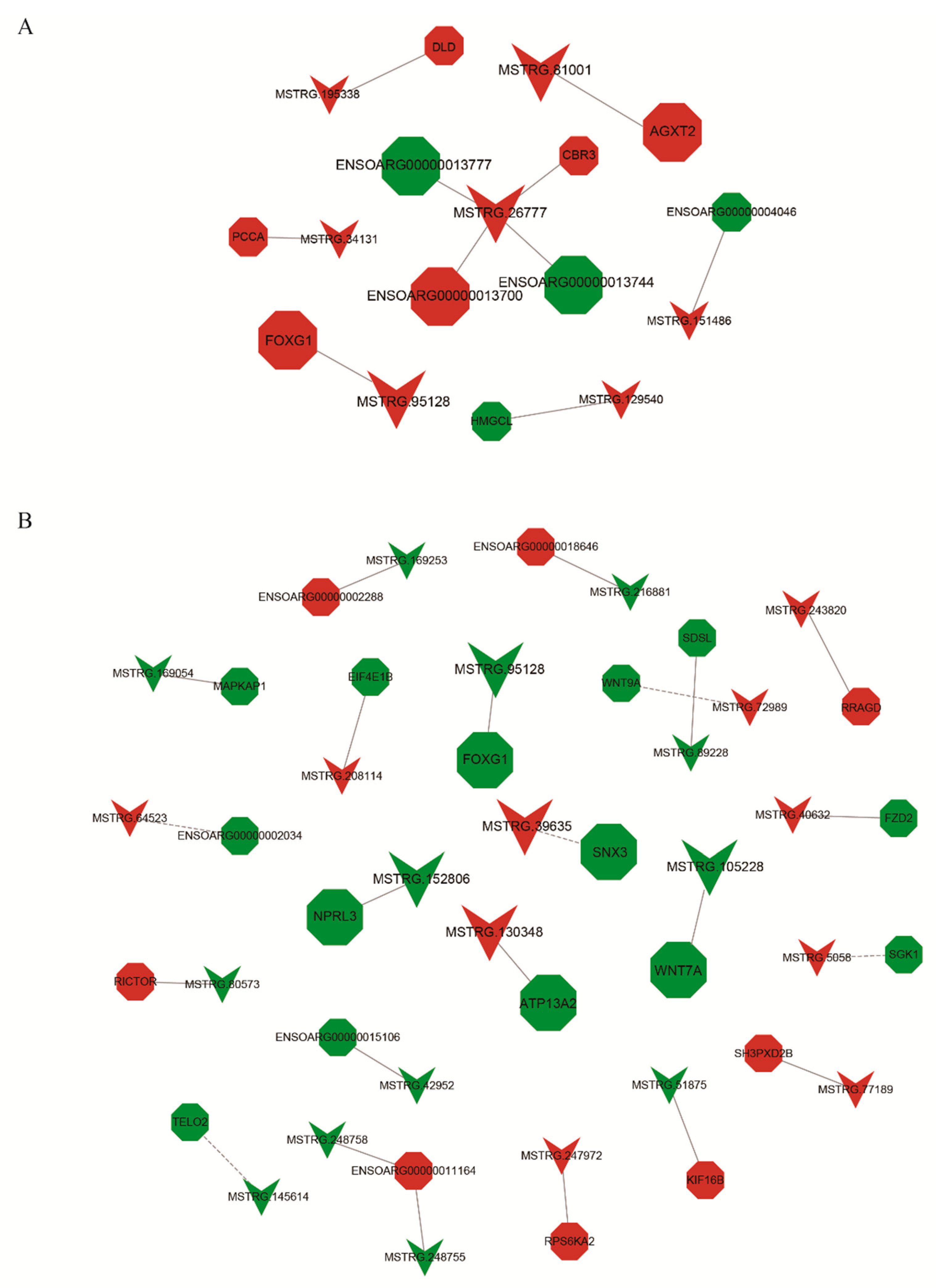
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Abreu, A.P.; Kaiser, U.B. Pubertal development and regulation. Lancet Diabetes Endocrinol. 2016, 4, 254–264. [Google Scholar] [CrossRef]
- Cao, C.Y.; Ding, Y.F.; Kong, X.J.; Feng, G.D.; Xiang, W.; Chen, L.; Yang, F.; Zhang, K.; Chu, M.X.; Wang, P.Q. Reproductive role of miRNA in the hypothalamic-pituitary axis. Mol. Cell. Neurosci. 2018, 88, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Plant, T.M. Neuroendocrine control of the onset of puberty. Front. Neuroendocr. 2015, 38, 73–88. [Google Scholar] [CrossRef]
- Messina, A.; Prevot, V. Hypothalamic microRNAs flip the switch for fertility. Oncotarget 2017, 8, 8993–8994. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.X.; Liu, Z.H.; Jiao, C.L.; He, Y.Q.; Fang, L.; Ye, S.C.; Chen, G.H.; Wang, J.Y. Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries). J. Anim. Sci. 2007, 85, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.X.; Yang, J.; Feng, T.; Cao, G.L.; Fang, L.; Di, R.; Huang, D.W.; Tang, Q.Q.; Ma, Y.H.; Li, K. GDF9 as a candidate gene for prolificacy of Small Tail Han sheep. Mol. Biol. Rep. 2011, 38, 5199–5204. [Google Scholar] [CrossRef]
- Fogarty, N.M. A review of the effects of the Booroola gene (FecB) on sheep production. Small Rumin. Res. 2009, 85, 75–84. [Google Scholar] [CrossRef]
- Davis, G.H.; Balakrishnan, L.; Ross, I.K.; Wilson, T.; Galloway, S.M.; Lumsden, B.M.; Hanrahan, J.P.; Mullen, M.; Mao, X.Z.; Wang, G.L. Investigation of the Booroola (FecB) and Inverdale (FecXI) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim. Reprod. Sci. 2006, 92, 87–96. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Ryu, Y.H.; Macdonald, P.M. RNA sequences required for the noncoding function of oskar RNA also mediate regulation of Oskar protein expression by bicoid stability factor. Dev. Biol. 2015, 407, 211–223. [Google Scholar] [CrossRef]
- Anguera, M.C.; Weiyuan, M.; Danielle, C.; Satoshi, N.; Kelleher, R.J.; Lee, J.T. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011, 7, e1002248. [Google Scholar] [CrossRef]
- Mandal, A.C.; Sarkar, M.; Bhattacharya, D.; Sen, P. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar]
