Integrating 16S rRNA Sequencing and LC–MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Housing, and Feeding
2.2. Rumen Fluid Sampling
2.3. DNA Extraction, Sequencing, and Diversity Analysis
2.4. Statistical Analysis
2.5. Non-Targeted Metabolomics Analysis
2.5.1. Sample Preparation and Analysis
2.5.2. Data and Statistical Analysis
3. Results
3.1. Sequencing Results
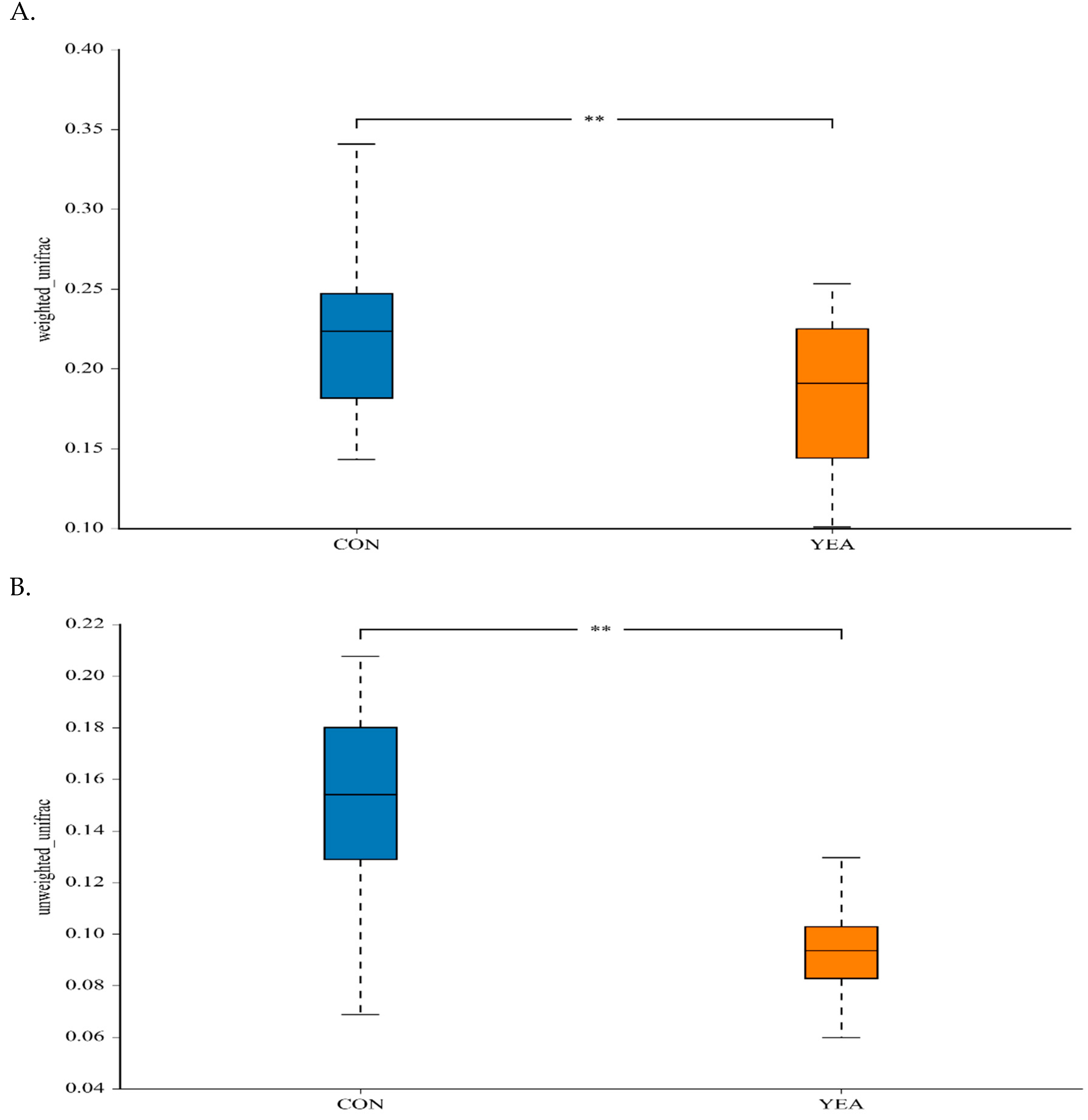
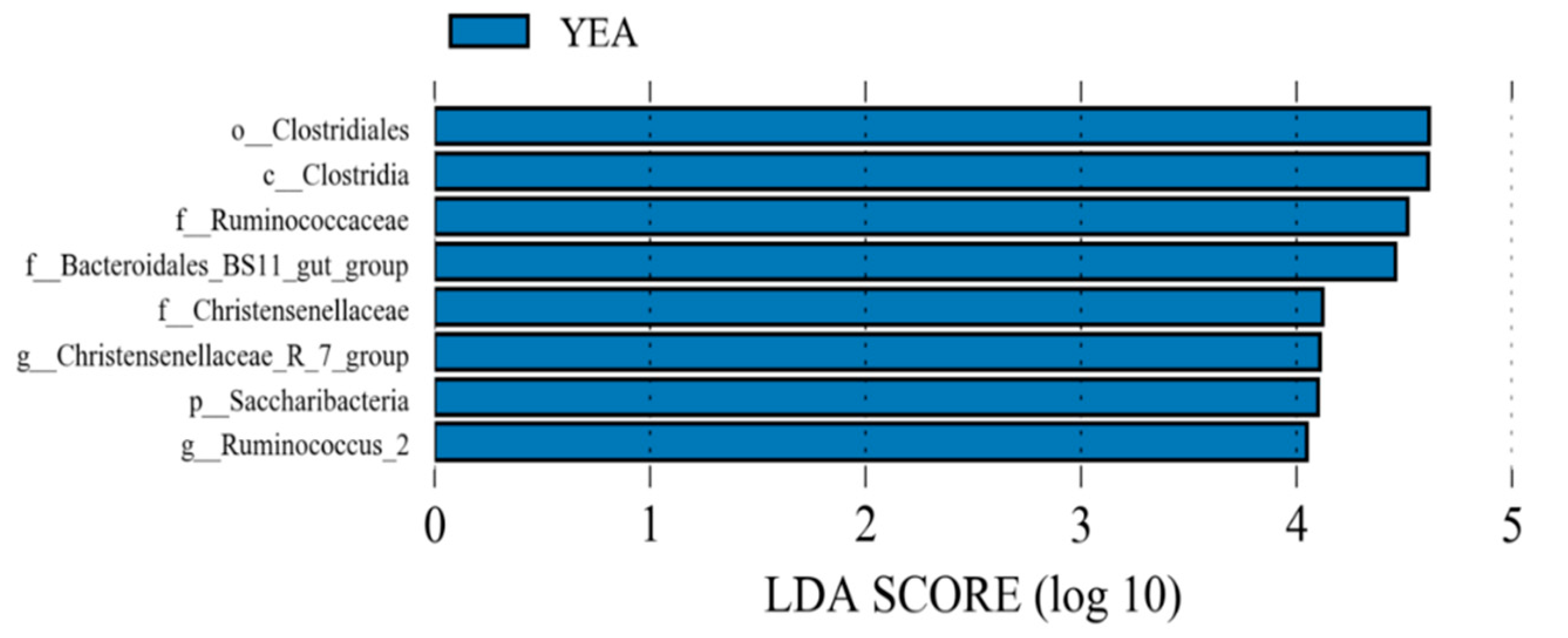
3.2. Diversity and Relative Abundance of Taxa
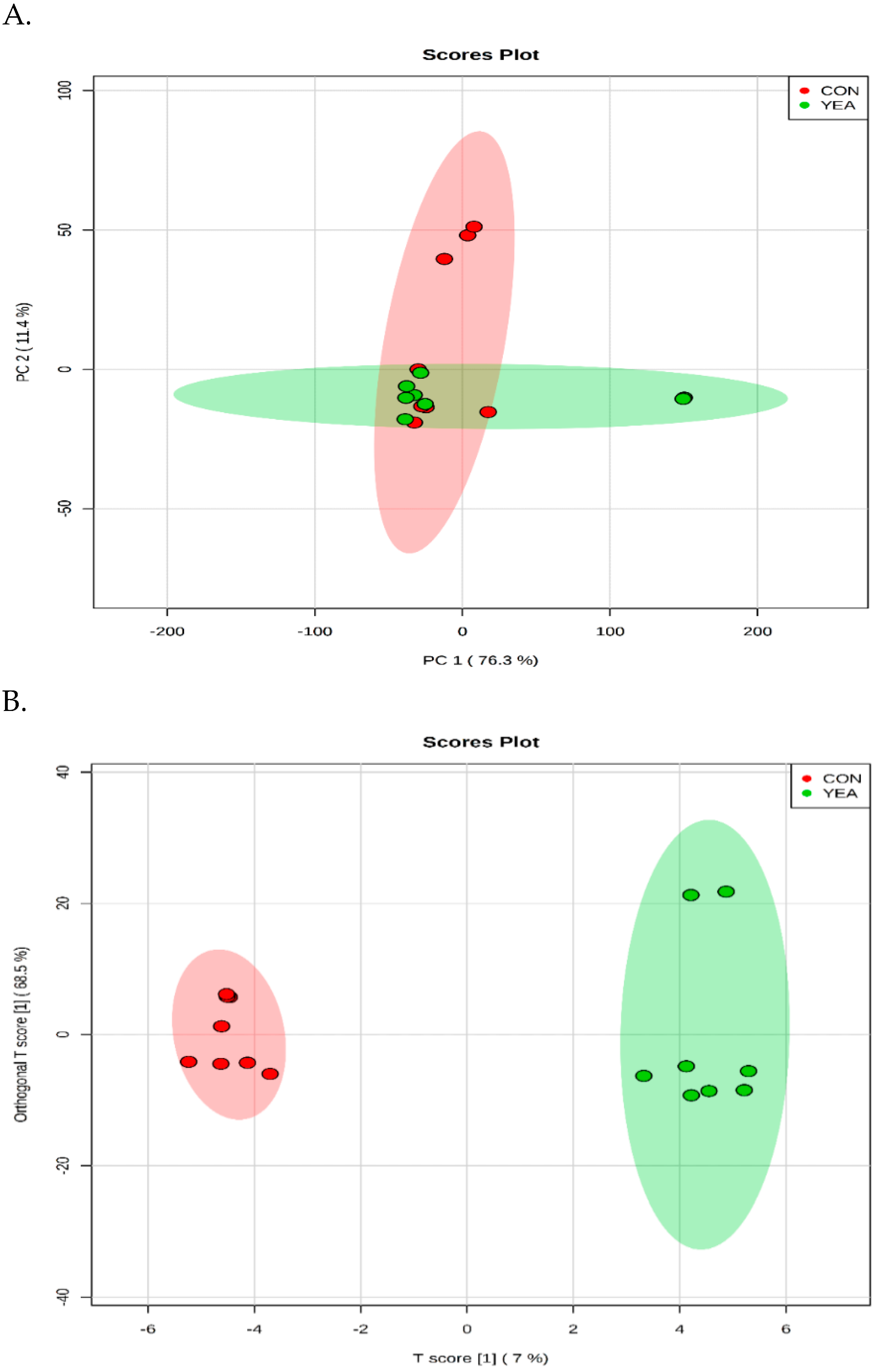
3.3. Rumen Fluid Metabolome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McAllister, T.A.; Beauchemin, K.A.; Alazzeh, A.Y.; Baah, J.; Teather, R.M.; Stanford, K. Review: The use of direct-fed microbials to mitigate pathogens and methane production in cattle. Can. J. Anim. Sci. 2011, 91, 193–211. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Chevaux, E.; Martin, C.; Forano, E. Use of yeast probiotics in ruminants: Effects and mechanisms of action on rumen pH, fibre degradation, and microbiota according to the diet. In Probiotic in Animals; Rigobelo, E.C., Ed.; InTech: Rijeka, Croatia, 2012; pp. 119–162. [Google Scholar]
- Jiang, Y.; Ogunade, I.M.; Qi, S.; Hackmann, T.J.; Staples, C.R.; Adesogan, A.T. Effects of the dose and viability of Saccharomyces cerevisiae. 1. Diversity of ruminal microbes as analyzed by Illumina MiSeq sequencing and quantitative PCR. J. Dairy Sci. 2017, 100, 325–342. [Google Scholar] [CrossRef] [PubMed]
- Chaucheyras-Durand, F.; Walker, N.D.; Bach, A. Effects of active dry yeasts on the rumen microbial ecosystem: Past, present, and future. Anim. Feed Sci. Technol. 2008, 145, 5–26. [Google Scholar] [CrossRef]
- McGinn, S.M.; Beauchemin, K.A.; Coates, T.; Colombatto, D. Methane emissions from beef cattle: Effect of monensin, sunflower oil, enzymes, yeast and fumaric acid. J. Anim. Sci. 2004, 82, 3346–3356. [Google Scholar] [CrossRef] [PubMed]
