Identification of Single Nucleotide Polymorphisms in Porcine MAOA Gene Associated with Aggressive Behavior of Weaned Pigs after Group Mixing
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Housing
2.2. Behavioral Assessment
2.3. Aggression Assessment
2.4. Genetic Polymorphism Analyses of MAOA Gene
2.4.1. Promoter Prediction of the Porcine MAOA Gene
2.4.2. Plasmid Construction
2.4.3. Cell Culture, Cell Transfection, and Luciferase Assays
2.4.4. Potential SNP Identification
2.5. Data Analyses
3. Results
3.1. The Difference of Behavioral Indicators between Aggressive and Docile Weaned Pigs after Mixing
3.2. Prediction and Identification of the Core Promoter in Porcine MAOA Gene
3.2.1. MAOA Promoter Prediction
3.2.2. MAOA Promoter Identification
3.3. Screening of SNPs in the Porcine MAOA Gene Associated with Aggression
3.3.1. MAOA Gene Polymorphism and Aggressive Behavior and Linkage Disequilibrium (LD) Analyses
3.3.2. Differences of Behavioral Indicators between Genotypes/Haplotypes of Four Linked SNPs
3.4. Promoter Activity Analyses of Porcine MAOA Gene
4. Discussion
4.1. The Selection of Aggressive and Docile Pigs after Mixing
4.2. Prediction and Identification of the Core Promoter in the Porcine MAOA Gene
4.3. Screening on SNPs in Porcine MAOA Gene Associated with Aggression
4.4. In Vitro Expression of Different Genotypes in the Porcine MAOA Gene Promoter Region
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Desire, S.; Turner, S.P.; D’Eath, R.B.; Doeschl-Wilson, A.B.; Lewis, C.R.; Roehe, R. Prediction of reduction in aggressive behaviour of growing pigs using skin lesion traits as selection criteria. Animal 2016, 10, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Verdon, M.; Rault, J.-L. Aggression in group housed sows and fattening pigs. In Advances in Pig Welfare; Elsevier: Amsterdam, The Netherlands, 2018; pp. 235–260. [Google Scholar]
- Williamson, C.M.; Lee, W.; Curley, J.P. Temporal dynamics of social hierarchy formation and maintenance in male mice. Anim. Behav. 2016, 115, 259–272. [Google Scholar] [CrossRef]
