Differential Expression of Circular RNAs in Polytocous and Monotocous Uterus during the Reproductive Cycle of Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. RNA Extraction, Library Construction, and RNA-Seq
2.3. Sequence Mapping and Circular RNAs (circRNA) Prediction
2.4. Differential Expression Analysis of circRNAs
2.5. Bioinformatics Analysis
2.6. Validation of the Expression of circRNAs
3. Results
3.1. Overview of circRNA Profiles in Polytocous and Monotocous Sheep Uteri
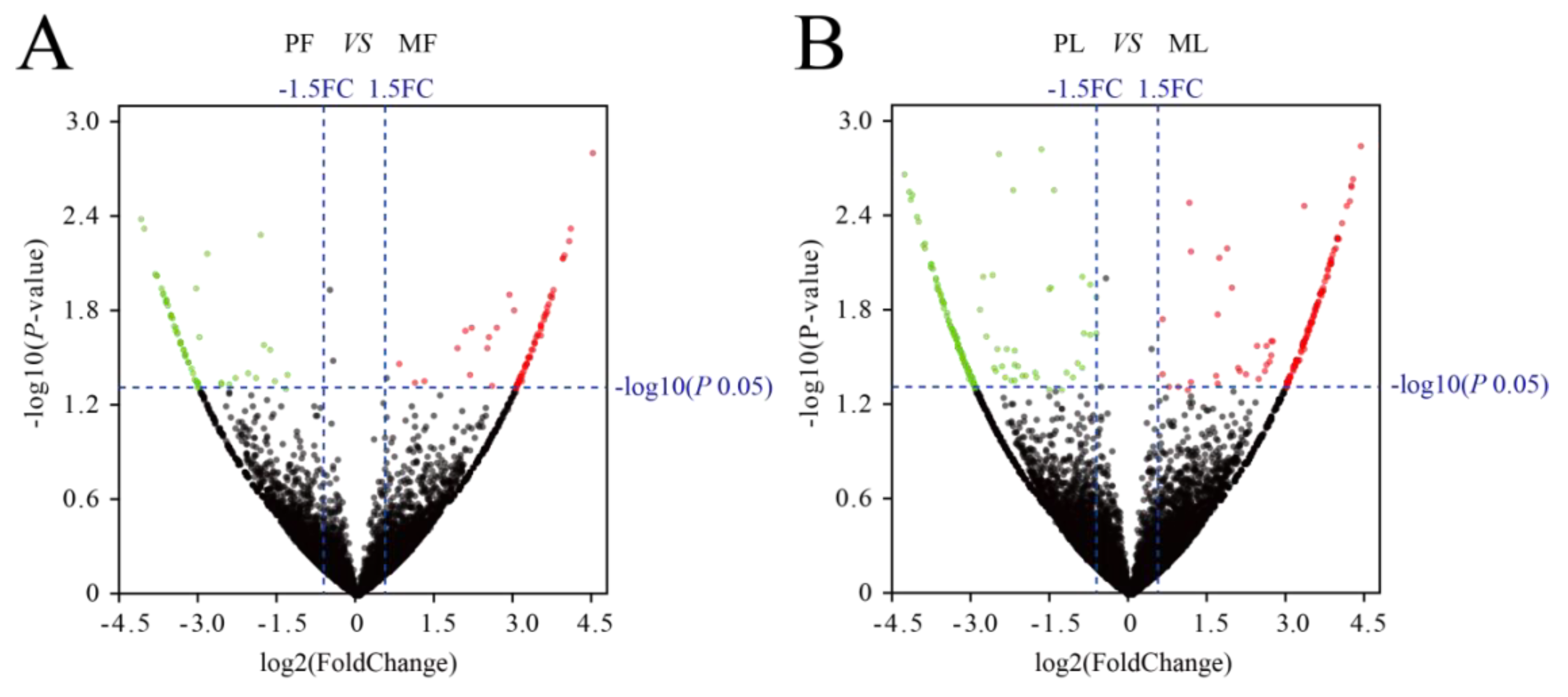
3.2. Differential Expression Analysis of circRNAs
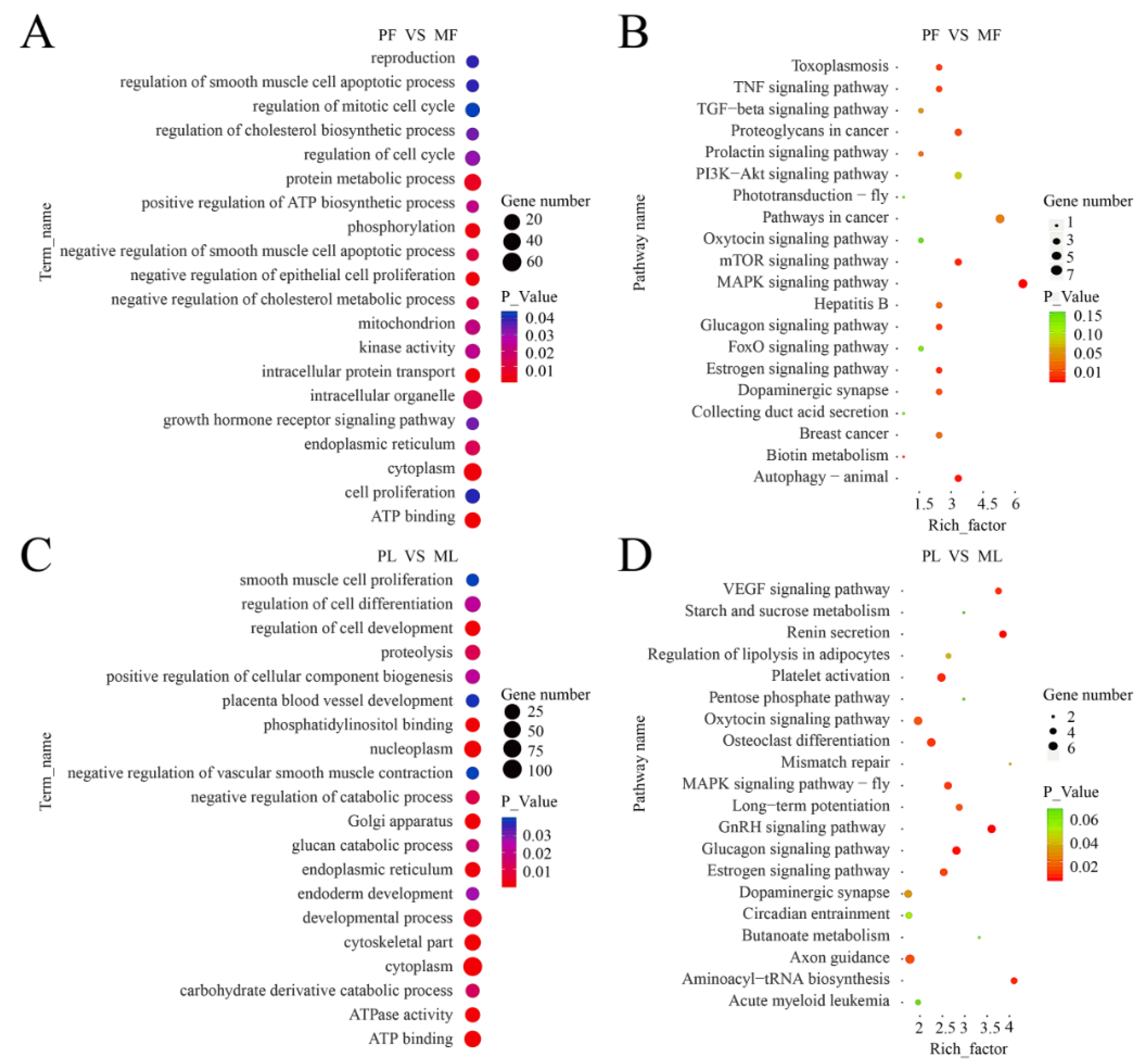
3.3. Gene Ontology (GO) and KEGG Pathway Enrichment Analyses
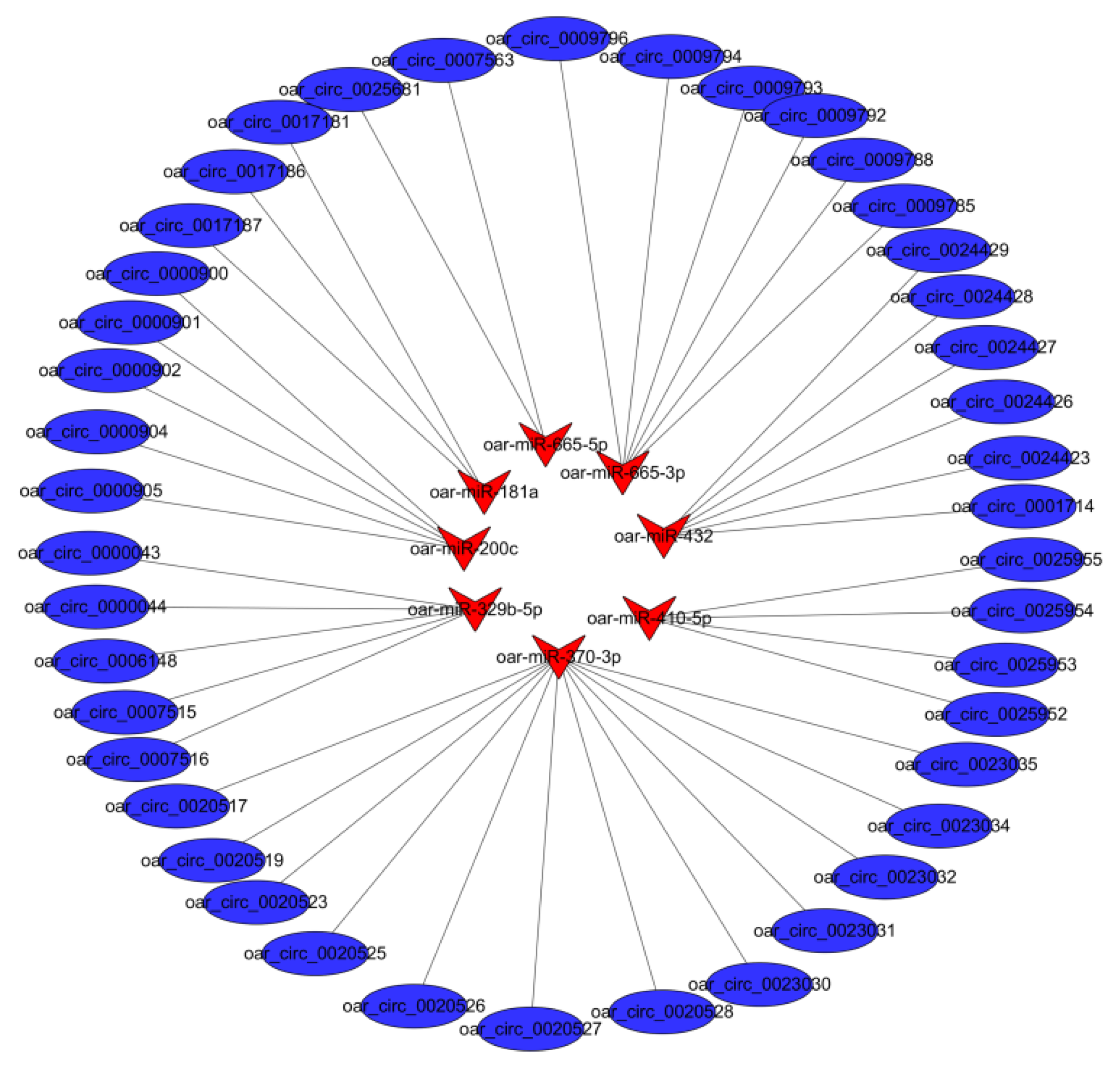
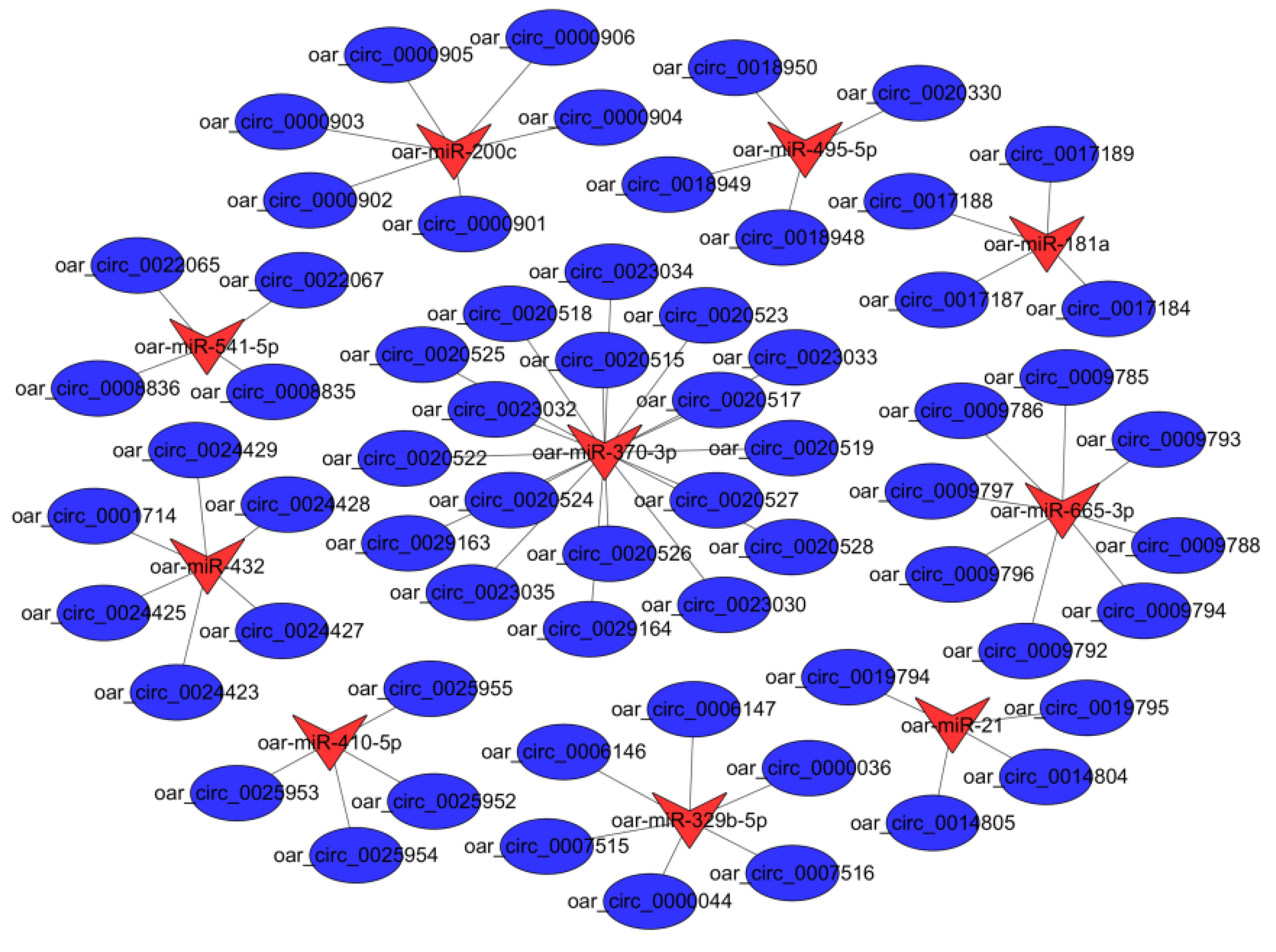
3.4. Target miRNAs of Differentially Expressed circRNAs in PG and MG
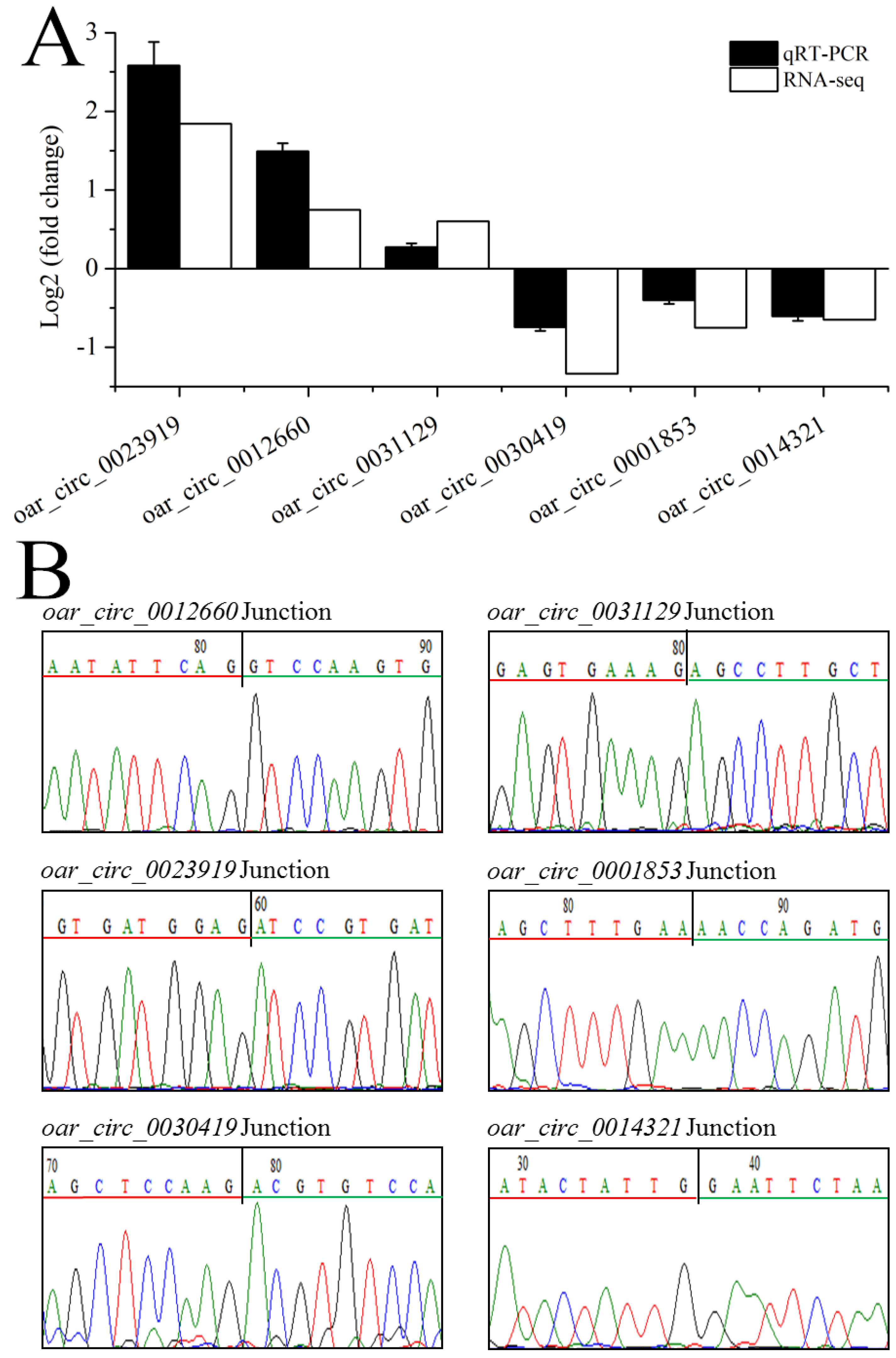
3.5. Validation of circRNA Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Miao, X.; Luo, Q.; Zhao, H.; Qin, X. Ovarian proteomic study reveals the possible molecular mechanism for hyperprolificacy of Small Tail Han sheep. Sci. Rep. 2016, 6, 27606. [Google Scholar] [CrossRef]
- Davis, G.H.; Montgomery, G.W.; Allison, A.J.; Kelly, R.W.; Bray, A.R. Segregation of a major gene influencing fecundity in progeny of Booroola sheep. N. Z. J. Agric. 1982, 67, 525–529. [Google Scholar] [CrossRef]
- Fogarty, N.M. A review of the effects of the Booroola gene (FecB) on sheep production. Small Rumin. Res. 2009, 85, 75–84. [Google Scholar] [CrossRef]
- Wilson, T.; Wu, X.Y.; Juenge, J.L.; Ross, I.K.; Lumsden, J.M.; Lord, E.A.; Dodds, K.G.; Walling, G.A.; McEwan, J.C.; Connell, A.R.; et al. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol. Reprod. 2001, 64, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.J.; MacDougall, C.; Campbell, B.K.; McNeilly, A.S.; Baird, D.T. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene. J. Endocrinol. 2001, 169, R1. [Google Scholar] [CrossRef]
- Mulsant, P.; Lecerf, F.; Fabre, S.; Schibler, L.; Monget, P.; Lanneluc, I.; Pisselet, C.; Riquet, J.; Monniaux, D.; Callebaut, I.; et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Mérino ewes. Proc. Natl. Acad. Sci. USA 2001, 98, 5104–5109. [Google Scholar] [CrossRef]