- McDonald, P.; Edwards, R.A.; Greenhalgh, J.F.D.; Morgan, C.A.; Sinclair, L.A.; Wilkinson, R.G. Food additives. In Animal Nutrition, 7th ed.; Greenhalgh, J.F.D., Morgan, C.A., Sinclair, L.A., Wilkinson, R.G., Eds.; Pearson Education Ltd.: Harlow, UK, 2011; pp. 594–607. [Google Scholar]
- Chaucheyras-Durand, F.; Durand, H. Probiotics in animal nutrition and health. Benef. Microbes 2010, 1, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Pinloche, E.; McEwan, N.; Marden, J.P.; Bayourthe, C.; Auclair, E.; Newbold, C.J. The effects of a probiotic yeast on the bacterial diversity and population structure in the rumen of cattle. PLoS ONE 2013, 8, e67824. [Google Scholar] [CrossRef] [PubMed]
- Karisa, B.K.; Thomson, J.; Wang, Z.; Li, C.; Montanholi, Y.R.; Miller, S.P.; Moore, S.S.; Plastow, G.S. Plasma metabolites associated with residual feed intake and other productivity performance traits in beef cattle. Livest. Sci. 2014, 165, 200–211. [Google Scholar] [CrossRef]
- Artegoitia, V.M.; Foote, A.P.; Lewis, R.M.; Freetly, H.C. Rumen fluid metabolomics analysis associated with feed efficiency on crossbred steers. Sci. Rep. 2017, 7, 2864. [Google Scholar] [CrossRef]
- Saleem, F.; Ametaj, B.N.; Bouatra, S.; Mandal, R.; Zebeli, Q.; Dunn, S.M.; Wishart, D.S. A metabolomics approach to uncover the effect of grain diets on rumen health in dairy cows. J. Dairy Sci. 2012, 95, 6606–6623. [Google Scholar] [CrossRef]
- Abarghuei, M.J.; Rouzbehan, Y.; Salem, A.Z.M.; Zamiri, M.J. Nitrogen balance, blood metabolites and milk fatty acid composition of dairy cows fed pomegranate-peel extract. Livest. Sci. 2014, 164, 72–80. [Google Scholar] [CrossRef]
- Ogunade, I.M.; Schweickart, H.; Andries, K.; Lay, J. Monensin alters the functional and metabolomics profile of rumen microbiota in beef cattle. Animals 2018, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Mukherjee, S.; Stamatis, D.; Bertsch, J.; Ovchinnikova, G.; Verezemska, O.; Isbandi, M.; Ali, R.; Sharma, K.; Kyrpides, N.C.; Reddy, T.B.K. Genomes OnLine Database (GOLD) v. 6: Data updates and feature enhancements. Nucleic Acids Res. 2017, 45, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez, P.A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 60. [Google Scholar] [CrossRef]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Muck, R.E.; Dickerson, J.T. Storage temperature effects on proteolysis in alfalfa silage. Trans. Asae. 1988, 31, 1005–1009. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Mitsuhiro, K.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using metaboAnalyst 3.0 for comprehensive metabolomics data analysis. Cur. Prot. Bioinform. 2016, 55, 14. [Google Scholar] [CrossRef] [PubMed]
- Bylesjo, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Tajima, K.; Aminov, R.I.; Nagamine, T.; Ogata, K.; Nakamura, M.; Matsui, H.; Benno, Y. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol. Ecol. 1999, 29, 159–169. [Google Scholar] [CrossRef]
- Nocek, J.E.; Holt, M.G.; Oppy, J. Effects of supplementation with yeast culture and enzymatically hydrolyzed yeast on performance of early lactation dairy cattle. J. Dairy Sci. 2011, 94, 4046–4056. [Google Scholar] [CrossRef] [PubMed]
- Chaucheyras-Durand, F.; Fonty, G. Establishment of cellulolytic bacteria and development of fermentative activities in the rumen of gnotobiotically-reared lambs receiving the microbial additive Saccharomyces cerevisiae CNCM I-1077. Reprod. Nutr. Dev. 2001, 41, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Marden, J.P.; Julien, C.; Monteils, V.; Auclair, E.; Moncoulon, R.; Bayourthe, C. How does live yeast differ from sodium bicarbonate to stabilize ruminal pH in high-yielding dairy cows? J. Dairy Sci. 2008, 91, 3528–3535. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, L.L.; Silva, J.R.M.; de Oliveira, B.M.L.; Dias Júnior, G.S.; Lopes, F.; Júnior, S.S.; de Fátima, O.Z.; Pereira, M.N. Diet digestibility and performance of dairy cows supplemented with live yeast. Sci. Agric. 2011, 68, 301–307. [Google Scholar] [CrossRef]