- Turner, S.P.; Nevison, I.M.; Desire, S.; Camerlink, I.; Roehe, R.; Ison, S.H.; Farish, M.; Jack, M.C.; D’Eath, R.B. Aggressive behaviour at regrouping is a poor predictor of chronic aggression in stable social groups. Appl. Anim. Behav. Sci. 2017, 191, 98–106. [Google Scholar] [CrossRef]
- Turner, S.P.; Farnworth, M.J.; White, I.M.S.; Brotherstone, S.; Mendl, M.; Knap, P.; Penny, P.; Lawrence, A.B. The accumulation of skin lesions and their use as a predictor of individual aggressiveness in pigs. Appl. Anim. Behav. Sci. 2006, 96, 245–259. [Google Scholar] [CrossRef]
- D’Eath, R.B.; Turner, S.P.; Kurt, E.; Evans, G.; Tholking, L.; Looft, H.; Wimmers, K.; Murani, E.; Klont, R.; Foury, A.; et al. Pigs’ aggressive temperament affects pre-slaughter mixing aggression, stress and meat quality. Animal 2010, 4, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Liang, T.; Fu, L.; Li, H.; Zhou, B. Behavioural genetic differences between Chinese and European pigs. J. Genet. 2017, 96, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Konig von Borstel, U.; Tonepohl, B.; Appel, A.K.; Voss, B.; Brandt, H.; Naderi, S.; Gauly, M. Suitability of traits related to aggression and handleability for integration into pig breeding programmes: Genetic parameters and comparison between Gaussian and binary trait specifications. PLoS ONE 2018, 13, e0204211. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, G.A.; Brown-Brandl, T.; Rempel, L.A.; Schneider, J.F.; Holl, J. Genetic analysis of behavior traits in swine production. Livest. Sci. 2013, 157, 28–37. [Google Scholar] [CrossRef]
- Dennis, R.L.; Chen, Z.Q.; Cheng, H.W. Serotonergic mediation of aggression in high and low aggressive chicken strains. Poult. Sci. 2008, 87, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Craig, I.W.; Halton, K.E. Genetics of human aggressive behaviour. Hum. Genet. 2009, 126, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Terranova, J.I.; Song, Z.; Larkin, T.E., 2nd; Hardcastle, N.; Norvelle, A.; Riaz, A.; Albers, H.E. Serotonin and arginine-vasopressin mediate sex differences in the regulation of dominance and aggression by the social brain. Proc. Natl. Acad. Sci. USA 2016, 113, 13233–13238. [Google Scholar] [CrossRef] [PubMed]
