Identification and Characterization of Long Noncoding RNAs in Ovine Skeletal Muscle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction
2.3. Library Construction and Deep Sequencing
2.4. Transcriptome Assembly
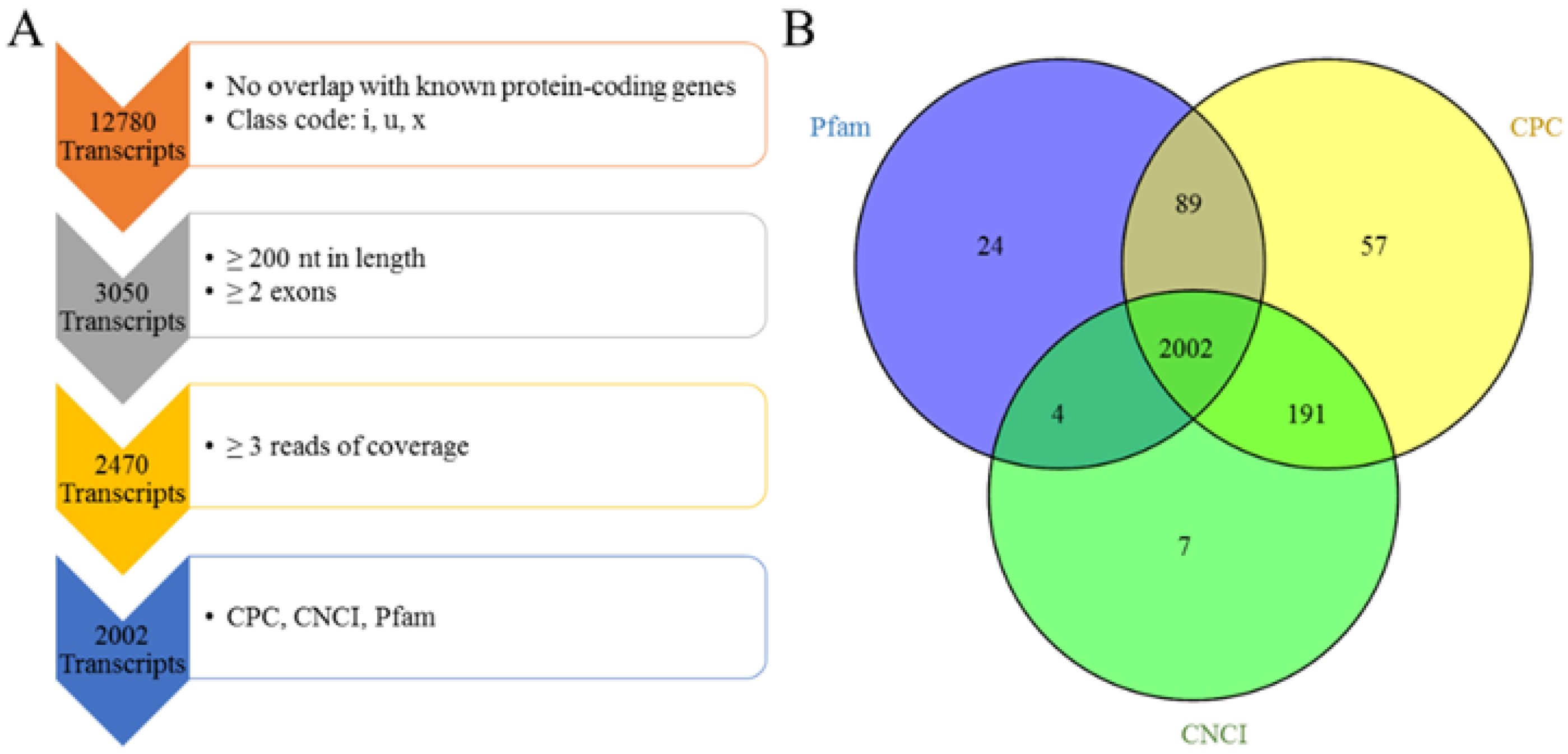
2.5. Pipeline for the Identification of Multiple-Exon LncRNA
2.6. Expression Analysis
2.7. Target Gene Prediction and Functional Enrichment Analysis
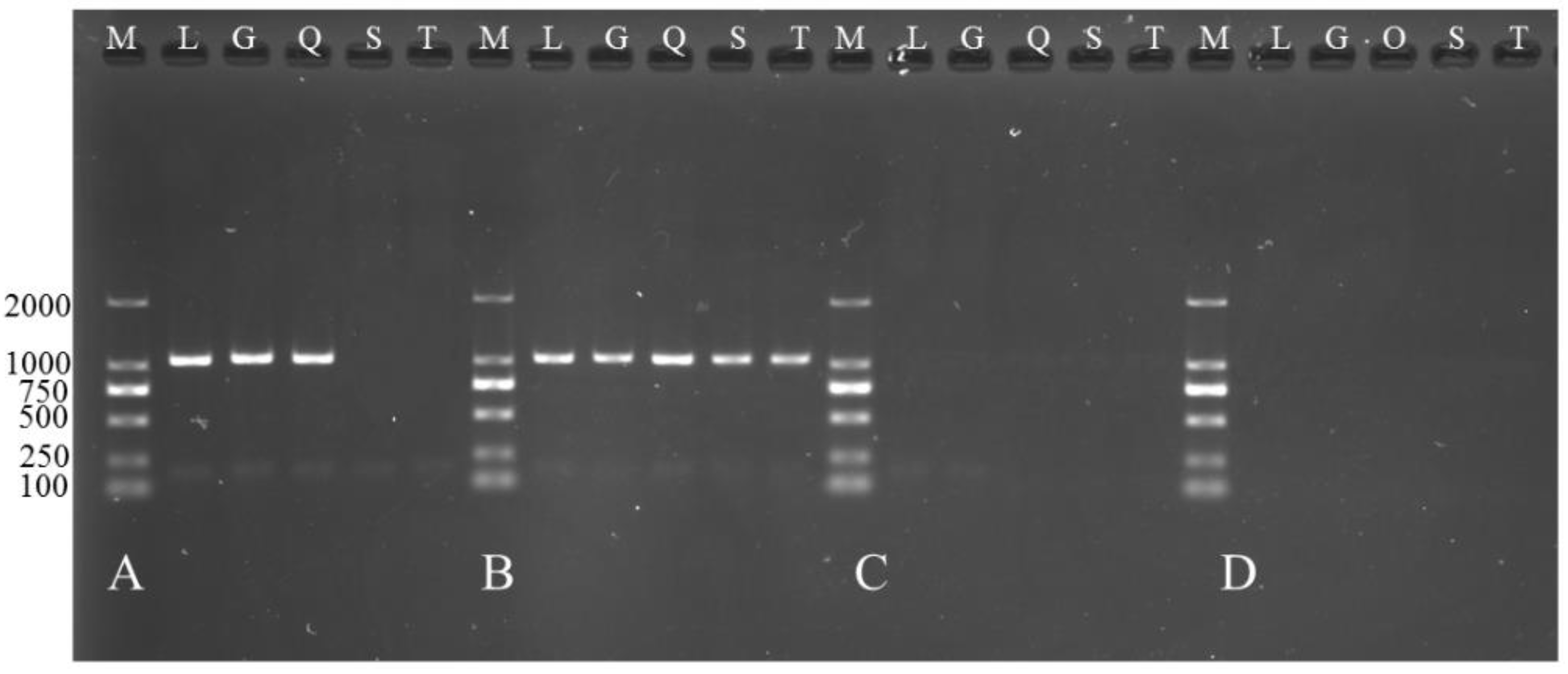
2.8. Functional Verification of TCONS_00102859 in Myoblasts
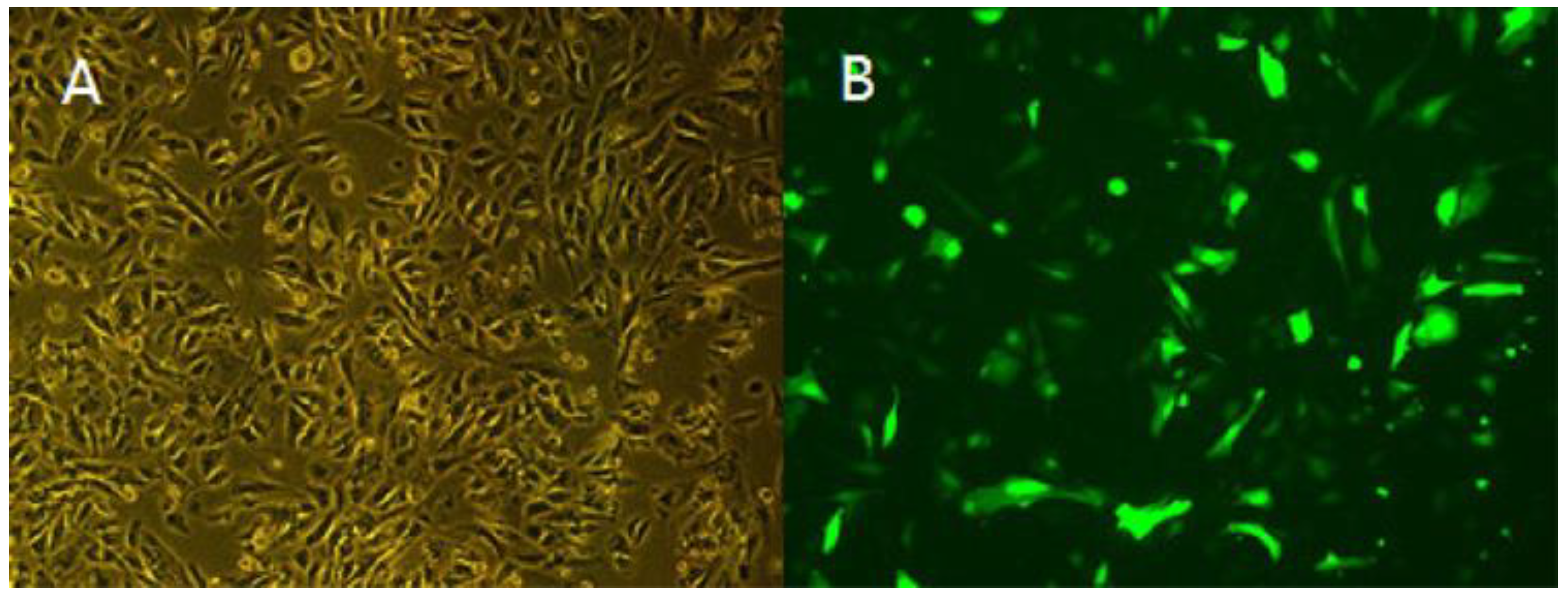
2.8.1. Cell Culture and Cell Transfection
2.8.2. Reverse Transcription and qPCR Analysis
3. Results
3.1. Overview of RNA Sequencing
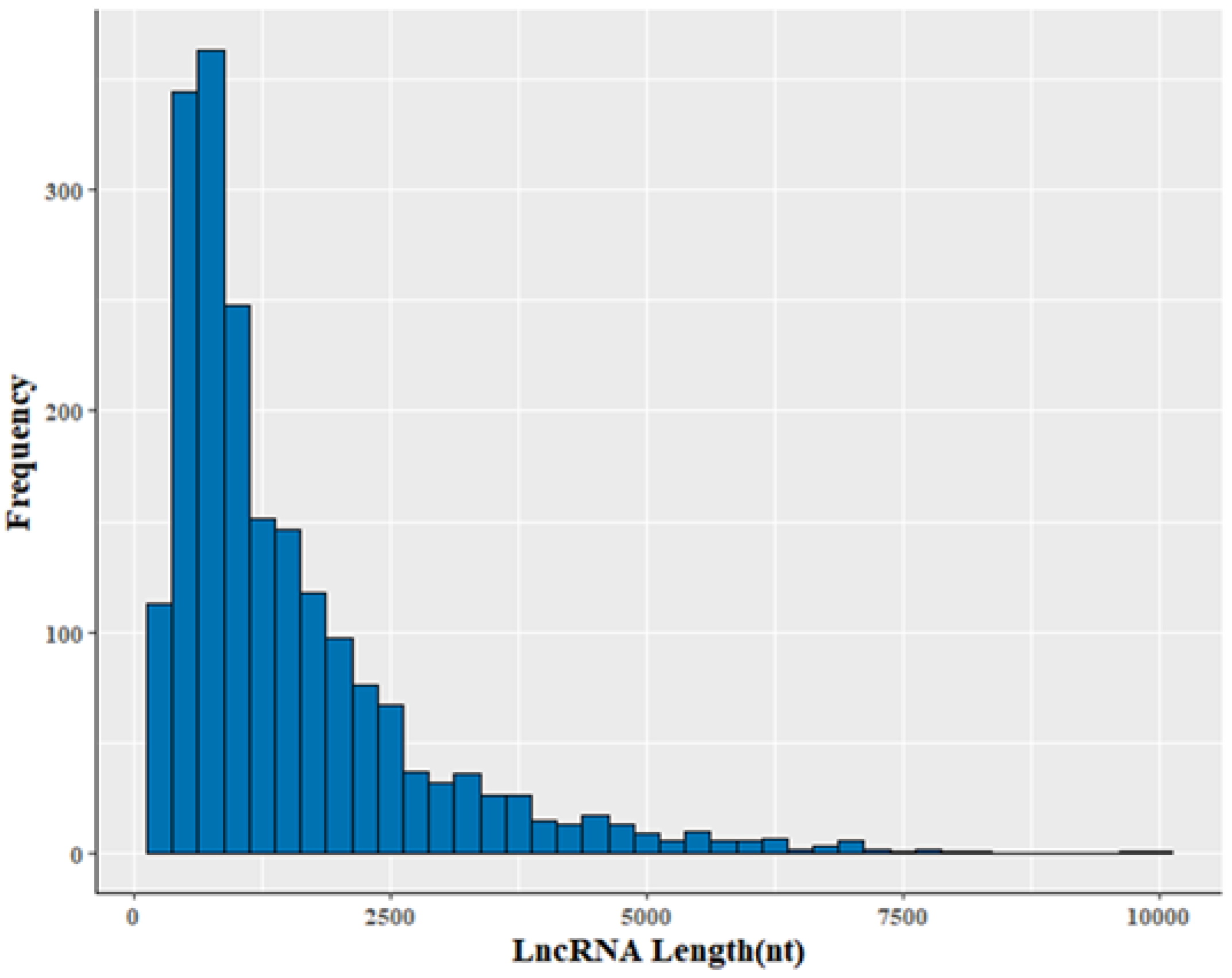
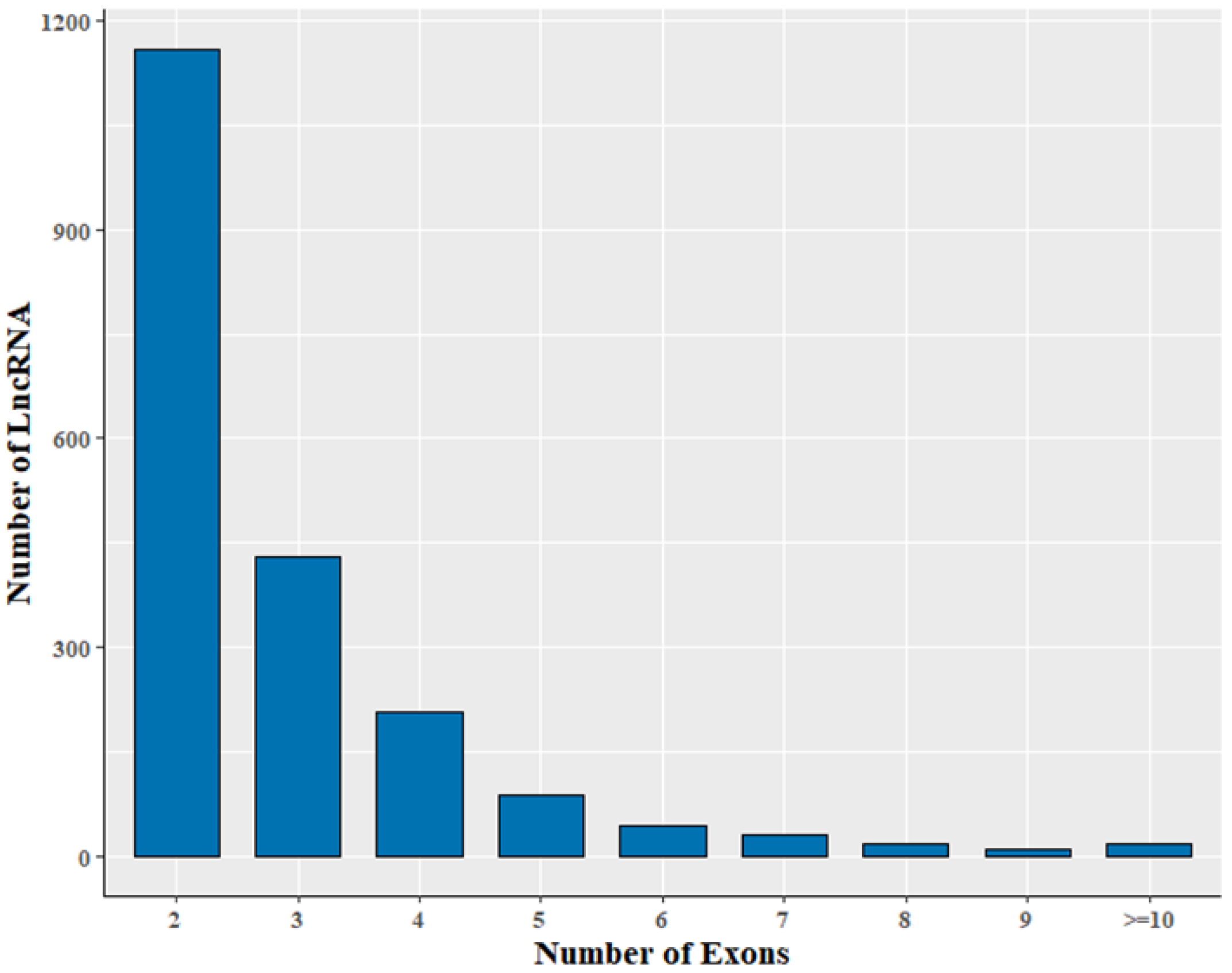
3.2. Genomic Information of LncRNA in Sheep Skeletal Muscle
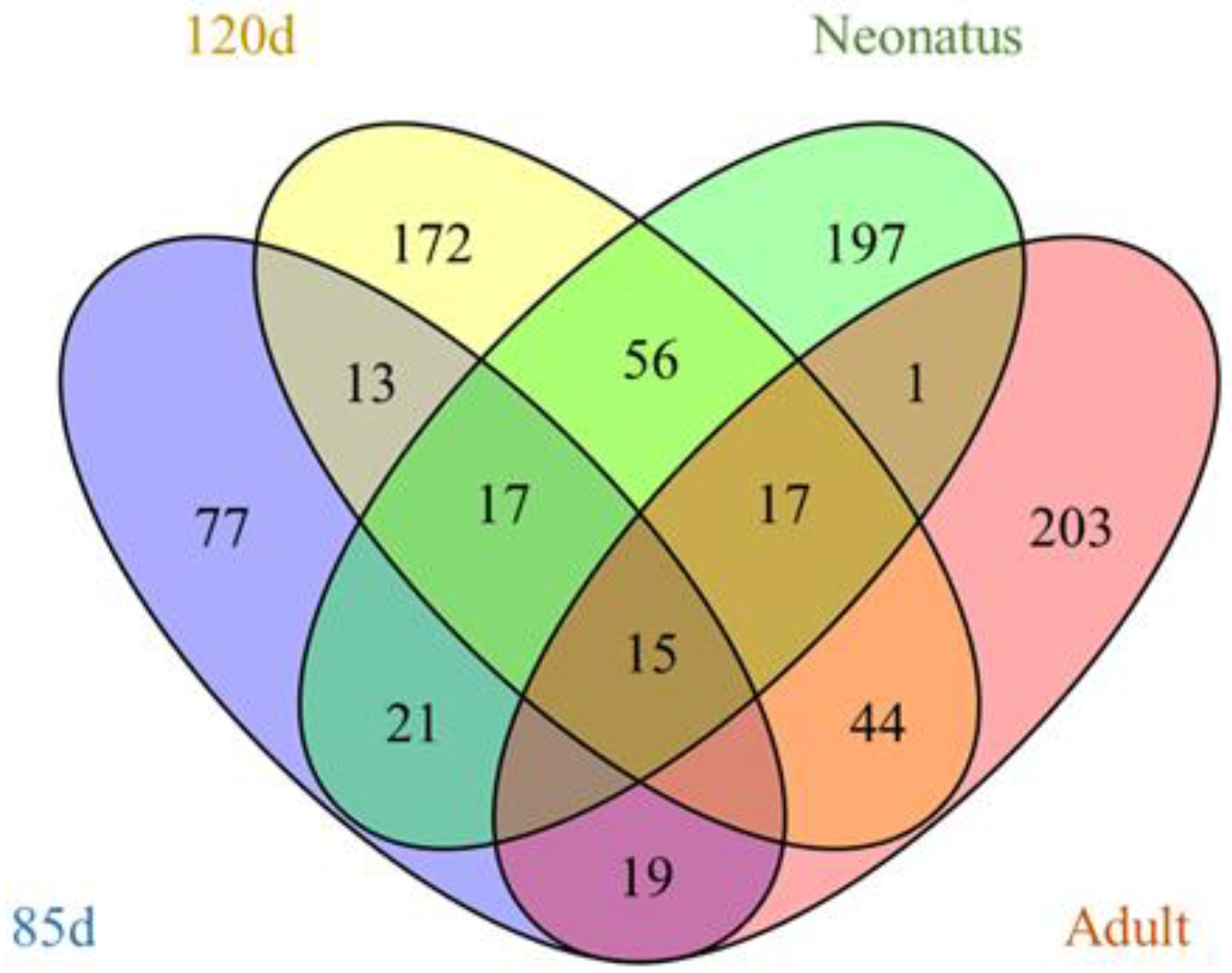
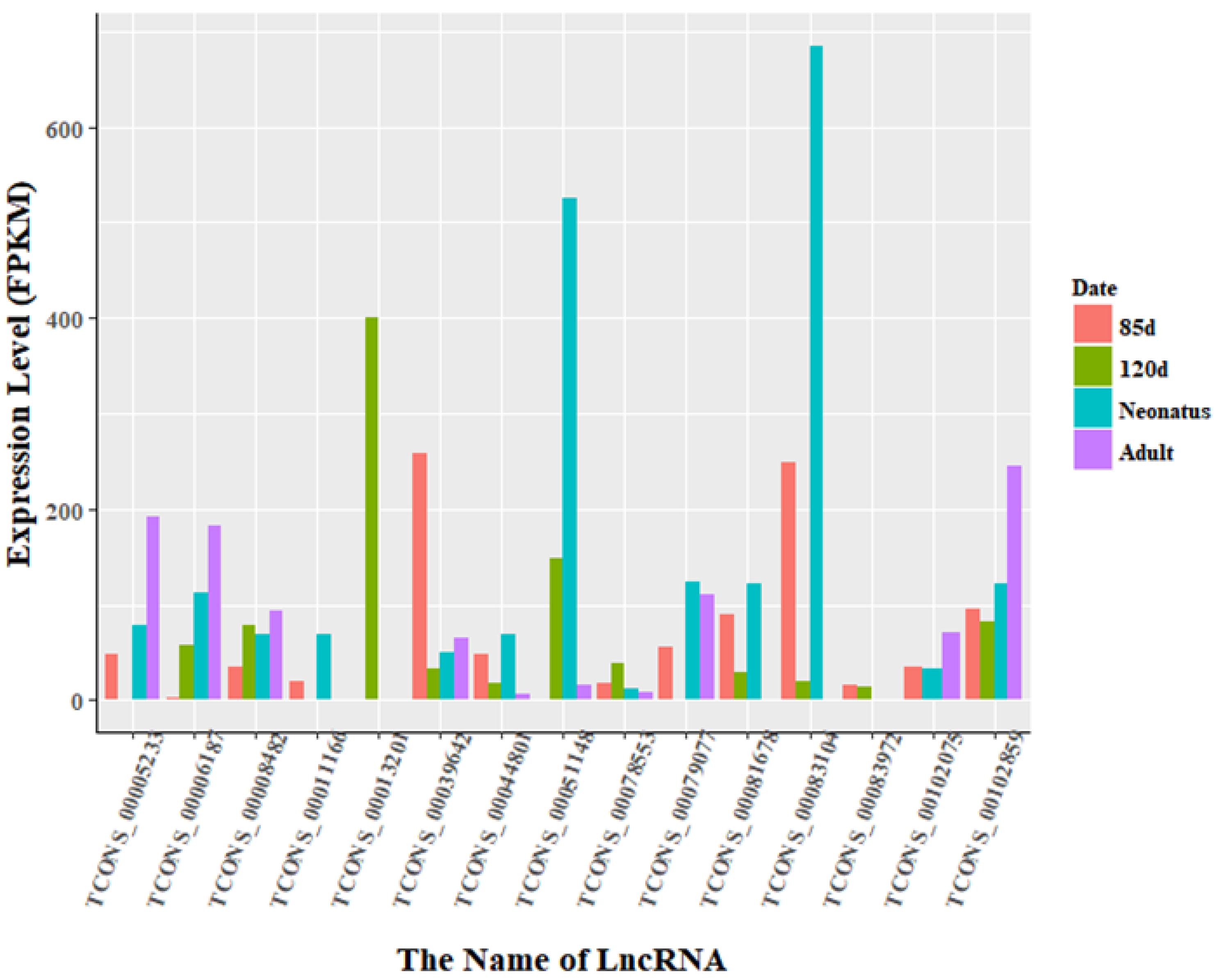
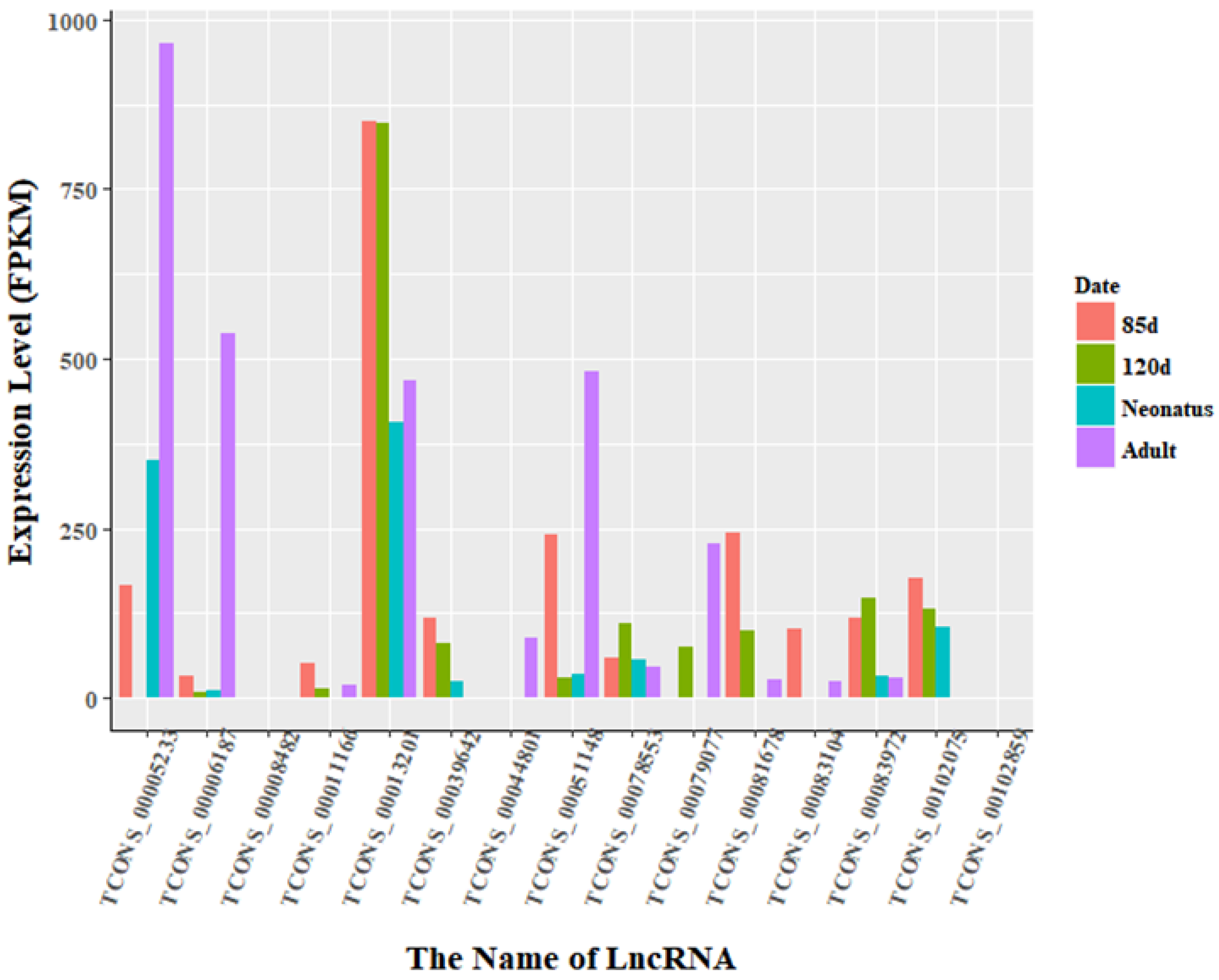
3.3. Identification of Differentially Expressed LncRNAs
3.4. Enrichment Analysis of Nearest Neighbor Genes of LncRNAs
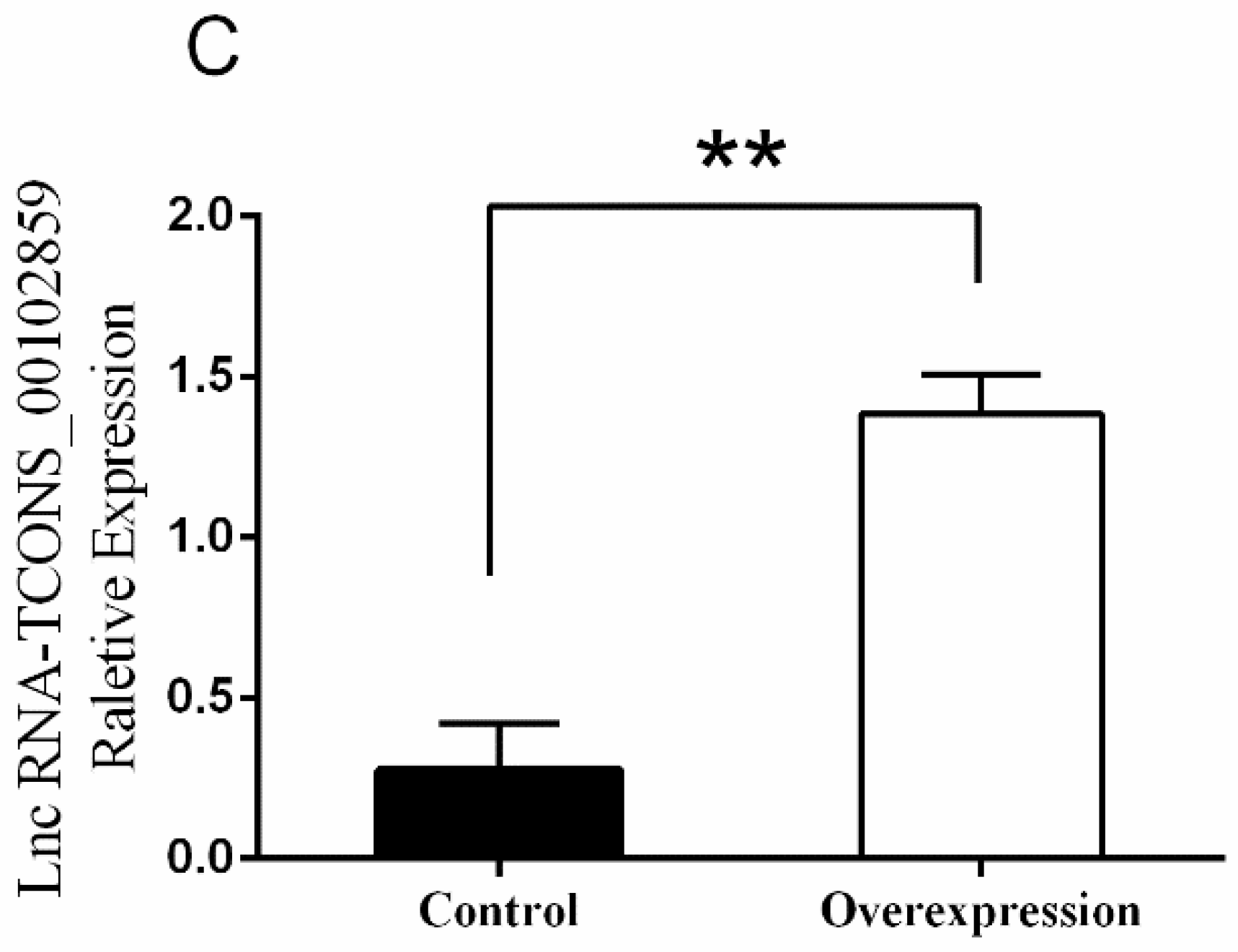