- Ferraretto, L.F.; Shaver, R.D.; Bertics, S.J. Effect of dietary supplementation with live-cell yeast at two dosages on lactation performance, ruminal fermentation, and total-tract nutrient digestibility in dairy cows. J. Dairy Sci. 2012, 95, 4017–4028. [Google Scholar] [CrossRef]
- Bertin, G.; Spring, P.; Fallon, R.; Earley, B. Benefits of yeast culture (Yea-Sacc®1026) supplementation on performance of bull calves. In Proceedings of the 21st Annual Symposium, Nutritional Biotechnology in the Feed and Food Industries, Lexington, KY, USA, 22–25 May 2005. [Google Scholar]
- Desnoyers, M.; Giger-Reverdin, S.; Bertin, G.; Duvaux-Ponter, C.; Sauvant, D. Meta-analysis of the influence of Saccharomyces cerevisiae supplementation on ruminal parameters and milk production of ruminants. J. Dairy Sci. 2009, 92, 1620–1632. [Google Scholar] [CrossRef]
- Finck, D.; Ribeiro, F.; Burdick, N.; Parr, S.; Carroll, J.; Young, T.; Bernhard, B.; Corley, J.; Estefan, A.; Rathmann, R. Yeast supplementation alters the performance and health status of receiving cattle. Prof. Anim. Sci. 2014, 30, 333–341. [Google Scholar] [CrossRef]
- Tong, J.; Zhang, H.; Yang, D.; Jiang, L.; Xiong, B. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS ONE 2018, 13, e0198225. [Google Scholar] [CrossRef] [PubMed]
- Drennan, M.J.; Moloney, A.P. Effect of yeast culture on of beef cattle fed on grass silage plus barley-based diet. Ir. J. Agric. Food Res. 1993, 32, 125–132. [Google Scholar]
- Moallem, U.; Lehrer, H.; Livshitz, L.; Zachut, M.; Yakoby, S. The effects of live yeast supplementation to dairy cows during the hot season on production, feed efficiency, and digestibility. J. Dairy Sci. 2009, 92, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Kim, S.W.; Kim, M.H.; Upadhaya, S.D.; Kam, D.K.; Ha, J.K. Direct-fed microbials for ruminant animals. Asian-Australas. J. Anim. Sci. 2010, 23, 1657–1667. [Google Scholar] [CrossRef]
- Williams, A.; Withers, S.E. Bacillus spp. in the rumen ecosystem. Hemicellulose depolymerases and glycosidehydrolases of Bacillus spp. and rumen isolates grown under anaerobic conditions. J. Appl. Microbiol. 1983, 55, 283–292. [Google Scholar]