- Buckholtz, J.W.; Meyer-Lindenberg, A. MAOA and the neurogenetic architecture of human aggression. Trends Neurosci. 2008, 31, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Popova, N.K. From genes to aggressive behavior: The role of serotonergic system. Bioessays 2006, 28, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Cases, O.; Seif, I.; Grimsby, J.; Gaspar, P.; Chen, K.; Pournin, S.; Muller, U.; Aguet, M.; Babinet, C.; Shih, J.C.; et al. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science 1995, 268, 1763–1766. [Google Scholar] [CrossRef] [PubMed]
- Shih, J.C.; Thompson, R.F. Monoamine oxidase in neuropsychiatry and behavior. Am. J. Hum. Genet. 1999, 65, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Brunner, H.G.; Nelen, M.; Breakefield, X.O.; Ropers, H.H.; van Oost, B.A. Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science 1993, 262, 578–580. [Google Scholar] [CrossRef] [PubMed]
- Ficks, C.A.; Waldman, I.D. Candidate genes for aggression and antisocial behavior: A meta-analysis of association studies of the 5HTTLPR and MAOA-uVNTR. Behav. Genet. 2014, 44, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, H.; Deater-Deckard, K.; Zhang, W. Interacting Effect of Catechol-O-Methyltransferase (COMT) and Monoamine Oxidase A (MAOA) Gene Polymorphisms, and Stressful Life Events on Aggressive Behavior in Chinese Male Adolescents. Front. Psychol. 2018, 9, 1079. [Google Scholar] [CrossRef] [PubMed]
- Chester, D.S.; DeWall, C.N.; Derefinko, K.J.; Estus, S.; Peters, J.R.; Lynam, D.R.; Jiang, Y. Monoamine oxidase A (MAOA) genotype predicts greater aggression through impulsive reactivity to negative affect. Behav. Brain Res. 2015, 283, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zheng, W.; Xue, Y.; Yao, W. Suhuai suckling piglet hindgut microbiome-metabolome responses to different dietary copper levels. Appl. Microbiol. Biotechnol. 2019, 103, 853–868. [Google Scholar] [CrossRef] [PubMed]
- Stukenborg, A.; Traulsen, I.; Puppe, B.; Presuhn, U.; Krieter, J. Agonistic behaviour after mixing in pigs under commercial farm conditions. Appl. Anim. Behav. Sci. 2011, 129, 28–35. [Google Scholar] [CrossRef]