- McBride, D.; Carre, W.; Sontakke, S.D.; Hogg, C.O.; Law, A.; Donadeu, F.X.; Clinton, M. Identification of miRNAs associated with the follicular-luteal transition in the ruminant ovary. Reproduction 2012, 144, 221–233. [Google Scholar] [CrossRef]
- Mihm, M.; Gangooly, S.; Muttukrishna, S. The normal menstrual cycle in women. Anim. Reprod. Sci. 2011, 124, 229–236. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.; Zhao, H.; Qin, X. Ovarian transcriptomic study reveals the differential regulation of miRNAs and lncRNAs related to fecundity in different sheep. Sci. Rep. 2016, 6, 35299. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.; Qin, X. Genome-wide transcriptome analysis in the ovaries of two goats identifies differentially expressed genes related to fecundity. Gene 2016, 582, 69–76. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, P. Understanding implantation window, a crucial phenomenon. J. Hum. Reprod. Sci. 2012, 5, 2–6. [Google Scholar] [PubMed]
- Kao, L.C.; Tulac, S.; Lobo, S.; Imani, B.; Yang, J.P.; Germeyer, A.; Osteen, K.; Taylor, R.N.; Lessey, B.A.; Giudice, L.C. Global gene profiling in human endometrium during the window of implantation. Endocrinology 2002, 143, 2119–2138. [Google Scholar] [CrossRef] [PubMed]
- Strassmann, B.I. The evolution of endometrial cycles and menstruation. Q. Rev. Biol. 1996, 71, 181–220. [Google Scholar] [CrossRef] [PubMed]
- Groebner, A.E.; Rubio-Aliaga, I.; Schulke, K.; Reichenbach, H.D.; Daniel, H.; Wolf, E.; Meyer, H.H.; Ulbrich, S.E. Increase of essential amino acids in the bovine uterine lumen during preimplantation development. Reproduction 2011, 141, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.E.; Gopichandran, N.; Picton, H.M.; Leese, H.J.; Orsi, N.M. Nutrient concentrations in murine follicular fluid and the female reproductive tract. Theriogenology 2005, 64, 992–1006. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W. Uterine protein secretions: Relationship to development of the conceptus. J. Anim. Sci. 1975, 41, 1376–1382. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wu, G.; Spencer, T.E.; Johnson, G.A.; Li, X.; Bazer, F.W. Select nutrients in the ovine uterine lumen. I. Amino acids, glucose, and ions in uterine lumenal flushings of cyclic and pregnant ewes. Biol. Reprod. 2009, 80, 86–93. [Google Scholar] [CrossRef]