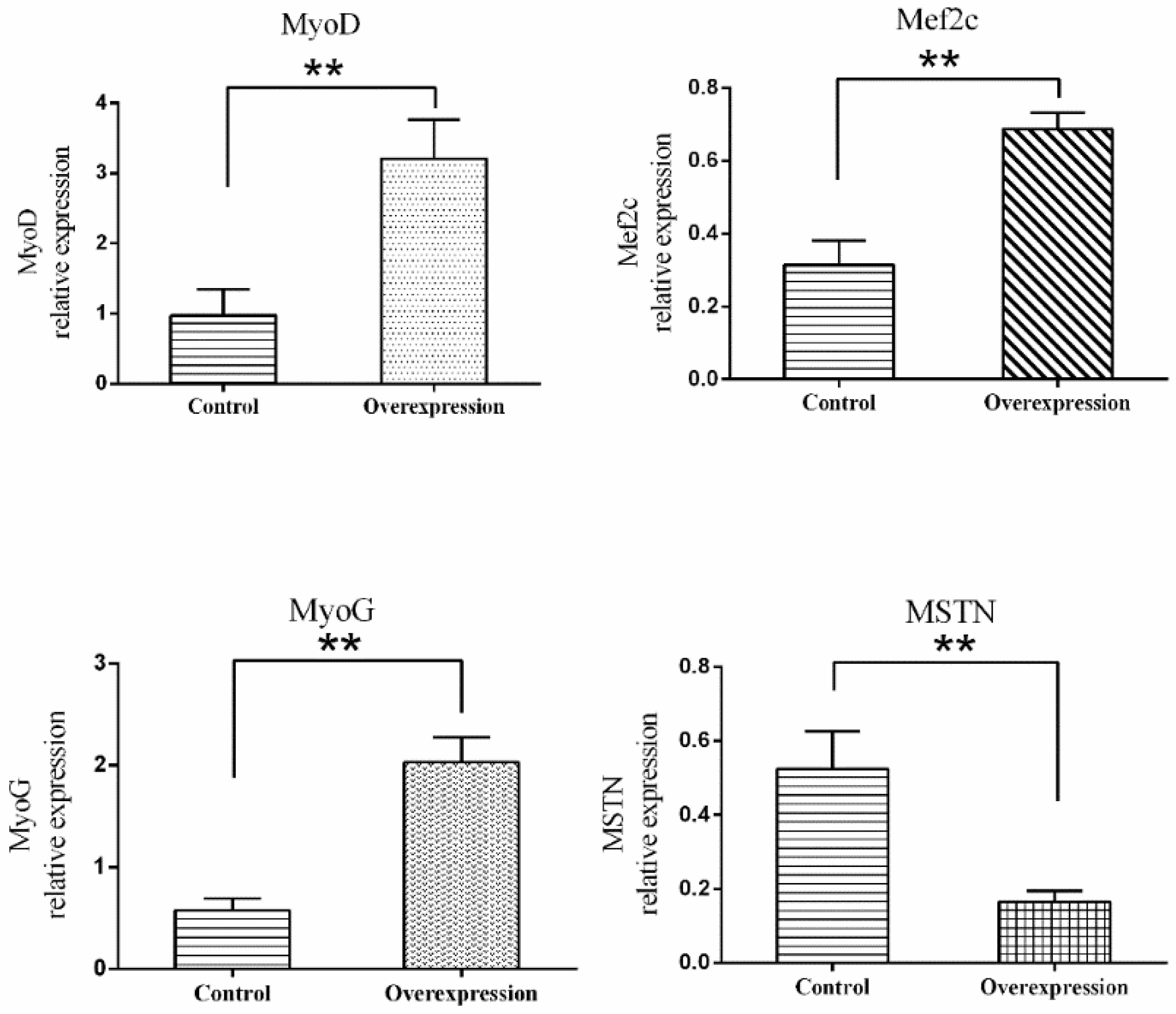
3.5. Overexpression of TCONS_00102859 in Skeletal Muscle and Its Effects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mattick, J.S.; Rinn, J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Gascoigne, D.K.; Cheetham, S.W.; Cattenoz, P.B.; Clark, M.B.; Amaral, P.P.; Taft, R.J.; Wilhelm, D.; Dinger, M.E.; Mattick, J.S. Pinstripe: A suite of programs for integrating transcriptomic and proteomic datasets identifies novel proteins and improves differentiation of protein-coding and non-coding genes. Bioinformatics 2012, 28, 3042–3050. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Russell, P.; Ingolia, N.T.; Weissman, J.S.; Lander, E.S. Ribosome profiling provides evidence that large non-coding RNAs do not encode proteins. Cell 2013, 154, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; An, J.; Wu, M.; Zheng, Q.; Xin, G.; Li, T.; Hu, P.; Lu, D. LncRNA hotair promotes human liver cancer stem cell malignant growth through downregulation of setd2. Oncotarget 2015, 6, 27847–27864. