The Hsp70 Gene Family in Boleophthalmus pectinirostris: Genome-Wide Identification and Expression Analysis under High Ammonia Stress
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Genome-Wide Identification of Hsp70 Genes in B. pectinirostris
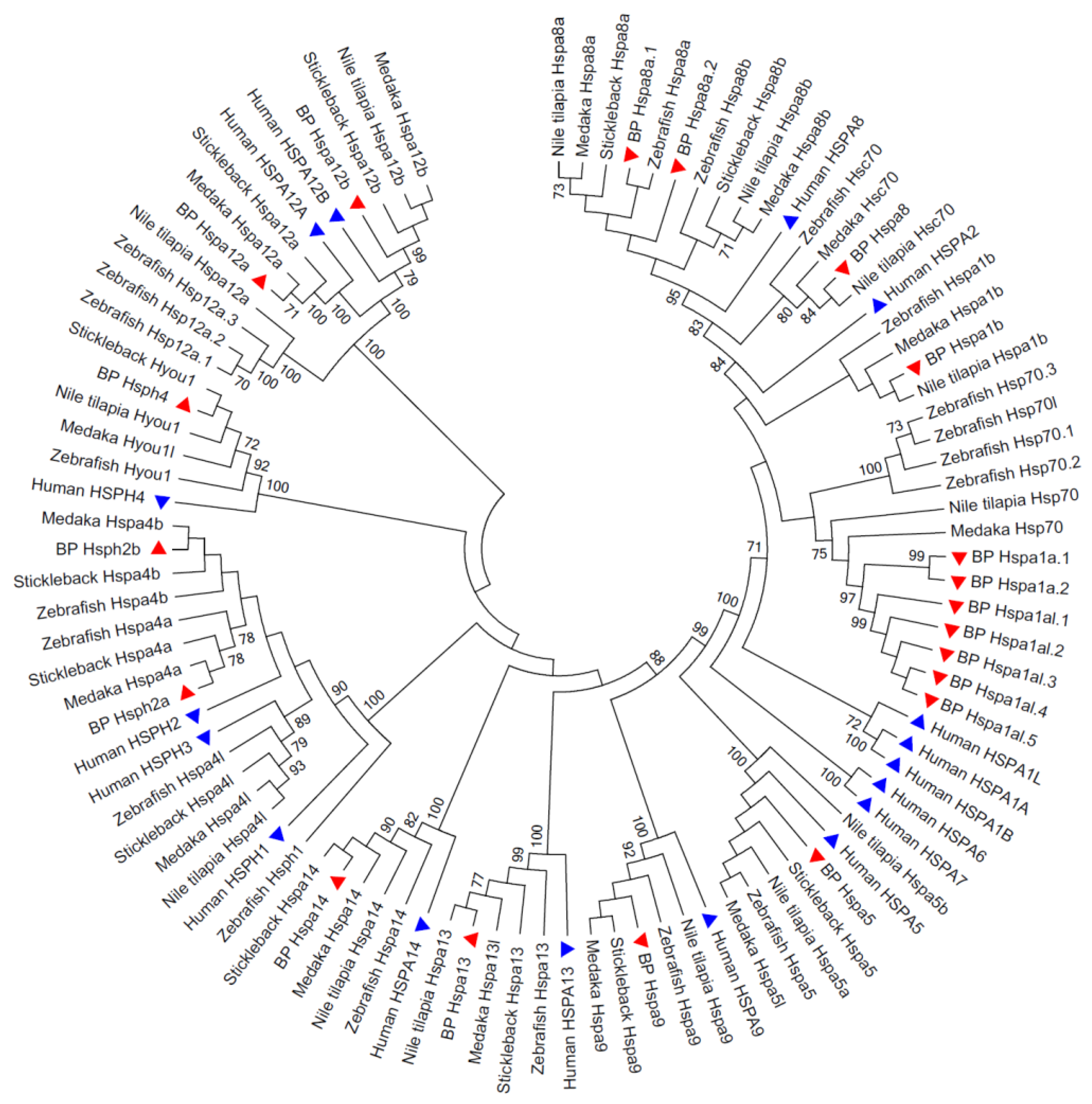
3.2. Phylogenetic Relationships of the Hsp70 Genes among Species
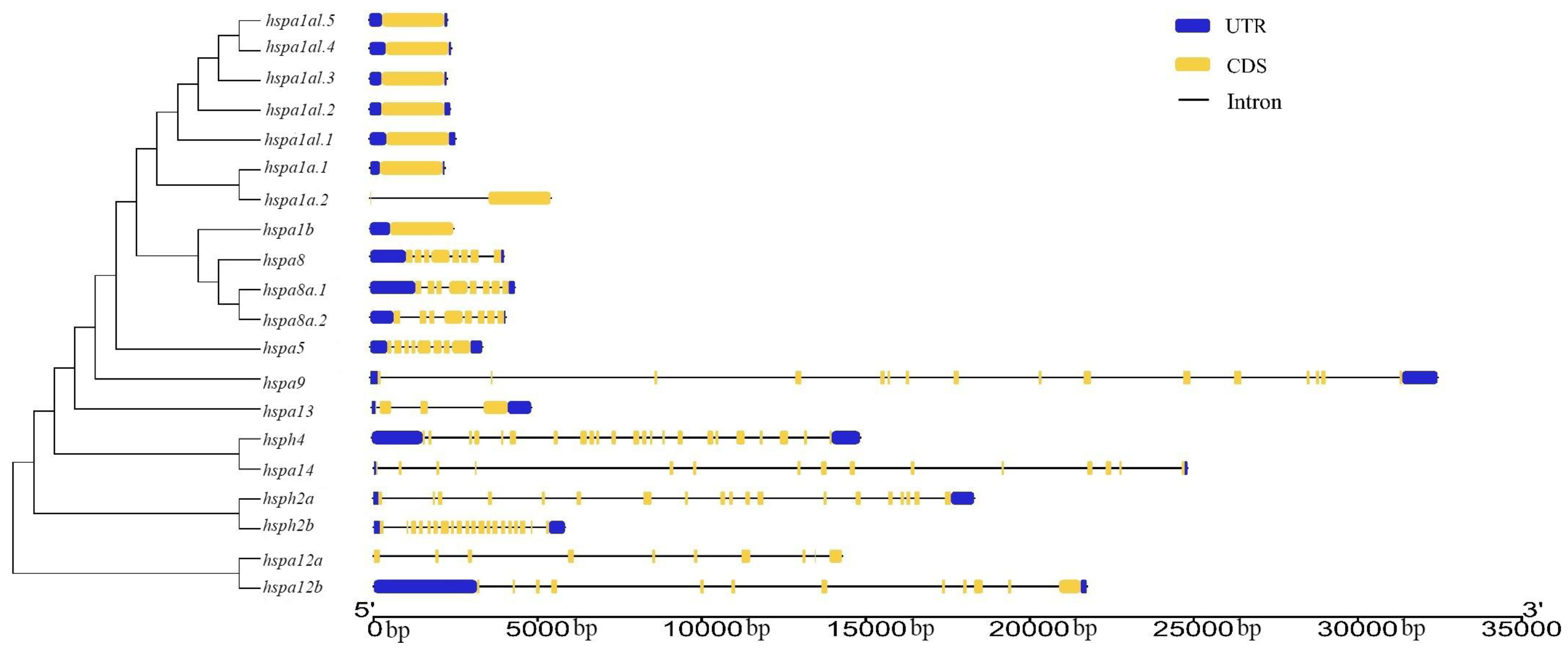
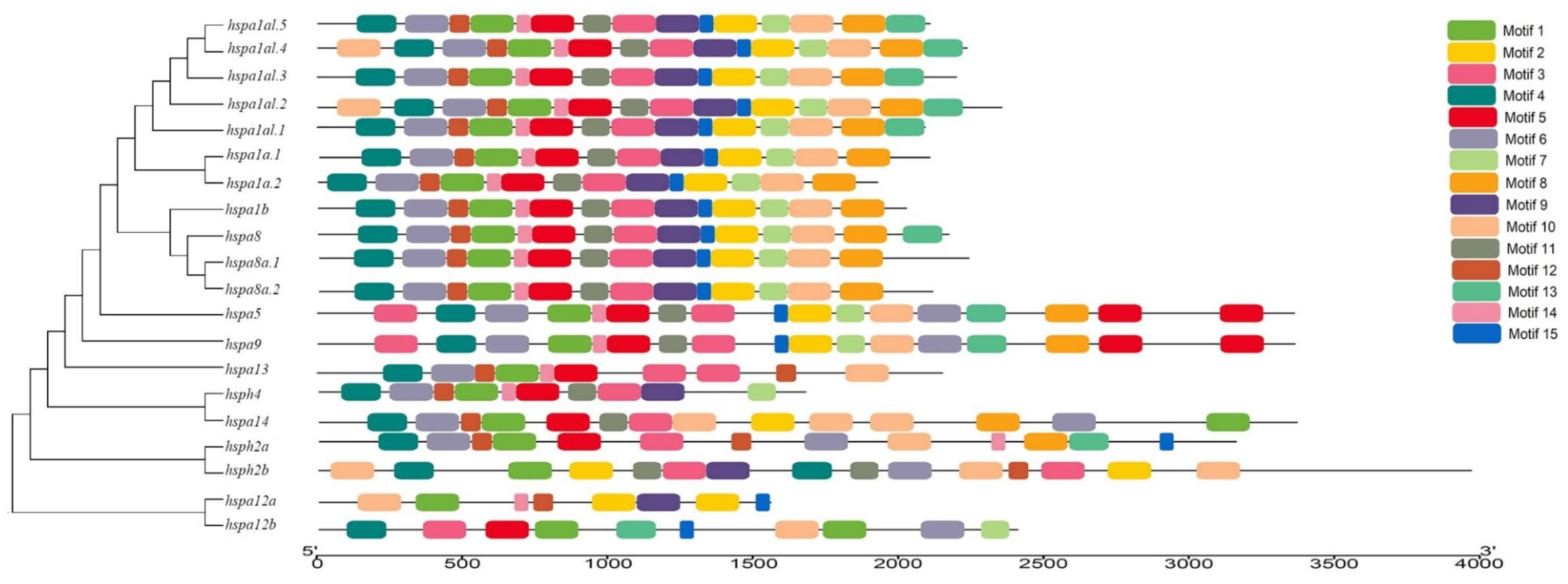
3.3. Gene Structure and Motif Analysis of Hsp70s
3.4. Expression Profiles of Hsp70 Genes in Different Tissues under HEA Stress
3.5. Selection on Duplicated Hsp70 Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Song, L.; Li, C.; Xie, Y.J.; Liu, S.K.; Zhang, J.R.; Yao, J.; Jiang, C.; Li, Y.; Liu, Z.J. Genome-wide identification of HSP70 genes in channel catfish and their regulated expression after bacterial infection. Fish Shellfish Immunol. 2016, 49, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Iwama, G.K.; Thomas, P.T.; Forsyth, R.B.; Vijayan, M.M. Heat shock protein expression in fish. Rev. Fish Biol. Fish. 1998, 8, 35–56. [Google Scholar] [CrossRef]
- Cheng, J.; Xun, X.; Kong, Y.; Wang, S.; Yang, Z.; Li, Y.; Kong, D.; Wang, Shi.; Zhang, L.; Hu, X.; et al. Hsp70 gene expansions in the scallop Patinopecten yessoensis genome and their expression regulation after exposure to the toxic dinoflagellate Alexandrium catenella. Fish Shellfish Immunol. 2016, 58, 266–273. [Google Scholar] [CrossRef]
- Arai, A.; Naruse, K.; Mitani, H.; Shima, A. Cloning and characterization of cDNAs for 70-kda heat-shock proteins (Hsp70) from two fish species of the genus Oryzias. Jpn. J. Genet. 1995, 70, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Deane, E.; Woo, N. Impact of heavy metals and organochlorines on HSP70 and HSC70 gene expression in black sea bream fibroblasts. Aquat. Toxicol. 2006, 79, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; He, J.Y.; Chi, C.F.; Shao, J. Differential HSP70 expression in Mytilus coruscus under various stressors. Gene 2014, 543, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.; Biemar, F.; Müller, F.; Iyengar, A.; Prunet, P.; Maclean, N.; Martiala, J.A.; Mullera, M. Cloning and expression analysis of an inducible HSP70 gene from tilapia fish. FEBS Lett. 2000, 474, 5–10. [Google Scholar] [CrossRef]