- Camerlink, I.; Arnott, G.; Farish, M.; Turner, S.P. Complex contests and the influence of aggressiveness in pigs. Anim. Behav. 2016, 121, 71–78. [Google Scholar] [CrossRef]
- O’Malley, C.I.; Wurtz, K.E.; Steibel, J.P.; Bates, R.O.; Ernst, C.W.; Siegford, J.M. Relationships among aggressiveness, fearfulness and finisher pigs response to humans in finisher pigs. Appl. Anim. Behav. Sci. 2018, 205, 194–201. [Google Scholar] [CrossRef]
- Figler, M.H.; Peeke, H.V.S.; Chang, E.S. Maternal aggression in American lobsters (Homarus americanusMilne-Edwards): Shelter-related encounters against non-maternal female conspecifics. Mar. Freshwat. Behav. Physiol. 2009, 30, 267–274. [Google Scholar] [CrossRef]
- Langbein, J.; Puppe, B. Analysing dominance relationships by sociometric methods—A plea for a more standardised and precise approach in farm animals. Appl. Anim. Behav. Sci. 2004, 87, 293–315. [Google Scholar] [CrossRef]
- Tuchscherer, M.; Puppe, B.; Tuchscherer, A.; Kanitz, E. Effects of social status after mixing on immune, metabolic, and endocrine responses in pigs. Physiol. Behav. 1998, 64, 353–360. [Google Scholar] [CrossRef]
- Albert, D.J.; Dyson, E.M.; Walsh, M.L.; Petrovic, D.M. Cohabitation with a female activates testosterone-dependent social aggression in male rats independently of changes in serum testosterone concentration. Physiol. Behav. 1988, 44, 735–740. [Google Scholar] [CrossRef]
- Turner, S.P. Breeding against harmful social behaviours in pigs and chickens: State of the art and the way forward. Appl. Anim. Behav. Sci. 2011, 134, 1–9. [Google Scholar] [CrossRef]
- Chen, G.L.; Vallender, E.J.; Miller, G.M. Functional characterization of the human TPH2 5’ regulatory region: Untranslated region and polymorphisms modulate gene expression in vitro. Hum. Genet. 2008, 122, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Juneja, M.; Ilm, K.; Schlag, P.M.; Stein, U. Promoter identification and transcriptional regulation of the metastasis gene MACC1 in colorectal cancer. Mol. Oncol. 2013, 7, 929–943. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.P.; Roehe, R.; D’Eath, R.B.; Ison, S.H.; Farish, M.; Jack, M.C.; Lundeheim, N.; Rydhmer, L.; Lawrence, A.B. Genetic validation of postmixing skin injuries in pigs as an indicator of aggressiveness and the relationship with injuries under more stable social conditions. J. Anim. Sci. 2009, 87, 3076–3082. [Google Scholar] [CrossRef] [PubMed]