- Paria, B.C.; Reese, J.; Das, S.K.; Dey, S.K. Deciphering the cross-talk of implantation: Advances and challenges. Science 2002, 296, 2185–2188. [Google Scholar] [CrossRef]
- Lim, H.; Ma, L.; Ma, W.G.; Maas, R.L.; Dey, S.K. Hoxa-10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Mol. Endocrinol. 1999, 13, 1005–1017. [Google Scholar] [CrossRef]
- Lijie, S.; Ruize, L.; Wei, C.; Mengjin, Z.; Xiaoping, L.; Shuhong, Z.; Mei, Y. Expression patterns of microRNAs in porcine endometrium and their potential roles in embryo implantation and placentation. PLoS ONE 2014, 9, e87867. [Google Scholar]
- Xia, H.F.; Jin, X.H.; Cao, Z.F.; Hu, Y.; Ma, X. MicroRNA expression and regulation in the uterus during embryo implantation in rat. FEBS J. 2014, 281, 1872–1891. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Bhosale, R.; Boudolf, V.; Cuevas, F.; Lu, R.; Eekhout, T.; Hu, Z.; van Isterdael, G.; Lambert, G.M.; Xu, F.; Nowack, M.K.; et al. A spatiotemporal DNA endoploidy map of the Arabidopsis root reveals roles for the endocycle in root development and stress adaptation. Plant Cell 2018, 30, 2330–2351. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Cai, X.; Wang, Q.; Wang, P.; Zhang, Y.; Cai, C.; Xu, Y.; Wang, K.; Zhou, Z.; Wang, C.; et al. Genome sequencing of the Australian wild diploid species Gossypium australe highlights disease resistance and delayed gland morphogenesis. Plant Biotechnol. J. 2019, 3, 13249. [Google Scholar] [CrossRef] [PubMed]
- La, Y.; Tang, J.; He, X.; Di, R.; Wang, X.; Liu, Q.; Zhang, L.; Zhang, X.; Zhang, J.; Hu, W.; et al. Identification and characterization of mRNAs and lncRNAs in the uterus of polytocous and monotocous Small Tail Han sheep (Ovis aries). Peer J. 2019, 7, e6938. [Google Scholar] [CrossRef]
- Veno, M.T.; Hansen, T.B.; Veno, S.T.; Clausen, B.H.; Grebing, M.; Finsen, B.; Holm, I.E.; Kjems, J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015, 16, 245. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Bente, F.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, X.; Che, S.; Cui, J.; Liu, Y.; An, X.; Cao, B.; Song, Y. CircRNA-9119 regulates the expression of prostaglandin-endoperoxide synthase 2 (PTGS2) by sponging miR-26a in the endometrial epithelial cells of dairy goat. Reprod. Fertil. Dev. 2018, 30, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, X.; Ma, Q.; Zhang, X.; Cao, Y.; Yao, Y.; You, S.; Wang, D.; Quan, R.; Hou, X.; et al. Genome-wide analysis of circular RNAs in prenatal and postnatal pituitary glands of sheep. Sci. Rep. 2017, 7, 16143. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, X.; Yao, Y.; Ma, Q.; Ni, W.; Zhang, X.; Cao, Y.; Hazi, W.; Wang, D.; Quan, R.; et al. Genome-wide analysis of circular RNAs in prenatal and postnatal muscle of sheep. Oncotarget 2017, 8, 97165–97177. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; You, S.; Yao, Y.; Liu, Z.J.; Hazi, W.; Li, C.Y.; Zhang, X.Y.; Hou, X.X.; Wei, J.C.; Li, X.Y.; et al. Expression profiles of circular RNAs in sheep skeletal muscle. Asian Australas. J. Anim. Sci. 2018, 31, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Bao, J.; Wang, Y.; Chen, W.; Zou, S.; Wu, T.; Wang, L.; Lv, X.; Gao, W.; Wang, B.; et al. Changes in circRNA expression profiles related to the antagonistic effects of Escherichia coli F17 in lamb spleens. Sci. Rep. 2018, 8, 14524. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Yang, W.W.; Liao, P.; Guo, Y.W.; Kumar, A.; Gao, W. Transcriptome analysis reveals differentially expressed ERF transcription factors associated with salt response in cotton. Plant Sci. 2019, 281, 72–81. [Google Scholar] [CrossRef]
- Wang, D.; Yang, C.; Long, D.; Zhu, J.; Wang, J.; Zhang, S. Comparative transcriptome analyses of drought-resistant and - susceptible Brassica napus L. and development of EST-SSR markers by RNA-Seq. J. Plant Biol. 2015, 58, 259–269. [Google Scholar] [CrossRef]
- Wang, P.; Yang, C.; Chen, H.; Luo, L.; Leng, Q.; Li, S.; Han, Z.; Li, X.; Song, C.; Zhang, X.; et al. Exploring transcription factors reveals crucial members and regulatory networks involved in different abiotic stresses in Brassica napus L. BMC Plant Biol. 2018, 18, 202. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, J.; Di, R.; Liu, Q.; Wang, X.; Gan, S.; Zhang, X.; Zhang, J.; Hu, W.; Chu, M. Comparative transcriptomics reveal key sheep (Ovis aries) hypothalamus lncRNAs that affect reproduction. Animals 2019, 9, 152. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Houtgast, E.J.; Sima, V.M.; Bertels, K.; Al-Ars, Z. Hardware acceleration of BWA-MEM genomic short read mapping for longer read lengths. Comput. Biol. Chem. 2018, 75, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Cheng, Y.; Zhang, C.; You, Q.; Shen, X.; Guo, W.; Jiao, Y. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 2017, 7, 5636. [Google Scholar] [CrossRef]