- Kritas, S.K.; Govaris, A.; Christodoulopoulos, G.; Burriel, A.R. Effect of Bacillus licheniformis and Bacillus subtilis supplementation of ewe’s feed on sheep milk production and young lamb mortality. J. Vet. Med. Ser. A 2006, 53, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Qiao, G.H.; Shan, A.S.; Ma, N.; Ma, Q.Q.; Sun, Z.W. Effect of supplemental bacillus cultures on rumen fermentation and milk yield in Chinese Holstein cows. J. Anim. Physiol. Anim. Nutr. 2009, 94, 429–436. [Google Scholar] [CrossRef]
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef]
- Magalhães, V.; Susca, F.; Lima, F.; Branco, A.; Yoon, I.; Santos, J. Effect of feeding yeast culture on performance, health, and immunocompetence of dairy calves. J. Dairy Sci. 2008, 91, 1497–1509. [Google Scholar] [CrossRef]
- Brewer, M.T.; Anderson, K.L.; Yoon, I.; Scott, M.F.; Carlson, S.A. Amelioration of salmonellosis in pre-weaned dairy calves fed Saccharomyces cerevisiae fermentation products in feed and milk replacer. Vet. Microbiol. 2014, 172, 248–255. [Google Scholar] [CrossRef]
- Ofek, I.; Mirelman, D.; Sharon, N. Adherence of Escherichia coli to human mucosal cells mediated by mannose receptors. Nature 1977, 265, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.B.; Seo, Y.M.; Kim, C.H.; Paik, I.K. Effect of dietary prebiotic supplementation on the performance, intestinal microflora, and immune response of broilers. Poult. Sci. 2011, 90, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Calsamiglia, S.; Castillejos, L.; Busquet, M. Alternatives to antimicrobial growth promoters in cattle. In Recent Advances in Animal Nutrition; Garnworthy, P.C., Wiseman, J., Eds.; Elsevier: Nottingham, UK, 2006; pp. 129–167. [Google Scholar]
- Robinson, P.H. Yeast Products for Growing and Lactating Ruminants: A Literature Summary of Impacts on Rumen Fermentation and Performance. 2010. Available online: http://animalscience.ucdavis.edu/faculty/robinson/Articles/FullText/pdf/Web200901.pdf (accessed on 10 October 2018).