- D’Eath, R.B.; Roehe, R.; Turner, S.P.; Ison, S.H.; Farish, M.; Jack, M.C.; Lawrence, A.B. Genetics of animal temperament: Aggressive behaviour at mixing is genetically associated with the response to handling in pigs. Animal 2009, 3, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Boyle, L.A.; Bjorklund, L. Effects of fattening boars in mixed or single sex groups and split marketing on pig welfare. Anim. Welfare 2007, 16, 259–262. [Google Scholar]
- Bongiorni, S.; Tilesi, F.; Bicorgna, S.; Iacoponi, F.; Willems, D.; Gargani, M.; D’Andrea, M.; Pilla, F.; Valentini, A. Promoter polymorphisms in genes involved in porcine myogenesis influence their transcriptional activity. BMC Genet. 2014, 15, 119. [Google Scholar] [CrossRef] [PubMed]
- Dennis, R.L.; Cheng, H.W. The dopaminergic system and aggression in laying hens. Poult. Sci. 2011, 90, 2440–2448. [Google Scholar] [CrossRef] [PubMed]
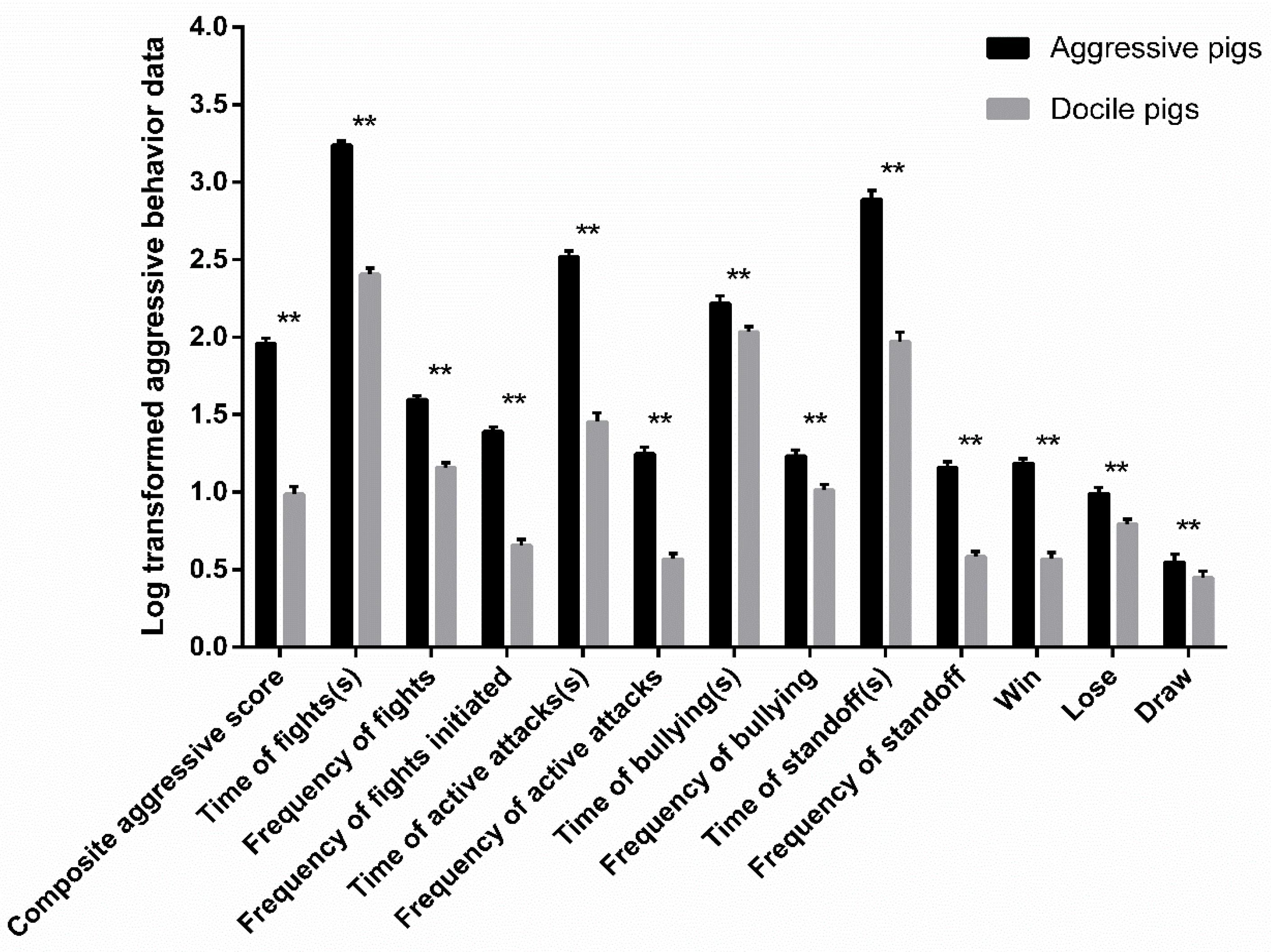
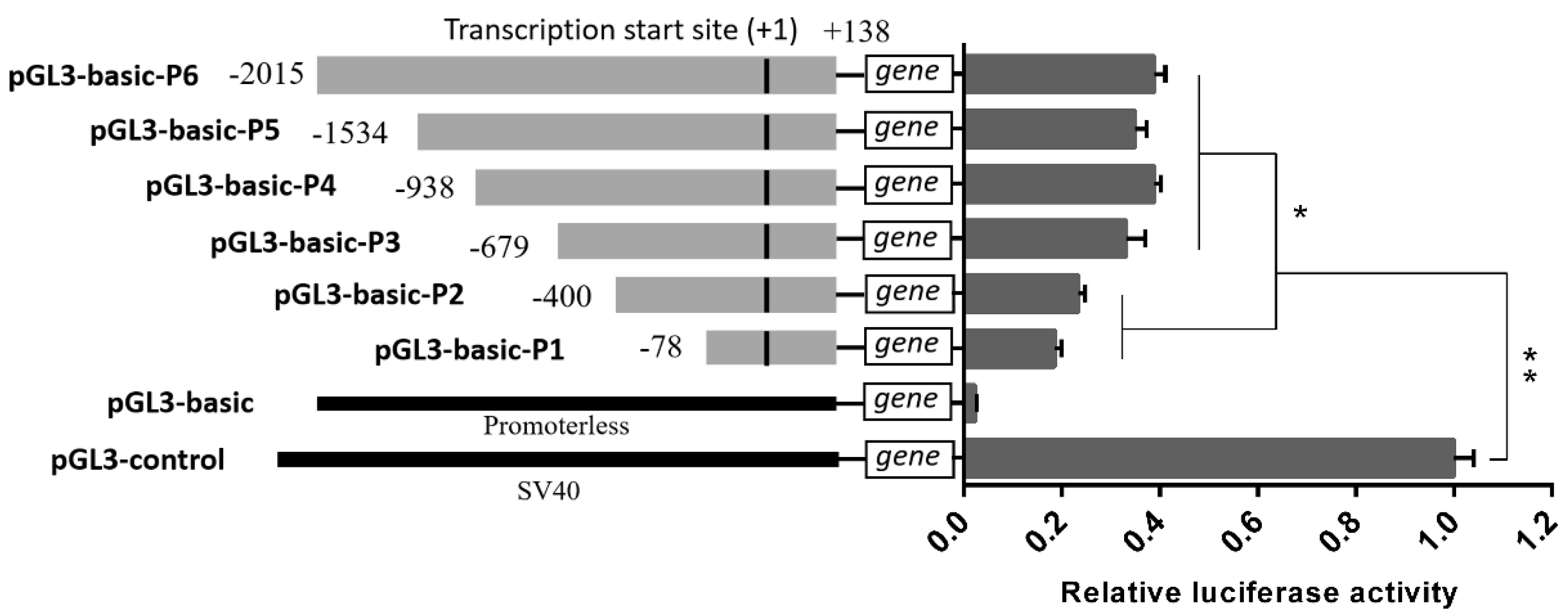
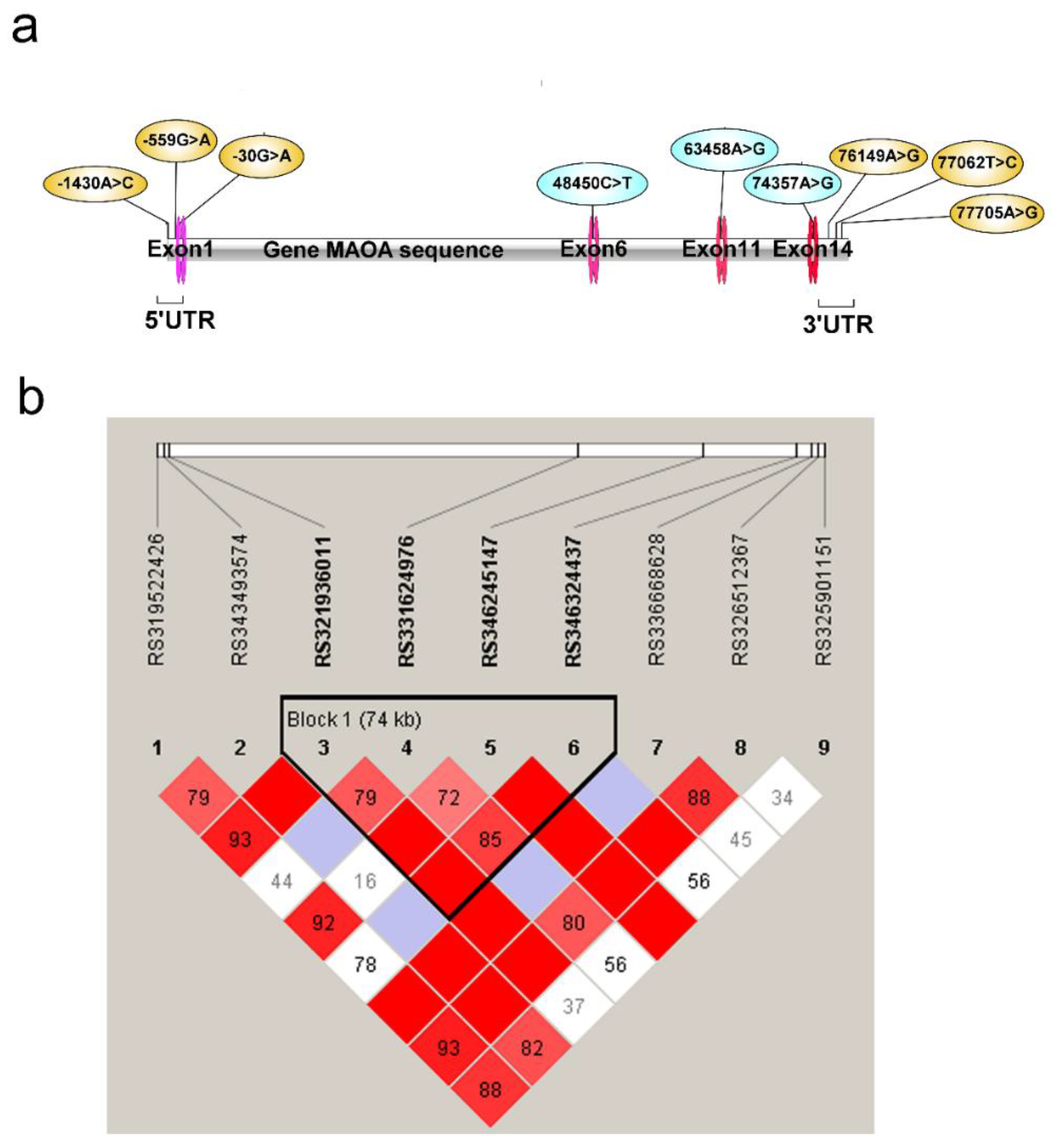
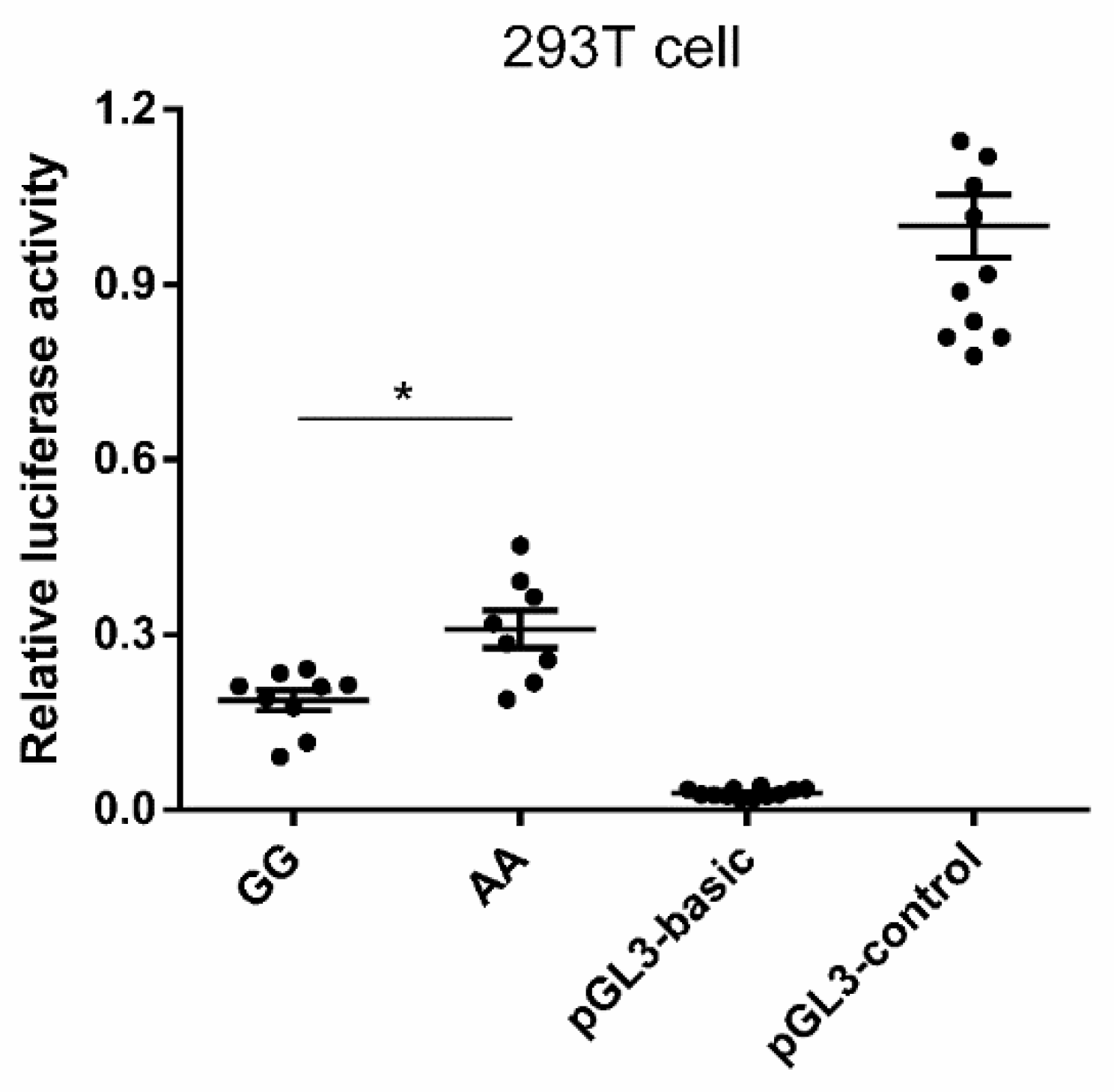
| Trait | Description |
|---|---|
| Fight | A vigorous biting, head-knocking, or a displacement with physical contact of two individuals for more than 3 s and intervening periods of at least 8 s, while the fight either was interrupted or the pigs showed other behaviors [22], including active attack, being bullied, and standoff. |
| Active Attack | In a fight, a pig give biting, pushing, chasing [23]. |
| Being bullied | When the recipient pig suffers from biting and head-knocking performed by the actor pig the recipient moves away without retaliation, it was identified as a being bullied [24]. |
| Standoff | If the two pigs stand side by side, shoulders by shoulders, and one pig throws his head to the head or neck of the other pig, there was an aggressive interaction with no dominance sign produced by either pair member at any time [25]. |