- Mulvey, B.B.; Olcese, U.; Cabrera, J.R.; Horabin, J.I. An interactive network of long non-coding RNAs facilitates the drosophila sex determination decision. Biochim. Biophys. Acta 2014, 1839, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.Y.; Luo, Q.M.; Zhao, H.J.; Qin, X.Y. Co-expression analysis and identification of fecundity-related long non-coding RNAs in sheep ovaries. Sci. Rep. 2016, 6, 39398. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.H.; Deng, M.T.; Fan, Y.X.; Yang, H.; Zhang, G.M.; Feng, X.Z.; Li, F.; Wang, D.; Wang, F.; Zhang, Y.L. Genome-wide analysis reveals extensive changes in lncRNAs during skeletal muscle development in Hu sheep. Genes 2017, 8, 191. [Google Scholar] [CrossRef]
- Gao, X.; Ye, J.; Yang, C.; Zhang, K.; Li, X.; Luo, L.; Ding, J.; Li, Y.; Cao, H.; Ling, Y. Screening and evaluating of long noncoding RNAs in the puberty of goats. BMC Genom. 2017, 18, 164. [Google Scholar] [CrossRef]
- Gao, X.; Ye, J.; Yang, C.; Luo, L.; Liu, Y.; Ding, J.; Zhang, Y.; Ling, Y.; Huang, W.; Zhang, X. RNA-seq analysis of lncRNA-controlled developmental gene expression during puberty in goat & rat. BMC Genet. 2018, 19, 19. [Google Scholar]
- Liu, Q.Y.; Hu, W.P.; He, X.Y.; Pan, Z.Y.; Guo, X.F.; Feng, T.; Cao, G.L.; Huang, D.W.; He, J.N.; Di, R.; et al. Establishment of high-throughput molecular detection methods for ovine high fecundity major gene FecB and their application. Acta Veterinaria Et Zootechnica Sinica 2017, 48, 39–51. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Wang, L.K.; Feng, Z.X.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Su, H.X.; Liu, L.; Zhang, Y.; Zhao, Y.L. Long Noncoding RNA NPCCAT1 promotes nasopharyngeal carcinoma progression via upregulating YY1. Biochimie 2018, 157, 184–194. [Google Scholar] [CrossRef]
- Hu, K.; Li, L.N.; Liao, Y.P.; Liang, M. LncRNA Gm2044 highly expresses in spermatocyte and inhibits Utf1 translation by interacting with Utf1 mRNA. Genes Genom. 2018, 40, 781–787. [Google Scholar] [CrossRef]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar] [CrossRef]
- Cabili, M.N.; Cole, T.; Loyal, G.; Magdalena, K.; Barbara, T.V.; Aviv, R.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef]
- Espey, L.L.; Yoshioka, S.; Russell, D.; Ujioka, T.; Vladu, B.; Skelsey, M.; Fujii, S.; Okamura, H.; Richards, J.S. Characterization of ovarian carbonyl reductase gene expression during ovulation in the gonadotropin-primed immature rat. Biol. Reprod. 2000, 62, 390–397. [Google Scholar] [CrossRef]
- Wintergalen, N.; Thole, H.H.; Galla, H.J.; Schlegel, W. Prostaglandin-E2 9-reductase from corpus luteum of pseudopregnant rabbit is a member of the aldo-keto reductase superfamily featuring 20 alpha-hydroxysteroid dehydrogenase activity. FEBS J. 2010, 234, 264–270. [Google Scholar] [CrossRef]