- Qin, C.; Zhao, D.; Gong, Q.; Qi, Z.; Zou, Y.; Yue, X.; Xie, B. Effects of pathogenic bacterial challenge after acute sublethal ammonia-N exposure on heat shock protein 70 expression in Botia reevesae. Fish Shellfish Immunol. 2013, 35, 1044–1047. [Google Scholar] [CrossRef]
- Kapoor, M.; Curle, C.A.; Runham, C. The hsp70 gene family of Neurospora crassa: Cloning, sequence analysis, expression, and genetic mapping of the major stress-inducible member. J. Bacteriol. 1995, 177, 212–221. [Google Scholar] [CrossRef]
- Werner-Washburne, M.; Craig, E.A. Expression of members of the Saccharomyces cerevisiae Hsp70 multigene family. Genome 1989, 31, 684–689. [Google Scholar] [CrossRef]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Gho, H.J.; Nguyen, M.X.; Kim, S.R.; An, G. Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Genom. 2013, 13, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.H.; Ma, X.; Zhai, Y.F.; Gong, Z.H.; Lu, M.H. Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Brocchieri, L.; Everly, C.D.M.; Macario, A.J.L. HSP70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.R.; Rush, S.J. Expression of heat shock genes (HSP70) in the mammalian brain: Distinguishing constitutively expressed and hyperthermia-inducible mRNA species. J. Neurosci. Res. 1990, 25, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.H.; Brenner, S. Short-range linkage relationships, genomic organisation and sequence comparisons of a cluster of five HSP70 genes in Fugu rubripes. Cell. Mol. Life Sci. 1999, 55, 668–678. [Google Scholar] [CrossRef]
- Cheng, C.H.; Yang, F.F.; Ling, R.Z.; Liao, S.A.; Miao, Y.T.; Ye, C.X.; Wang, A.L. Effects of ammonia exposure on apoptosis, oxidative stress and immune response in pufferfish (Takifugu obscurus). Aquat. Toxicol. 2015, 164, 61–71. [Google Scholar] [CrossRef]