- Dias, A.G.; Freitas, J.A.; Micai, B.; Greco, L.F.; Santos, J.E.P. Effect of supplemental yeast culture and dietary starch content on rumen fermentation and digestion in dairy cows. J. Dairy Sci. 2018, 101, 201–221. [Google Scholar] [CrossRef]
- Evans, E.; Patterson, R.J.; Clark, N. Case study: Effects of a supplemental enhanced yeast product on digestion and milk production in dairy cows. Prof. Anim. Sci. 2012, 28, 682–688. [Google Scholar] [CrossRef]
- Erasmus, L.J.; Botha, P.M.; Kistner, A. Effect of yeast culture supplement on production, rumen fermentation, and duodenal nitrogen flow in dairy cows. J. Dairy Sci. 1992, 75, 3056–3065. [Google Scholar] [CrossRef]
- Garrity, G.; Staley, J.T.; Boone, D.R.; De Vos, P.; Goodfellow, M.; Rainey, F.A. Volume 2, The Proteobacteria. In Bergey’s Manual of Systematic Bacteriology; Springer Science & Business Media: New York, NY, USA, 2006. [Google Scholar]
- Serna, C.L.; Rodríguez, S.A. Lactic acid production by a strain of Lactococcus lactis subs lactis isolated from sugar cane plants. Electron. J. Biotechnol. 2006, 9. [Google Scholar] [CrossRef]
- Vos, P.; Garrity, G.; Jones, D.; Krieg, N.R.; Ludwig, W.; Rainey, F.A.; Schleifer, K.H.; Whitman, W.B. Volume 3, The Firmicutes. In Bergey’s Manual of Systematic Bacteriology; Springer Science & Business Media: New York, NY, USA, 2009. [Google Scholar]
- Yang, L.Y.; Chen, J.; Cheng, X.L.; Xi, D.M.; Yang, S.L.; Deng, W.D.; Mao, H.M. A phylogenetic analysis of 16S rRNA gene sequences reveals rumen bacterial diversity in Yaks (Bos grunniens). Mol. Biol. Rep. 2010, 37, 553–562. [Google Scholar] [CrossRef]
- Matthies, C.; Evers, S.; Ludwig, W.; Schink, B. Anaerovorax odorimutans gen. nov., sp. nov., a putrescine-fermenting, strictly anaerobic bacterium. Int. J. Syst. Evol. Microbiol. 2000, 50, 1591–1594. [Google Scholar] [CrossRef] [PubMed]
- Hristov, A.N.; Varga, G.; Cassidy, T.; Long, M.; Heyler, K.; Karnati, S.K.R.; Corl, B.; Hovde, C.J.; Yoon, I. Effect of Saccharomyces cerevisiae fermentation product on ruminal fermentation and nutrient utilization in dairy cows. J. Dairy Sci. 2010, 93, 682–692. [Google Scholar] [CrossRef]
- Al Ibrahim, R.M.; Kelly, A.K.; O’Grady, L.; Gath, V.P.; McCarney, C.; Mulligan, F.J. The effect of body condition score at calving and supplementation with Saccharomyces cerevisiae on milk production, metabolic status, and rumen fermentation of dairy cows in early lactation. J. Dairy Sci. 2010, 93, 5318–5328. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Fonty, G. Influence of a probiotic yeast (Saccharomyces cerevisiae CNCM I-1077) on microbial colonization and fermentations in the rumen of newborn lambs. Microb. Ecol. Health Dis. 2002, 14, 30–36. [Google Scholar] [CrossRef]
- Kamel, H.E.M.; Sekine, J.; El Waziry, A.M.; Yacount, M.H.M. Effects of Saccharomyces cerevisiae on the synchronization of organic matter and nitrogen degradation kinetics and microbial nitrogen synthesis in sheep fed Barseem hay (Trifolium alexandrinum). Small Rumin. Res. 2004, 52, 211–216. [Google Scholar] [CrossRef]