- Murdoch, W.J.; Farris, M.L. Prostaglandin E2-9-ketoreductase activity of preovulatory ovine follicles. J. Anim. Sci. 1988, 66, 2924–2929. [Google Scholar] [CrossRef]
- Patel, F.A.; Funder, J.W.; Challis, J.R.G. Mechanism of cortisol/progesterone antagonism in the regulation of 15-hydroxyprostaglandin dehydrogenase activity and messenger ribonucleic acid levels in human chorion and placental trophoblast cells at term. J. Clin. Endocrinol. Metab. 2003, 88, 2922–2933. [Google Scholar] [CrossRef]
- Wermuth, B. Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to prostaglandin 9-ketoreductase and xenobiotic ketone reductase. J. Biol. Chem. 1981, 256, 1206–1213. [Google Scholar]
- Gonzalez-Covarrubias, V.; Ghosh, D.; Lakhman, S.S.; Pendyala, L.; Blanco, J.G. A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and NADPH binding affinity. Drug Metab. Dispos. 2007, 35, 973–980. [Google Scholar] [CrossRef]
- Mittelman-Smith, M.A.; Rudolph, L.M.; Mohr, M.A.; Micevych, P.E. Rodent models of non-classical progesterone action regulating ovulation. Front. Endocrinol. 2017, 8, 165. [Google Scholar] [CrossRef]
- Ikonomov, O.C.; Diego, S.; Takeshi, I.; Tadaomi, T.; Assia, S. Sac3 is an insulin-regulated phosphatidylinositol 3,5-bisphosphate phosphatase: Gain in insulin responsiveness through Sac3 down-regulation in adipocytes. J. Biol. Chem. 2009, 284, 23961–23971. [Google Scholar] [CrossRef]
- Sliwowska, J.H.; Fergani, C.; Gawałek, M.; Skowronska, B.; Fichna, P.; Lehman, M.N. Insulin: Its role in the central control of reproduction. Physiol. Behav. 2014, 133, 197–206. [Google Scholar] [CrossRef]
- Harterink, M.; Port, F.; Lorenowicz, M.J.; Mcgough, I.J.; Silhankova, M.; Betist, M.C.; Weering, J.R.T.V.; Heesbeen, R.G.H.P.V.; Middelkoop, T.C.; Basler, K. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor wntless and is required for Wnt secretion. Nat. Cell Biol. 2011, 13, 914–923. [Google Scholar] [CrossRef]
- Pons, V.; Luyet, P.P.; Morel, E.; Abrami, L.; Fg, V.D.G.; Parton, R.G.; Gruenberg, J. Hrs and SNX3 functions in sorting and membrane invagination within multivesicular bodies. PLoS Biol. 2008, 6, e214. [Google Scholar] [CrossRef]
- Hideaki, M.; Fumiaki, S.; Shigeto, S.; Masato, K.; Yosuke, T.; Shinji, S.; Manabu, F.; Hidefumi, I.; Yoshihito, T.; Norihito, U. ATP13A2 deficiency induces a decrease in cathepsin D activity, fingerprint-like inclusion body formation, and selective degeneration of dopaminergic neurons. FEBS Lett. 2013, 587, 1316–1325. [Google Scholar]