- Randall, D.J.; Tsui, T.K. Ammonia toxicity in fish. Mar. Pollut. Bull. 2002, 45, 17–23. [Google Scholar] [CrossRef]
- Hegazi, M.M.; Attia, Z.I.; Ashour, O.A. Oxidative stress and antioxidant enzymes in liver and white muscle of Nile tilapia juveniles in chronic ammonia exposure. Aquat. Toxicol. 2010, 99, 118–125. [Google Scholar] [CrossRef]
- Murthy, C.R.; Rama Rao, K.V.; Bai, G.; Norenberg, M.D. Ammonia-induced production of free radicals in primary cultures of rat astrocytes. J. Neurosci. Res. 2001, 66, 282–288. [Google Scholar] [CrossRef]
- Jin, J.; Wang, Y.; Wu, Z.; Hergazy, A.; Lan, J.; Zhao, L.; Liu, X.; Chen, N.; Lin, L. Transcriptomic analysis of liver from grass carp (Ctenopharyngodon idellus) exposed to high environmental ammonia reveals the activation of antioxidant and apoptosis pathways. Fish Shellfish Immunol. 2017, 63, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Samali, A.; Orrenius, S. Triggering and modulation of apoptosis by oxidative stress. Free Radic. Biol. Med. 2000, 29, 323–333. [Google Scholar] [CrossRef]
- Padmini, E.; Usha, R.M. Impact of seasonal variation on HSP70 expression quantitated in stressed fish hepatocytes. Comp. Biochem. Physiol. B 2008, 151, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Hangzo, H.; Banerjee, B.; Saha, S.; Saha, N. Ammonia stress under high environmental ammonia induces Hsp70 and Hsp90 in the mud eel, Monopterus cuchia. Fish Physiol. Biochem. 2017, 43, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Takegaki, T. Threatened fishes of the world: Boleophthalmus pectinirostris (Linnaeus 1758) (Gobiidae). Environ. Biol. Fish. 2008, 81, 373–374. [Google Scholar] [CrossRef]
- Han, Y.L.; Yang, W.X.; Long, L.L.; Sheng, Z.; Zhou, Y.; Zhao, Y.Q.; Wang, Y.F.; Zhu, Z.Q. Molecular cloning, expression pattern, and chemical analysis of heat shock protein 70 (HSP70) in the mudskipper Boleophthalmus pectinirostris: Evidence for its role in regulating spermatogenesis. Gene 2016, 575, 331–338. [Google Scholar] [CrossRef] [PubMed]