- Quan, G.; Li, J. Circular RNAs: Biogenesis, expression and their potential roles in reproduction. J. Ovarian Res. 2018, 11, 9. [Google Scholar] [CrossRef]
- Talhouarne, G.J.; Gall, J.G. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 2014, 20, 1476–1487. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, X.; Wu, X.; Guo, H.; Hu, Y.; Tang, F.; Huang, Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015, 16, 148. [Google Scholar] [CrossRef]
- Lin, X.; Han, M.; Cheng, L.; Chen, J.; Zhang, Z.; Shen, T.; Wang, M.; Wen, B.; Ni, T.; Han, C. Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells. RNA Biol. 2016, 13, 1011–1024. [Google Scholar] [CrossRef]
- Dong, W.W.; Li, H.M.; Qing, X.R.; Huang, D.H.; Li, H.G. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci. Rep. 2016, 6, 39080. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Lu, Y.; Rui, C.; Qian, Y.; Cai, M.; Jia, R. Potential Significance of Circular RNA in Human Placental Tissue for Patients with Preeclampsia. Cell Physiol. Biochem. 2016, 39, 1380–1390. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Yan, L.; Hu, B.; Fan, X.; Ren, Y.; Li, R.; Lian, Y.; Yan, J.; Li, Q.; Zhang, Y.; et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016, 17, 130. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Huang, J.; Yuan, S.; Zhou, S.; Yan, W.; Shen, W.; Chen, Y.; Xia, X.; Luo, A.; Zhu, D.; et al. Circular RNA expression profiling of human granulosa cells during maternal aging reveals novel transcripts associated with assisted reproductive technology outcomes. PLoS ONE 2017, 12, e0177888. [Google Scholar] [CrossRef] [PubMed]
- Rhinehart, E.M. Mechanisms linking energy balance and reproduction: Impact of prenatal environment. Horm. Mol. Biol. Clin. Investig. 2016, 25, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Härter, C.J.; Lima, L.D.; Hgo, S.; Castagnino, D.S.; Rivera, A.R.; Resende, K.T.; Iama, T. Energy and protein requirements for maintenance of dairy goats during pregnancy and their efficiencies of use. J. Anim. Sci. 2017, 95, 4181–4193. [Google Scholar] [CrossRef]
- Schneider, J.E. Energy balance and reproduction. Physiol. Behav. 2004, 81, 289–317. [Google Scholar] [CrossRef]