- Lahav, M.; Meidan, R.; Amsterdam, A.; Gebauer, H.; Lindner, H.R. Intracellular distribution of cathepsin D in rat corpora lutea in relation to reproductive state and the action of prostaglandin F2alpha and prolactin. J. Endocrinol. 1977, 75, 317–324. [Google Scholar] [CrossRef]
- Akopyan, T.N.; Barchudaryan, N.A.; Karabashyan, L.V.; Arutunyan, A.A.; Lajtha, A.; Galoyan, A.A. Hypothalamic cathepsin D: Assay and isoenzyme composition. J. Neurosci. Res. 1979, 4, 365–370. [Google Scholar] [CrossRef]
- Brunetti, L.; Orlando, G.; Michelotto, B.; Ragazzoni, E.; Vacca, M. Leptin stimulates prostaglandin E2 and F2alpha, but not nitric oxide production in neonatal rat hypothalamus. Eur. J. Pharmacol. 1999, 369, 299–304. [Google Scholar] [CrossRef]
- Salais-Lopez, H.; Agustin-Pavon, C.; Lanuza, E.; Martinez-Garcia, F. The maternal hormone in the male brain: Sexually dimorphic distribution of prolactin signalling in the mouse brain. PLoS ONE 2018, 13, e0208960. [Google Scholar] [CrossRef]
- Downing, J.A.; Joss, J.; Scaramuzzi, R.J. A mixture of the branched chain amino acids leucine, isoleucine and valine increases ovulation rate in ewes when infused during the late luteal phase of the oestrous cycle: An effect that may be mediated by insulin. J. Endocrinol. 1995, 145, 315–323. [Google Scholar] [CrossRef]
- Kim, J.; Burghardt, R.C.; Wu, G.; Johnson, G.A.; Spencer, T.E.; Bazer, F.W. Select nutrients in the ovine uterine lumen. IX. Differential effects of arginine, leucine, glutamine, and glucose on interferon Tau, Ornithine Decarboxylase, and Nitric Oxide Synthase in the ovine conceptus. Biol. Reprod. 2011, 84, 1139–1147. [Google Scholar] [CrossRef]
- Laís Rosa, V. A leucine-rich diet modulates the tumor-induced down-regulation of the MAPK/ERK and PI3K/Akt/mTOR signaling pathways and maintains the expression of the ubiquitin-proteasome pathway in the placental tissue of NMRI mice. Biol. Reprod. 2015, 92, 1–8. [Google Scholar]
- Parr, B.A.; Mcmahon, A.P. Sexually dimorphic development of the mammalian reproductive tract requires Wnt-7a. Nature 1998, 395, 707–710. [Google Scholar] [CrossRef]
- Jane, V.; Alexandra, G.; Laura, L. Wnt regulation of progenitor maturation in the cortex depends on Shh or fibroblast growth factor 2. J. Neurosci. 2003, 23, 5919–5927. [Google Scholar]
- Prajerova, I.; Honsa, P.; Chvatal, A.; Anderova, M. Distinct effects of sonic hedgehog and Wnt-7a on differentiation of neonatal neural stem/progenitor cells in vitro. Neuroscience 2010, 171, 693–711. [Google Scholar] [CrossRef]