- You, X.X.; Chen, J.M.; Bian, C.; Yi, Y.H.; Ruan, Z.Q.; Li, J.; Zhang, X.H.; Yu, H.; Xu, J.M.; Shi, Q. Transcriptomic evidence of adaptive tolerance to high environmental ammonia in mudskippers. Genomics 2018, 110, 404–413. [Google Scholar] [CrossRef]
- You, X.; Bian, C.; Zan, Q.; Xu, X.; Liu, X.; Chen, J.; Wang, J.; Qiu, Y.; Li, W.; Zhang, X.; et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5, 5594. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Xia, R.; Chen, H.; He, Y.H. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018, 2018, 289660. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Buske, F.A.; Boden, M.; Bauer, D.C.; Bailey, T.L. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics 2010, 26, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Ou, W.; Mao, X.; Huang, C.; Tie, W.; Yan, Y.; Ding, Z.; Wu, C.; Xia, Z.; Wang, W.; Zhou, S.; et al. Genome-wide identification and expression analysis of the KUP family under abiotic stress in cassava (Manihot esculenta Crantz). Front. Physiol. 2018, 9, 17. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Kulmuni, J.; Wurm, Y.; Pamilo, P. Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 2013, 110, 538–547. [Google Scholar] [CrossRef]
- Evgen’ev, M.B.; Garbuz, D.G.; Zatsepina, O.G. Heat Shock Proteins and Whole Body Adaptation to Extreme Environments; Springer: Dordrecht, The Netherlands, 2014; pp. 127–128. ISBN 978-94-017-9234-9. [Google Scholar]
- Metzger, D.C.H.; Hemmer-Hansen, J.; Schulte, P.M. Conserved structure and expression of hsp70 paralogs in teleost fishes. Comp. Biochem. Phys. D 2016, 18, 10–20. [Google Scholar] [CrossRef]
- Currie, S.; Bagatto, B.; Demille, M.; Learner, A.; Leblanc, D.; Marks, C.; Ong, K.; Parker, J.; Templeman, N.; Tufts, B.L.; et al. Metabolism, nitrogen excretion, and heat shock proteins in the central mudminnow (Umbra limi), a facultative air-breathing fish living in a variable environment. Can. J. Zool. 2010, 88, 43–58. [Google Scholar] [CrossRef]