- Raga, F.; Casan, E.M.; Kruessel, J.S.; Wen, Y.; Huang, H.Y.; Nezhat, C.; Polan, M.L. Quantitative gonadotropin-releasing hormone gene expression and immunohistochemical localization in human endometrium throughout the menstrual cycle. Biol. Reprod. 1998, 59, 661–669. [Google Scholar] [CrossRef]
- Limonta, P.; Marelli, M.M.; Moretti, R.; Marzagalli, M.; Fontana, F.; Maggi, R. GnRH in the human female reproductive axis. Vitam. Horm. 2018, 107, 27–66. [Google Scholar]
- Moenter, S.M.; Caraty, A.; Locatelli, A.; Karsch, F.J. Pattern of gonadotropin-releasing hormone (GnRH) secretion leading up to ovulation in the ewe: Existence of a preovulatory GnRH surge. Endocrinology 1991, 129, 1175–1182. [Google Scholar] [CrossRef]
- Rance, N.E. Menopause and the human hypothalamus: Evidence for the role of kisspeptin/neurokinin B neurons in the regulation of estrogen negative feedback. Peptides 2009, 30, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.S.; MacCalman, C.D.; Leung, P.C. Differential effects of gonadotropin-releasing hormone I and II on the urokinase-type plasminogen activator/plasminogen activator inhibitor system in human decidual stromal cells in vitro. J. Clin. Endocrinol. Metab. 2003, 88, 3806–3815. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.M.; Huang, H.Y.; Lee, C.L.; Soong, Y.K.; Leung, P.C.; Wang, H.S. Gonadotropin-releasing hormone type II (GnRH-II) agonist regulates the motility of human decidual endometrial stromal cells: Possible effect on embryo implantation and pregnancy. Biol. Reprod. 2015, 92, 98. [Google Scholar] [CrossRef] [PubMed]
- Grazul-Bilska, A.T.; Bairagi, S.; Kraisoon, A.; Dorsam, S.T.; Reyaz, A.; Navanukraw, C.; Borowicz, P.P.; Reynolds, L.P. Placental development during early pregnancy in sheep: Nuclear estrogen and progesterone receptor mRNA expression in the utero-placental compartments. Domest. Anim. Endocrinol. 2019, 66, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Boos, A.; Kohtes, J.; Stelljes, A.; Zerbe, H.; Thole, H.H. Immunohistochemical assessment of progesterone, oestrogen and glucocorticoid receptors in bovine placentomes during pregnancy, induced parturition, and after birth with or without retention of fetal membranes. J. Reprod. Fertil. 