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Gao, Y. Increased expression of lncRNA BANCR and its prognostic significance in human hepatocellular carcinoma. World J. Surg. Oncol. 2015, 14, 1–8. [Google Scholar]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X. Human colorectal cancer-specific ccat1-l lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Arun, G.; Mao, Y.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.; Wu, J.; Zhang, C. The lncRNA malat1 is dispensable for mouse development but its transcription plays a cis -regulatory role in the adult. Cell Rep. 2012, 2, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.D.; Zhang, E.B.; You, L.H.; Wang, N.; Wang, L.T.; Jin, F.Y.; Zhu, Y.N.; Cao, L.H.; Yuan, Q.X.; De, W. Downregulation of lncRNA tug1 affects apoptosis and insulin secretion in mouse pancreatic β cells. Cell. Physiol. Biochem. 2015, 35, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Koufariotis, L.T.; Chen, Y.P.P.; Chamberlain, A.; Jagt, C.V.; Hayes, B.J. A catalogue of novel bovine long noncoding RNA across 18 tissues. PLoS ONE 2015, 10, e0141225. [Google Scholar] [CrossRef] [PubMed]
- Billerey, C.; Boussaha, M.; Esquerré, D.; Rebours, E.; Djari, A.; Meersseman, C.; Klopp, C.; Gautheret, D.; Rocha, D. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genom. 2014, 15, 499. [Google Scholar] [CrossRef] [PubMed]
- Ran, M.; Chen, B.; Li, Z.; Wu, M.; Liu, X.; He, C.; Zhang, S.; Li, Z. Systematic identification of long non-coding RNAs in immature and mature porcine testes. Biol. Reprod. 2016, 94, 77. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, S.; Liu, X.; Liu, H.; Hu, T.; Qiu, X.; Zhang, J.; Lei, M. Analyses of long non-coding RNA and mrna profiling using rna sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 2016, 6, 20238. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, E.L.; Belhocine, M.; Dao, L.T.; Puthier, D.; Spicuglia, S. Functions of lncRNA in development and diseases. Med. Sci. 2014, 30, 790–796. [Google Scholar]
- Paralkar, V.R.; Mishra, T.; Luan, J.; Yao, Y.; Kossenkov, A.V.; Anderson, S.M.; Dunagin, M.; Pimkin, M.; Gore, M.; Sun, D. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood 2014, 123, 1927–1937. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Mu, Y.; Ma, L.; Wang, C.; Tang, Z.; Yang, S.; Zhou, R.; Hu, X.; Li, M.H.; Li, K. Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Sci. Rep. 2015, 5, 8957. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.Y.; Lee, J.C.; Chang, Y.T.; Hou, M.F.; Huang, H.W.; Liaw, C.C.; Chang, H.W. Long noncoding RNAs-related diseases, cancers, and drugs. Scientificworldjournal 2013, 2013, 943539. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, S.; Wu, R.; Zhou, X.; Zhu, D.; Zhang, Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics 2012, 99, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, Y.; Bao, X.; Zhu, X.; Kwok, Y.K.; Sun, K.; Chen, X.; Huang, Y.; Jauch, R.; Esteban, M.A. LncRNA dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015, 25, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Ballarino, M.; Cazzella, V.; D’Andrea, D.; Grassi, L.; Bisceglie, L.; Cipriano, A.; Santini, T.; Pinnarò, C.; Morlando, M.; Tramontano, A. Novel long noncoding RNAs (lncRNAs) in myogenesis: A miR-31 overlapping LncRNA transcript controls myoblast differentiation. Mol. Cell. Biol. 2015, 35, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Ren, H. Histological and transcriptome-wide level characteristics of fetal myofiber hyperplasia during the second half of gestation in texel and Ujumqin sheep. BMC Genom. 2011, 12, 411. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.L.; Dodds, K.G.; Bain, W.E.; Greer, G.J.; Mclean, N.J.; Mclaren, R.J.; Galloway, S.M.; Stijn, T.C.V.; Mcewan, J.C. Investigations into the GDF8 g+6723G-A polymorphism in New Zealand Texel sheep. J. Anim. Sci. 2009, 87, 1856. [Google Scholar] [CrossRef] [PubMed]
- Clop, A.; Marcq, F.; Takeda, H.; Pirottin, D.; Tordoir, X.; Bibé, B.; Bouix, J.; Caiment, F.; Elsen, J.M.; Eychenne, F. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006, 38, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, F.; Wei, C.; Sheng, X.; Ren, H.; Xu, L.; Lu, J.; Liu, J.; Zhang, L.; Du, L. Identification and characterization of the mirna transcriptome of ovis aries. PLoS ONE 2013, 8, e58905. [Google Scholar] [CrossRef] [PubMed]