- Yamashita, M.; Hirayoshi, K.; Nagata, K. Characterization of multiple members of the HSP70 family in platyfish culture cells: Molecular evolution of stress protein HSP70 in vertebrates. Gene 2004, 336, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Benli, A.C.; Köksal, G.; Ozkul, A. Sublethal ammonia exposure of Nile tilapia (Oreochromis niloticus L.): Effects on gill, liver and kidney histology. Chemosphere 2008, 72, 1355–1358. [Google Scholar] [CrossRef]
- Peng, K.W.; Chew, S.F.; Lim, C.B.; Kuah, S.S.L.; Kok, W.K.; Ip, Y.K. The mudskippers Periophthalmodon schlosseri and Boleophthalmus boddaerti can tolerate environmental NH3, concentrations of 446 and 36 µM, respectively. Fish Physiol. Biochem. 1998, 19, 59–69. [Google Scholar] [CrossRef]
- Campbell, J.W.; Aster, P.L.; Vorhaben, J.E. Mitochondrial ammoniagenesis in liver of the channel catfish Ictalurus punctatus. Am. J. Physiol. 1983, 244, R709–R717. [Google Scholar] [CrossRef] [PubMed]
- Ip, Y.K.; Chew, S.F.; Leong, I.A.; Jin, Y.; Lim, C.B.; Wu, R.S. The sleeper Bostrichthys sinensis (family Eleotridae) stores glutamine and reduces ammonia production during aerial exposure. J. Comp. Physiol. B 2001, 171, 357–367. [Google Scholar] [PubMed]
- Miron, D.D.S.; Moraes, B.; Becker, A.G.; Crestani, M.; Spanevello, R.; Loro, V.L.; Baldisserotto, B. Ammonia and pH effects on some metabolic parameters and gill histology of silver catfish, Rhamdia quelen (Heptapteridae). Aquaculture 2008, 277, 192–196. [Google Scholar] [CrossRef]
- Thurston, R.V.; Phillips, G.R.; Russo, R.C.; Hinkins, S.M. Increased toxicity of ammonia to rainbow trout (Salmo gairdneri) resulting from reduced concentrations of dissolved oxygen. Can. J. Fish. Aquat. Sci. 1981, 38, 983–988. [Google Scholar] [CrossRef]
- Alabaster, J.S.; Shurben, D.G.; Knowles, G. The effect of dissolved oxygen and salinity on the toxicity of ammonia to smolts of salmon, Salmo salar L. J. Fish. Biol. 1979, 15, 705–712. [Google Scholar] [CrossRef]