2000, 120, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Pastore, M.B.; Jobe, S.O.; Jayanth, R.; Magness, R.R. Estrogen receptor-α and estrogen receptor-β in the uterine vascular endothelium during pregnancy: Functional implications for regulating uterine blood flow. Semin. Reprod. Med. 2012, 30, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.X.; Owiny, J.; Zhang, Q.; Ma, X.H.; Nathanielsz, P.W. Regulation of the estrogen receptor and its messenger ribonucleic acid in the ovariectomized sheep myometrium and endometrium: The role of estradiol and progesterone. Biol. Reprod. 1996, 55, 762–768. [Google Scholar] [PubMed]
- Kim, S.H.; Pohl, O.; Chollet, A.; Gotteland, J.P.; Fairhurst, A.D.; Bennett, P.R.; Terzidou, V. Differential Effects of Oxytocin Receptor Antagonists, Atosiban and Nolasiban, on Oxytocin Receptor-Mediated Signaling in Human Amnion and Myometrium. Mol. Pharmacol. 2017, 91, 403–415. [Google Scholar] [CrossRef]
- Langendijk, P.; Bouwman, E.G.; Schams, D.; Soede, N.M.; Kemp, B. Effects of different sexual stimuli on oxytocin release, uterine activity and receptive behavior in estrous sows. Theriogenology 2003, 59, 849–861. [Google Scholar] [CrossRef]
- Domino, M.; Pawlinski, B.; Gajewska, M.; Jasinski, T.; Sady, M.; Gajewski, Z. Uterine EMG activity in the non-pregnant sow during estrous cycle. BMC Vet. Res. 2018, 14, 176. [Google Scholar] [CrossRef]
- Twombly, V.; Blackman, R.K.; Jin, H.; Graff, J.M.; Padgett, R.W.; Gelbart, W.M. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development 1996, 122, 1555–1565. [Google Scholar] [PubMed]
- Elvin, J.A.; Yan, C.; Matzuk, M.M. Oocyte-expressed TGF-beta superfamily members in female fertility. Mol. Cell Endocrinol. 2000, 159, 1–5. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.; Zhao, H.; Qin, X. Co-expression analysis and identification of fecundity-related long non-coding RNAs in sheep ovaries. Sci. Rep. 2016, 6, 39398. [Google Scholar] [CrossRef] [PubMed]
- Tribulo, P.; Balzano-Nogueira, L.; Conesa, A.; Siqueira, L.G.; Hansen, P.J. Changes in the uterine metabolome of the cow during the first 7 days after estrus. Mol. Reprod. Dev. 2018, 86, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Romero, J.J.; Liebig, B.E.; Broeckling, C.D.; Prenni, J.E.; Hansen, T.R. Pregnancy-induced changes in metabolome and proteome in ovine uterine flushings. Biol. Reprod. 