- Iffland II, P.H.; Baybis, M.; Barnes, A.E.; Leventer, R.J.; Lockhart, P.J.; Crino, P.B. DEPDC5 and NPRL3 modulate cell size, filopodial outgrowth, and localization of mTOR in neural progenitor cells and neurons. Neurobiol. Dis. 2018, 114, 184–193. [Google Scholar] [CrossRef]
- Barrett, E.; Ross, R.P.; O’toole, P.W.; Fitzgerald, G.F.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef]
- Davila, A.M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.H.; Sanz, Y.; Tomé, D. Re-print of “Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host”. Pharmacol. Res. 2013, 69, 114–126. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Reviews. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef]
- Jackson, G.L.; Kuehl, D. Gamma-aminobutyric acid (GABA) regulation of GnRH secretion in sheep. Reprod. Suppl. 2002, 59, 15–24. [Google Scholar]
- Moore, A.M.; Abbott, G.; Mair, J.; Prescott, M.; Campbell, R.E. Mapping GABA and glutamate inputs to gonadotrophin-releasing hormone neurones in male and female mice. J. Neuroendocrinol. 2018, 30, e12657. [Google Scholar] [CrossRef]
- Piet, R.; Kalil, B.; Mclennan, T.; Porteous, R.; Czieselsky, K.; Herbison, A.E. Dominant neuropeptide co-transmission in kisspeptin-GABA regulation of GnRH neuron firing driving ovulation. J. Neurosci. 2018, 38, 6310–6322. [Google Scholar] [CrossRef]
- Manfredi-Lozano, M.; Roa, J.; Tena-Sempere, M. Connecting metabolism and gonadal function: Novel central neuropeptide pathways involved in the metabolic control of puberty and fertility. Front. Neuroendocrinol. 2018, 48, 37–49. [Google Scholar] [CrossRef]
- Goodman, R.L.; Maltby, M.J.; Millar, R.P.; Hileman, S.M.; Nestor, C.C.; Whited, B.; Tseng, A.S.; Coolen, L.M.; Lehman, M.N. Evidence that dopamine acts via kisspeptin to hold GnRH pulse frequency in check in anestrous ewes. Endocrinology 2012, 153, 5918–5927. [Google Scholar] [CrossRef]
- Nestor, C.C.; Bedenbaugh, M.N.; Hileman, S.M.; Coolen, L.M.; Lehman, M.N.; Goodman, R.L. Regulation of GnRH pulsatility in ewes. Reproduction 2018, 156, R83–R99. [Google Scholar] [CrossRef]
- Kim, H.S.; Yumkham, S.; Choi, J.H.; Son, G.H.; Kim, K.; Ryu, S.H.; Suh, P.G. Serotonin stimulates GnRH secretion through the c-Src-PLC gamma 1 pathway in GT1-7 hypothalamic cells. J. Endocrinol. 2006, 190, 581. [Google Scholar] [CrossRef]