- Ao, J.; Mu, Y.; Xiang, L.; Fan, D.; Feng, M.; Zhang, S.; Shi, Q.; Zhu, L.; Li, T.; Ding, Y.; et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015, 11, e1005118. [Google Scholar] [CrossRef]
- Huang, L.; Mivechi, N.F.; Moskophidis, D. Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: Analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Mol. Cell Biol. 2001, 21, 8575–8591. [Google Scholar] [CrossRef] [PubMed]

| No. | Gene Name | Gen Accession Number | Protein Accession Number | CDS * Length (bp) | Protein Length (aa) | Hsp70 Domain Location (aa) | Domain Feature |
|---|---|---|---|---|---|---|---|
| 1 | hspa1al.5 | XM_020933256.1 | XP_020788915.1 | 1917 | 638 | 2-638 | HSPA1-2_6-8-like_NBD |
| 2 | hspa1al.4 | XM_020933257.1 | XP_020788916.1 | 1917 | 638 | 2-638 | HSPA1-2_6-8-like_NBD |
| 3 | hspa1al.1 | XM_020933255.1 | XP_020788914.1 | 1917 | 638 | 2-638 | HSPA1-2_6-8-like_NBD |
| 4 | hspa1al.2 | XM_020933253.1 | XP_020788912.1 | 1917 | 638 | 2-638 | HSPA1-2_6-8-like_NBD |
| 5 | hspa1al.3 | XM_020933254.1 | XP_020788913.1 | 1917 | 638 | 2-638 | HSPA1-2_6-8-like_NBD |
| 6 | hspa1a.1 | XM_020932913.1 | XP_020788572.1 | 1917 | 638 | 2-638 | HSPA1-2_6-8-like_NBD |
| 7 | hspa1a.2 | XM_020933010.1 | XP_020788669.1 | 1920 | 639 | 4-639 | HSPA1-2_6-8-like_NBD |
| 8 | hspa1b | XM_020929942.1 | XP_020785601.1 | 1917 | 638 | 2-638 | HSPA1-2_6-8-like_NBD |
| 9 | hspa8 | XM_020937593.1 | XP_020793252.1 | 1968 | 655 | 1-612 | HSPA1-2_6-8-like_NBD |
| 10 | hspa8a.1 | XM_020934606.1 | XP_020790265.1 | 1953 | 650 | 1-614 | HSPA1-2_6-8-like_NBD |
| 11 | hspa8a.2 | XM_020934610.1 | XP_020790269.1 | 1953 | 650 | 1-613 | HSPA1-2_6-8-like_NBD |
| 12 | hspa5 | XM_020925086.1 | XP_020780745.1 | 1956 | 651 | 28-633 | HSPA5-like_NBD |
| 13 | hspa9 | XM_020929023.1 | XP_020784682.1 | 2043 | 680 | 53-658 | HSPA9-like_NBD |
| 14 | hspa13 | XM_020926778.1 | XP_020782437.1 | 1314 | 437 | 12-429 | HSPA13-like_NBD |
| 15 | hspa14 | XM_020940290.1 | XP_020795949.1 | 1524 | 507 | 2-376 | HSPA14-like_NBD |