| Win/lose/draw | A pig showing a submissive manner, such as stopping fighting, turning away from an attack, trying to flee or was displaced from the location, it was defined as a loser, the other pig in the fight was defined as a winner [26,27]. If there was no clear outcome, the fight was designated as a draw [22]. |
| SNPs | Position | Region | SNP Site | Allele | Allele Frequency | MAF | HW P | χ2 | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| (Aggressive/Docile Pigs) | |||||||||
| rs319522426 | ChrX: 38,929,022 | 5’UTR | A > C | A | 0.55/0.35 | 0.458 | 0.3672 | 8.083 | 0.004 ** |
| (−1430 A or C) | C | 0.45/0.65 | |||||||
| rs343493574 | ChrX: 38,929,893 | 5’UTR | G > A | G | 0.72/0.65 | 0.289 | 0.8452 | 1.038 | 0.308 |
| (−559 G or A) | A | 0.28/0.35 | |||||||
| rs321936011 | ChrX: 38,930,422 | 5’UTR | G > A | G | 0.75/0.65 | 0.295 | 0.6978 | 4.389 | 0.036 * |
| (−30 G or A) | A | 0.25/0.35 | |||||||
| rs331624976 | ChrX: 38,978,901 | Intron 5 | C > T | C | 0.94/0.84 | 0.109 | 0.0268 | 10.046 | 0.002 ** |
| (+48,450 C or T) | T | 0.06/0.16 | |||||||
| rs346245147 | ChrX: 38,993,909 | Intron 11 | A > G | A | 0.63/0.47 | 0.425 | 0.9756 | 10.332 | 0.001 ** |
| (+63,458 A or G) | G | 0.37/0.53 | |||||||
| rs346324437 | ChrX: 39,004,808 | Intron 14 | A > G | A | 0.94/0.84 | 0.108 | 0.9197 | 9.813 | 0.002 ** |
| (+74,357 A or G) | G | 0.06/0.16 | |||||||
| rs336668628 | ChrX: 39,006,600 | 3’UTR | A > G | A | 0.84/0.81 | 0.184 | 0.7396 | 0.258 | 0.611 |
| (+76,149 A or G) | G | 0.16/0.19 | |||||||
| rs326512367 | ChrX: 39,007,513 | 3’UTR | T > C | T | 0.82/0.63 | 0.284 | 0.7494 | 8.841 | 0.003 ** |
| (+77,062 T or C) | C | 0.18/0.37 | |||||||
| rs325901151 | ChrX: 39,008,156 | 3’UTR | A > G | A | 0.88/0.79 | 0.152 | 0.0002 | 2.818 | 0.093 |
| (+77,705 A or G) | G | 0.12/0.21 |
| SNP | Geno-Type | n (Barrows/Gilts) | CAS | Duration of Fights (s) | Frequency of Fights Initiated | Duration of Active Attacks (s) | Frequency of Active Attacks | Duration of Standoff (s) | Frequency of Standoff | Win |
|---|---|---|---|---|---|---|---|---|---|---|
| rs321936011 | GG | 114 (82/32) | 4.32 ± 0.11 a | 7.11 ± 0.10 a | 3.00 ± 0.10 | 5.72 ± 0.11 a | 2.68 ± 0.10 ab | 6.54 ± 0.10 a | 2.28 ± 0.10 b | 2.51 ± 0.10 |