- Yoon, S.H.; Mukerjea, R.; Robyt, J.F. Specificity of yeast (Saccharomyces cerevisiae) in removing carbohydrates by fermentation. Carbohydr. Res. 2003, 338, 1127–1132. [Google Scholar] [CrossRef]
- Simpson, G.L.W.; Ortwerth, B.J. The non-oxidative degradation of ascorbic acid at physiological conditions. Biochim. Biophys. Acta 2000, 1501, 12–24. [Google Scholar] [CrossRef]
- Isihida, Y.; Shirafiji, H.; Kida, M.; Yoneda, M. Studies on the guanosine degrading system in bacterial cell. III Preparation and properties of guanosine deaminase. Agric. Biol. Chem. 1969, 33, 384–390. [Google Scholar]
- Li, T.; Chiang, J.Y. Regulation of bile acid and cholesterol metabolism by PPARs. PPAR Res. 2009, 2009, 501739. [Google Scholar] [CrossRef] [PubMed]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.D.; Kawas, J.R.; Mahgoub, O.G. Fibre digestion and utilization in goats. Small Rumin. Res. 2005, 60, 45–52. [Google Scholar] [CrossRef]
- Williams, P.E.V.; Tait, C.A.G.; Innes, G.M.; Newbold, C.J. Effects of the inclusion of yeast culture (Saccharomyces cerevisiae plus growth medium) in the diet of dairy cows on milk yield and forage degradation and fermentation patterns in the rumen of steers. J. Anim. Sci. 1991, 69, 3016–3302. [Google Scholar] [CrossRef]
- Callaway, E.S.; Martin, S.A. Effects of a Saccharomyces cerevisiae culture on ruminal bacteria that utilize lactate and digest cellulose. J. Dairy Sci. 1997, 80, 2035–2044. [Google Scholar] [CrossRef]
- Martin, S.A.; Nisbet, D.J. Effect of direct-fed microbials on rumen microbial fermentation. J. Dairy Sci. 1992, 75, 1736–1744. [Google Scholar] [CrossRef]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, T.D. Metabolomics: The final frontier? Genome Med. 2012, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Aretz, I.; Meierhofer, D. Advantages and Pitfalls of mass spectrometry based metabolome profiling in systems biology. Int. J. Mol. Sci. 2016, 17, 632. [Google Scholar] [CrossRef] [PubMed]
| Item | Red Clover/Orchard Grass Hay Mixture | Concentrate Supplement 1 |
|---|---|---|
| Dry matter (%) | 92.6 | 89.3 |
| Neutral detergent fiber (% DM) | 58.9 | 45.3 |
| Acid detergent fiber (% DM) | 40.2 | 24.4 |
| Crude protein (% DM) | 11.4 | 14.3 |
| Ether extract (% DM) | NA 2 | 2.44 |
| Starch (% DM) | NA 2 | 23.6 |
| Item | Treatment 1 | SEM | p-Value | |
|---|---|---|---|---|
| CON | YEA | |||
| Ruminococcaceae NK4A214 group | 3.27 | 4.99 | 0.40 | 0.01 |
| Candidatus_Saccharimonas | 3.35 | 5.81 | 0.61 | 0.01 |
| Christensenellaceae R-7 group | 4.80 | 7.29 | 0.68 | 0.03 |
| Bacteroidales BS11 gut group * | 1.11 | 2.36 | 0.21 | 0.01 |
| Ruminococcaceae UCG-010 | 0.67 | 1.00 | 0.16 | 0.01 |
| Ruminococcus 2 | 1.53 | 4.01 | 0.93 | 0.03 |
| Anaerovorax | 0.44 | 0.72 | 0.08 | 0.01 |
| Lachnoclostridium | 0.22 | 0.00 | 0.09 | 0.04 |
| Lachnoclostridium 5 | 0.35 | 0.02 | 0.11 | 0.04 |
| Lachnospiraceae UCG-008 | 0.10 | 0.21 | 0.01 | 0.02 |
| Ruminococcaceae UCG-005 | 0.21 | 0.30 | 0.02 | 0.02 |