| 16 | hsph2a | XM_020925033.1 | XP_020780692.1 | 2508 | 835 | 2-384 | HSPA4_NBD |
| 17 | hsph2b | XM_020927815.1 | XP_020783474.1 | 2487 | 828 | 2-384 | HSPA4_NBD |
| 18 | hsph4 | XM_020934583.1 | XP_020790242.1 | 2928 | 975 | 30-416 | HYOU1-like_NBD |
| 19 | hspa12b | XM_020920454.1 | XP_020776113.1 | 2091 | 696 | 68-535 | HSPA12B_like_NBD |
| 20 | hspa12a | XM_020937725.1 | XP_020793384.1 | 1549 | 515 | 1-379 | HSPA12A_like_NBD |
| No. | Homo sapiens | Danio rerio | Boleophthalmus pectinirostris |
|---|---|---|---|
| 1 | HSPA1A | hsp70.1; hsp70.2; hsp70.3; hsp70-like | hspa1a.1; hspa1a.2; hspa1al.1; hspa1al.2; hspa1al.3; hspa1al.4; hspa1al.5 |
| 2 | HSPA1B | hspa1b | hspa1b |
| 3 | HSPA1L | - | - |
| 4 | HSPA2 | - | - |
| 5 | HSPA5 | hspa5 | hspa5 |
| 6 | HSPA6 | - | - |
| 7 | HSPA7 | - | - |
| 8 | HSPA8 | hspa8a; hspa8b; hsc70 | hspa8a.1; hspa8a.2; hspa8 |
| 9 | HSPA9 | hspa9 | hspa9 |
| 10 | HSPA12A | hspa12a.1; hspa12a.2; hspa12a.3 | hspa12a |
| 11 | HSPA12B | - | hspa12b |
| 12 | HSPA13 | hspa13 | hspa13 |
| 13 | HSPA14 | hspa14 | hspa14 |
| 14 | HSPH1 | hsph1 | - |
| 15 | HSPH2 | hspa4a; hspa4b | hsph2a; hsph2b |
| 16 | HSPH3 | hspa4l | - |
| 17 | HSPH4 | hyou1 | hsph4 |
| Gene\Tissue | Liver | Gill | ||||
|---|---|---|---|---|---|---|
| log2FC | 0 h | 72 h | log2FC | 0 h | 72 h | |
| hspa1a.2 | - | 0 | 0 | −6.6 | 1.94 | 0.02 |
| hspa1a.1 | −1.75 | 7.68 | 2.29 | −4.02 | 20.63 | 1.27 |
| hspa1al.1 | −1.2 | 49.14 | 21.39 | −3.54 | 37.57 | 3.23 |
| hspa1al.2 | −2.17 | 59.1 | 13.13 | −3.72 | 64.8 | 4.91 |
| hspa1al.3 | - | 0 | 0.63 | −2.21 | 1.62 | 0.35 |
| hspa1al.4 | −2.13 | 54.67 | 12.45 | −4.14 | 75.78 | 4.31 |
| hspa1al.5 | −1.28 | 8.48 | 3.49 | - | 0 | 0.67 |
| hspa1b | −0.32 | 1.24 | 0.99 | −1.04 | 1.46 | 0.71 |
| hspa8 | 2.92 | 3.06 | 23.11 | 0.01 | 23.32 | 23.51 |
| hspa8a.1 | −1.35 | 3483.05 | 1368.19 | 0.22 | 2486.01 | 2889.56 |
| hspa8a.2 | 0.64 | 14.15 | 22.14 | 0.61 | 38.26 | 58.47 |
| hspa5 | 0.46 | 227.06 | 312.16 | −1.61 | 363.63 | 118.77 |
| hspa9 | 0.21 | 48.79 | 56.41 | −0.03 | 60.68 | 59.34 |