| AG | 52 (1/51) | 4.24 ± 0.17 a | 7.29 ± 0.17 a | 2.87 ± 0.17 | 5.52 ± 0.17 a | 2.97 ± 0.17 a | 6.99 ± 0.17 b | 2.88 ± 0.17 a | 2.62 ± 0.17 | |
| AA | 34 (21/13) | 3.61 ± 0.17 b | 6.60 ± 0.17 b | 2.66 ± 0.17 | 4.86 ± 0.17 b | 2.38 ± 0.17 b | 5.88 ± 0.18 c | 2.04 ± 0.17 b | 2.21 ± 0.17 | |
| rs331624976 | CC | 168 (94/24) | 4.26 ± 0.08 a | 7.15 ± 0.08 a | 2.97 ± 0.08 | 5.61 ± 0.08 a | 2.77 ± 0.08 a | 6.66 ± 0.08 a | 2.49 ± 0.08 a | 2.50 ± 0.08 |
| TC | 20 (2/18) | 4.13 ± 0.24 a | 6.90 ± 0.24 ab | 2.61 ± 0.23 | 5.49 ± 0.24 a | 2.63 ± 0.24 a | 6.24 ± 0.24 ab | 2.14 ± 0.24 ab | 2.61 ± 0.23 | |
| TT | 13 (8/5) | 3.20 ± 0.28 b | 6.34 ± 0.28 b | 2.4 ± 0.28 | 4.54 ± 0.28 b | 1.72 ± 0.28 b | 5.65 ± 0.28 b | 1.69 ± 0.28 b | 2.25 ± 0.28 | |
| rs346245147 | AA | 83 (61/22) | 4.51 ± 0.12 a | 7.31 ± 0.12 a | 3.17 ± 0.12 a | 5.89 ± 0.12 a | 2.92 ± 0.13 a | 6.78 ± 0.12 a | 2.50 ± 0.13 | 2.77 ± 0.12 a |
| GA | 53 (5/48) | 3.97 ± 0.16 b | 7.07 ± 0.16 ab | 2.68 ± 0.16 b | 5.26 ± 0.16 b | 2.70 ± 0.16 ab | 6.68 ± 0.16 a | 2.56 ± 0.16 | 2.33 ± 0.16 b | |
| GG | 63 (36/27) | 3.88 ± 0.13 b | 6.76 ± 0.13 b | 2.68 ± 0.13 b | 5.24 ± 0.13 b | 2.38 ± 0.13 b | 6.17 ± 0.13 b | 2.19 ± 0.13 | 2.17 ± 0.13 b | |
| rs346324437 | AA | 168 (94/73) | 4.26 ± 0.08 a | 7.15 ± 0.08 a | 2.97 ± 0.08 a | 5.61 ± 0.08 a | 2.77 ± 0.08 a | 6.67 ± 0.08 a | 2.49 ± 0.08 a | 2.49 ± 0.08 a |
| GA | 22 (1/21) | 4.05 ± 0.23 a | 6.83 ± 0.23 ab | 2.59 ± 0.23 ab | 5.41 ± 0.23 a | 2.54 ± 0.23 ab | 6.14 ± 0.23 ab | 2.08 ± 0.23 ab | 2.72 ± 0.23 a | |
| GG | 12 (9/3) | 3.23 ± 0.29 b | 6.34 ± 0.29 b | 2.33 ± 0.29 b | 4.56 ± 0.29 b | 1.81 ± 0.29 b | 5.70 ± 0.29 b | 1.71 ± 0.29 b | 1.79 ± 0.29 b |
| Haplotypes | GCAA | ACGA | GCGA | ATGG | ACGG/GTAA |
|---|---|---|---|---|---|
| n (Barrows/Gilts) | 82 (60/22) | 55 (12/43) | 29 (19/10) | 22 (9/13) | 6 (2/4) |
| Composite aggressive score | 4.51 ± 0.12 a | 4.08 ± 0.15 b | 3.81 ± 0.19 bc | 3.46 ± 0.22 c | 3.95 ± 0.41 abc |
| Duration of fights (s) | 7.33 ± 0.12 a | 7.12 ± 0.14 ab | 6.66 ± 0.19 bc | 6.54 ± 0.22 c | 6.41 ± 0.41 bc |