- Kittel, A.; Müller, F.; König, J.; Mieth, M.; Sticht, H.; Zolk, O.; Kralj, A.; Heinrich, M.R.; Fromm, M.F.; Maas, R. Alanine-glyoxylate aminotransferase 2 (AGXT2) polymorphisms have considerable impact on methylarginine and β-aminoisobutyrate metabolism in healthy volunteers. PLoS ONE 2014, 9, e88544. [Google Scholar] [CrossRef]
- Cardounel, A.J.; Zweier, J.L. Endogenous methylarginines regulate neuronal nitric-oxide synthase and prevent excitotoxic injury. J. Biol. Chem. 2002, 277, 33995–34002. [Google Scholar] [CrossRef]
- Bellefontaine, N.; Hanchate, N.K.; Parkash, J.; Campagne, C.; De Seranno, S.; Clasadonte, J.; d’Anglemont de Tassigny, X.; Prevot, V. Nitric oxide as key mediator of neuron-to-neuron and endothelia-to-glia communication involved in the neuroendocrine control of reproduction. Neuroendocrinology 2011, 93, 74–89. [Google Scholar] [CrossRef]
- Romanelli, R.G.; Barni, T.; Maggi, M.; Luconi, M.; Failli, P.; Pezzatini, A.; Pelo, E.; Torricelli, F.; Crescioli, C.; Ferruzzi, P.; et al. Expression and function of gonadotropin-releasing hormone (GnRH) receptor in human olfactory GnRH-secreting neurons: An autocrine GnRH loop underlies neuronal migration. J. Biol. Chem. 2004, 279, 117–126. [Google Scholar] [CrossRef]
- Brancaccio, M.; Pivetta, C.; Granzotto, M.; Filippis, C.; Mallamaci, A. Emx2 and Foxg1 inhibit gliogenesis and promote neuronogenesis. Stem Cells 2010, 28, 1206–1218. [Google Scholar] [CrossRef]
- Dastidar, S.G.; Landrieu, P.M.Z.; D’Mello, S.R. FoxG1 promotes the survival of postmitotic neurons. J. Neurosci. 2011, 31, 402–413. [Google Scholar] [CrossRef]
- Seoane, J.; Le, H.V.; Shen, L.; Anderson, S.A.; Massagué, J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 2004, 117, 211–223. [Google Scholar] [CrossRef]
| Group | Sheep (Earmark) | Litter Size | Ovulation Rate | ||
|---|---|---|---|---|---|
| First Parity | Second Parity | Third Parity | |||
| Polytocous group | 4177 | 3 | 3 | 3 | 3 |
| 4775 | 2 | 3 | 3 | 3 | |
| 211 | 3 | 3 | 3 | 3 | |
| 265 | 2 | 3 | 3 | 3 | |
| 446 | 3 | 3 | 3 | 3 | |
| 12 | 3 | 3 | 3 | 3 | |
| Monotocous group | 4053 | 1 | 1 | 1 | 1 |
| 4205 | 1 | 1 | 1 | 1 | |
| 3048 | 1 | 1 | 1 | 1 | |
| 4298 | 1 | 1 | 1 | 1 | |
| 3390 | 1 | 1 | 1 | 1 | |
| 4282 | 1 | 1 | 1 | 1 | |
| Items | Total Reads | Mapped Reads | Mapping Rate | UnMapped Reads | MultiMap Reads | MultiMap Rate |
|---|---|---|---|---|---|---|
| PF_H1 | 122,545,762 | 112,451,378 | 91.76% | 10,094,384 | 5,101,242 | 4.16% |
| PF_H2 | 119,960,438 | 111,318,118 | 92.80% | 8,642,320 | 4,914,792 | 4.10% |