2017, 97, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, J.; Wang, H.; Li, X.; Yang, Q.; Li, H.; Chang, Y. Identification and characterization of circRNAs in Pyrus betulifolia Bunge under drought stress. PLoS ONE 2018, 13, e0200692. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, M.; Fan, Y.; Li, S.; Lai, Z.; Huang, Y.; Lan, X.; Lei, C.; Chen, H.; Dang, R. Identification and characterization of circular RNAs in Qinchuan cattle testis. R. Soc. Open Sci. 2018, 5, 180413. [Google Scholar] [CrossRef]

| Gene Name | Primer Sequence (5′–3′) | Product Size (bp) |
|---|---|---|
| oar_circ_0030419 | F: CCCATCCCCGGCACGTCCA R: GTCCCGTCCTCACTGCACTCG | 120 |
| oar_circ_0012660 | F: AAACCTGGCACGTACGCAGA R: AGGTTGACGGCATACATCAGC | 199 |
| oar_circ_0023919 | F: CTTCCTTCAGACAGAATGCAC R: CGAGCTCTCCAATATTGTCAC | 169 |
| oar_circ_0031129 | F: GAGTGAATAAAGATGTCATCCGA R: ACCAGAATATTCAGAAATGTGCC | 147 |
| oar_circ_0014321 | F: GAAACTGCACACAGATAACCA R: ACATGCTGGATCTAAAACCAC | 160 |
| oar_circ_0001853 | F: AAGTCACTTAAAACGCACCT R: TAAATTACTGTTCTCCGCTTC | 180 |
| β-Actin | F:CCAACCGTGAGAAGATGACC R:CCCGAGGCGTACAGGGACAG | 97 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
La, Y.; Tang, J.; Di, R.; Wang, X.; Liu, Q.; Zhang, L.; Zhang, X.; Zhang, J.; Hu, W.; Chu, M. Differential Expression of Circular RNAs in Polytocous and Monotocous Uterus during the Reproductive Cycle of Sheep. Animals 2019, 9, 797. https://doi.org/10.3390/ani9100797
La Y, Tang J, Di R, Wang X, Liu Q, Zhang L, Zhang X, Zhang J, Hu W, Chu M. Differential Expression of Circular RNAs in Polytocous and Monotocous Uterus during the Reproductive Cycle of Sheep. Animals. 2019; 9(10):797. https://doi.org/10.3390/ani9100797
Chicago/Turabian StyleLa, Yongfu, Jishun Tang, Ran Di, Xiangyu Wang, Qiuyue Liu, Liping Zhang, Xiaosheng Zhang, Jinlong Zhang, Wenping Hu, and Mingxing Chu. 2019. "Differential Expression of Circular RNAs in Polytocous and Monotocous Uterus during the Reproductive Cycle of Sheep" Animals 9, no. 10: 797. https://doi.org/10.3390/ani9100797
APA StyleLa, Y., Tang, J., Di, R., Wang, X., Liu, Q., Zhang, L., Zhang, X., Zhang, J., Hu, W., & Chu, M. (2019). Differential Expression of Circular RNAs in Polytocous and Monotocous Uterus during the Reproductive Cycle of Sheep. Animals, 9(10), 797. https://doi.org/10.3390/ani9100797