| Frequency of fights | 3.62 ± 0.12 a | 3.49 ± 0.14 ab | 3.18 ± 0.19 b | 3.38 ± 0.21 ab | 2.86 ± 0.41 ab |
| Frequency of fights initiated | 3.15 ± 0.12 a | 2.91 ± 0.15 ab | 2.46 ± 0.19 b | 2.47 ± 0.22 b | 2.25 ± 0.41 b |
| Duration of active attacks (s) | 5.90 ± 0.12 a | 5.33 ± 0.15 b | 5.26 ± 0.20 bc | 4.82 ± 0.22 c | 5.34 ± 0.41 abc |
| Frequency of active attacks | 2.91 ± 0.12 a | 2.86 ± 0.15 a | 2.03 ± 0.19 bc | 1.92 ± 0.21 b | 2.22 ± 0.41 abc |
| Frequency of being bullied | 2.99 ± 0.12 ac | 3.16 ± 0.14 a | 2.54 ± 0.19 b | 3.05 ± 0.21 abc | 2.28 ± 0.41 bc |
| Duration of standoff (s) | 6.82 ± 0.12 a | 6.72 ± 0.14 a | 6.00 ± 0.19 b | 5.95 ± 0.21 b | 5.29 ± 0.41 b |
| Frequency of standoff | 2.51 ± 0.12 a | 2.73 ± 0.15 a | 1.88 ± 0.19 b | 1.81 ± 0.21 b | 1.58 ± 0.41 b |
| Win | 2.74 ± 0.12 a | 2.44 ± 0.14 a | 1.67 ± 0.19 b | 2.35 ± 0.22 a | 2.06 ± 0.41 ab |
| Draw | 1.36 ± 0.12 a | 1.39 ± 0.14 a | 1.67 ± 0.19 a | 0.22 ± 0.23 c | −0.75 ± 0.42 b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, R.; Chu, Q.; Shen, C.; Tong, X.; Gao, S.; Liu, X.; Zhou, B.; Schinckel, A.P. Identification of Single Nucleotide Polymorphisms in Porcine MAOA Gene Associated with Aggressive Behavior of Weaned Pigs after Group Mixing. Animals 2019, 9, 952. https://doi.org/10.3390/ani9110952
Chen R, Chu Q, Shen C, Tong X, Gao S, Liu X, Zhou B, Schinckel AP. Identification of Single Nucleotide Polymorphisms in Porcine MAOA Gene Associated with Aggressive Behavior of Weaned Pigs after Group Mixing. Animals. 2019; 9(11):952. https://doi.org/10.3390/ani9110952
Chicago/Turabian StyleChen, Ruonan, Qingpo Chu, Chunyan Shen, Xian Tong, Siyuan Gao, Xinpeng Liu, Bo Zhou, and Allan P. Schinckel. 2019. "Identification of Single Nucleotide Polymorphisms in Porcine MAOA Gene Associated with Aggressive Behavior of Weaned Pigs after Group Mixing" Animals 9, no. 11: 952. https://doi.org/10.3390/ani9110952
APA StyleChen, R., Chu, Q., Shen, C., Tong, X., Gao, S., Liu, X., Zhou, B., & Schinckel, A. P. (2019). Identification of Single Nucleotide Polymorphisms in Porcine MAOA Gene Associated with Aggressive Behavior of Weaned Pigs after Group Mixing. Animals, 9(11), 952. https://doi.org/10.3390/ani9110952