| Bacillus | 0.22 | 0.00 | 0.09 | 0.03 |
| Metabolites | RT 1 | FC 2 | p-Value |
|---|---|---|---|
| 4-cyclohexenedione | 8.76 | 1.21 | 0.01 |
| Methoxybenzoic acid | 6.02 | 0.62 | 0.03 |
| Threonic acid | 0.61 | 0.38 | 0.03 |
| 2-acetoxy-6-pentadecylbenzoic acid | 7.06 | 0.32 | 0.05 |
| Methyl β-d-glucopyranoside | 8.76 | 1.26 | 0.07 |
| Lauroylcarnitine | 8.13 | 0.55 | 0.07 |
| Xanthosine | 2.05 | 0.14 | 0.08 |
| Deoxycholic acid | 7.01 | 0.38 | 0.09 |
| Metabolites | Metabolic Pathway |
|---|---|
| Christensenellaceae R-7 group | |
| Hypoxanthine | Purine metabolism |
| Hydroquinone | Riboflavin metabolism |
| Guanine | Purine metabolism |
| Glucose-1-phosphate | Glycolysis or gluconeogenesis, pentose and glucuronate interconversions, starch and sucrose metabolism, galactose metabolism, amino sugar and nucleotide sugar metabolism |
| Citrulline | Arginine and proline metabolism |
| Choline | Glycerophospholipid metabolism, glycine, serine and threonine metabolism, |
| 5-hydroxyindole-3-acetic acid | Tryptophan metabolism |
| Uncultured bacterium (Bacteroidales BS11 gut group) | |
| Glucose-1-phosphate | Glycolysis or gluconeogenesis, pentose and glucuronate interconversions, starch and sucrose metabolism, galactose metabolism, amino sugar and nucleotide sugar metabolism |
| Citrulline | Arginine and proline metabolism |
| Choline | glycerophospholipid metabolism, glycine, serine and threonine metabolism |
| Alanine-valine | Aminoacyl-tRNA biosynthesis, valine, leucine and isoleucine biosynthesis, selenoamino acid metabolism, alanine, aspartate and glutamate metabolism |
| Candidatus Saccharimonas | |
| Spermidine | Beta-alanine metabolism, glutathione metabolism, arginine and proline metabolism |
| Item | Treatment 1 | SEM | p-Value | |
|---|---|---|---|---|
| CON | YEA | |||
| Acetate (mM) | 54.6 | 57.9 | 1.09 | 0.01 |
| Propionate (mM) | 24.9 | 26.5 | 0.81 | 0.18 |
| Butyrate (mM) | 11.2 | 12.5 | 0.68 | 0.36 |
| Lactate (mM) | 1.16 | 0.94 | 0.51 | 0.67 |
| Ammonia-N (mM) | 3.87 | 3.07 | 0.16 | 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogunade, I.; Schweickart, H.; McCoun, M.; Cannon, K.; McManus, C. Integrating 16S rRNA Sequencing and LC–MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle. Animals 2019, 9, 28. https://doi.org/10.3390/ani9010028
Ogunade I, Schweickart H, McCoun M, Cannon K, McManus C. Integrating 16S rRNA Sequencing and LC–MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle. Animals. 2019; 9(1):28. https://doi.org/10.3390/ani9010028
Chicago/Turabian StyleOgunade, Ibukun, Hank Schweickart, Megan McCoun, Kyle Cannon, and Christina McManus. 2019. "Integrating 16S rRNA Sequencing and LC–MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle" Animals 9, no. 1: 28. https://doi.org/10.3390/ani9010028
APA StyleOgunade, I., Schweickart, H., McCoun, M., Cannon, K., & McManus, C. (2019). Integrating 16S rRNA Sequencing and LC–MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle. Animals, 9(1), 28. https://doi.org/10.3390/ani9010028