| PF_H3 | 126,498,382 | 116,661,576 | 92.22% | 9,836,806 | 4,706,940 | 3.72% |
| PL_H1 | 125,264,852 | 116,254,380 | 92.81% | 9,010,472 | 5,134,340 | 4.10% |
| PL_H2 | 96,925,068 | 90,129,243 | 92.99% | 6,795,825 | 4,194,309 | 4.33% |
| PL_H3 | 129,389,274 | 120,052,226 | 92.78% | 9,337,048 | 5,431,242 | 4.20% |
| MF_H1 | 120,588,746 | 111,485,736 | 92.45% | 9,103,010 | 4,572,173 | 3.79% |
| MF_H2 | 123,669,806 | 114,267,242 | 92.40% | 9,402,564 | 5,236,126 | 4.23% |
| MF_H3 | 127,179,196 | 117,301,094 | 92.23% | 9,878,102 | 6,219,896 | 4.89% |
| ML_H1 | 122,378,740 | 113,525,900 | 92.77% | 8,852,840 | 4,388,514 | 3.59% |
| ML_H2 | 121,575,158 | 112,668,594 | 92.67% | 8,906,564 | 4,770,077 | 3.92% |
| ML_H3 | 124,279,134 | 114,588,456 | 92.20% | 9,690,678 | 4,778,914 | 3.85% |
| LncRNA | Primer Sequence | Product Size (bp) |
|---|---|---|
| ENSOARG00000025579 | CCTCTGAAGCTGCGTGTGTA | 241 |
| TGGGGAGTGATTGAAGTCGG | ||
| ENSOARG00000025327 | AGTCCTTGTTCTGCTTGGGG | 176 |
| GCTTGACCATCTGCTCACCT | ||
| ENSOARG00000026466 | CCACAGCAAACTCAACGACC | 157 |
| TGCCATCAGAGAAAGAGCCG | ||
| MSTRG.158551 | AGGAAAGGCTGATGGTGGTG | 238 |
| CCTCGGGCTTTGTCTCCATT | ||
| MSTRG.235458 | TGATGGGAGGTTAGCTGGGA | 169 |
| TACGCACGGTTTGGTTGGTA | ||
| MSTRG.135103 | GGAGGTAGAGGGCAAAAGGT | 197 |
| GGGGAAGCAGAAACACAAGG | ||
| β-actin | CCAACCGTGAGAAGATGACC | 97 |
| CCCGAGGCGTACAGGGACAG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Tang, J.; Di, R.; Liu, Q.; Wang, X.; Gan, S.; Zhang, X.; Zhang, J.; Hu, W.; Chu, M. Comparative Transcriptomics Reveal Key Sheep (Ovis aries) Hypothalamus LncRNAs that Affect Reproduction. Animals 2019, 9, 152. https://doi.org/10.3390/ani9040152
Zhang Z, Tang J, Di R, Liu Q, Wang X, Gan S, Zhang X, Zhang J, Hu W, Chu M. Comparative Transcriptomics Reveal Key Sheep (Ovis aries) Hypothalamus LncRNAs that Affect Reproduction. Animals. 2019; 9(4):152. https://doi.org/10.3390/ani9040152
Chicago/Turabian StyleZhang, Zhuangbiao, Jishun Tang, Ran Di, Qiuyue Liu, Xiangyu Wang, Shangquan Gan, Xiaosheng Zhang, Jinlong Zhang, Wenping Hu, and Mingxing Chu. 2019. "Comparative Transcriptomics Reveal Key Sheep (Ovis aries) Hypothalamus LncRNAs that Affect Reproduction" Animals 9, no. 4: 152. https://doi.org/10.3390/ani9040152
APA StyleZhang, Z., Tang, J., Di, R., Liu, Q., Wang, X., Gan, S., Zhang, X., Zhang, J., Hu, W., & Chu, M. (2019). Comparative Transcriptomics Reveal Key Sheep (Ovis aries) Hypothalamus LncRNAs that Affect Reproduction. Animals, 9(4), 152. https://doi.org/10.3390/ani9040152