| hsph2a | 0.48 | 3.53 | 4.92 | −0.52 | 9.02 | 6.27 |
| hsph2b | −0.45 | 20.68 | 15.15 | −0.77 | 64.33 | 37.78 |
| hsph4 | −0.54 | 29.33 | 20.23 | −1 | 21.98 | 11.01 |
| hspa12a | - | 0.05 | 0.06 | - | 0.41 | 0.27 |
| hspa12b | 0.74 | 0.37 | 0.62 | 0 | 2.91 | 2.91 |
| hspa13 | −0.11 | 14.39 | 13.32 | −0.58 | 16.85 | 11.24 |
| hspa14 | 0.62 | 15.69 | 24.16 | 0.73 | 29.67 | 49.24 |
| Model | −InL a | 2ΔInL | df | Parameters | Positively Selected Sites |
|---|---|---|---|---|---|
| M1a | 3122.75 | - | - | p0 = 0.9724, p1 = 0.0276, ω0 = 0.024, ω1 = 1.00 | - |
| M2a | 3113.92 | - | - | p0 = 0.9927, p1 = 0, p2 = 0.0073, ω0 = 0.0318, ω1 = 1.00, ω2 = 28.2246 | 2 (0.982); 3 (0.925); 5 (0.910); 6 (0.936) |
| M7 | 3124.70 | - | - | p = 0.0569, q = 0.9137 | - |
| M8 | 3114.02 | - | - | p0 = 0.9927, p1 = 0.0073, p = 3.3214, q = 99.00, ω = 28.2314 | 2 (0.983); 3 (0.942); 5 (0.938); 6 (0.951); 100 (0.510) |
| M1a vs. M2a b | 17.66 c | 2 | - | - | |
| M7 vs. M8 b | 21.36 c | 2 | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Z.; Sun, S.; Gao, T.; Han, Z. The Hsp70 Gene Family in Boleophthalmus pectinirostris: Genome-Wide Identification and Expression Analysis under High Ammonia Stress. Animals 2019, 9, 36. https://doi.org/10.3390/ani9020036
Deng Z, Sun S, Gao T, Han Z. The Hsp70 Gene Family in Boleophthalmus pectinirostris: Genome-Wide Identification and Expression Analysis under High Ammonia Stress. Animals. 2019; 9(2):36. https://doi.org/10.3390/ani9020036
Chicago/Turabian StyleDeng, Zhaochao, Shanxiao Sun, Tianxiang Gao, and Zhiqiang Han. 2019. "The Hsp70 Gene Family in Boleophthalmus pectinirostris: Genome-Wide Identification and Expression Analysis under High Ammonia Stress" Animals 9, no. 2: 36. https://doi.org/10.3390/ani9020036
APA StyleDeng, Z., Sun, S., Gao, T., & Han, Z. (2019). The Hsp70 Gene Family in Boleophthalmus pectinirostris: Genome-Wide Identification and Expression Analysis under High Ammonia Stress. Animals, 9(2), 36. https://doi.org/10.3390/ani9020036