- Ponjavic, J.; Oliver, P.L.; Lunter, G.; Ponting, C.P. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009, 5, e1000617. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q. Long noncoding RNAs with enhancer-like function in human cells. Med. Sci. M/S 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; Wang, J. WEGO: A web tool for plotting go annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 27, 29–34. [Google Scholar] [CrossRef]
- Engström, P.G.; Suzuki, H.; Ninomiya, N.; Akalin, A.; Sessa, L.; Lavorgna, G.; Brozzi, A.; Luzi, L.; Tan, S.L.; Yang, L. Complex loci in human and mouse genomes. PLoS Genet. 2006, 2, e47. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarianamiri, M.E.; Ru, K.; Soldà, G.; Simons, C. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008, 18, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.Y.; Nusse, R. The WNT signaling pathway in development and disease. Ann. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [PubMed]
- Cadigan, K.M.; Nusse, R. Wnt signaling: A common theme in animal development. Genes Dev. 1997, 11, 3286–3305. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, W.K.; Gao, W.S.; Shen, Y.; Ding, W.Y. Function of tgf-beta and p38 MAKP signaling pathway in osteoblast differentiation from rat adipose-derived stem cells. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 1611–1619. [Google Scholar] [PubMed]
- Takada, F.; Vander Woude, D.L.; Tong, H.Q.; Thompson, T.G.; Watkins, S.C.; Kunkel, L.M.; Beggs, A.H. Myozenin: An alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc. Natl. Acad. Sci. USA 2001, 98, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Frey, N.; Richardson, J.A.; Olson, E.N. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 14632–14637. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Gonzalez, O.; Ramagli, L.; Xu, J.; Siciliano, M.J.; Bachinski, L.L.; Roberts, R. Identification and characterization of a novel gene (C4orf5) located on human chromosome 4q with specific expression in cardiac and skeletal muscle. Genomics 2000, 70, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Chin, E.R.; Olson, E.N.; Richardson, J.A.; Yang, Q.; Humphries, C.; Shelton, J.M.; Wu, H.; Zhu, W.; BasselDuby, R.; Williams, R.S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998, 12, 2499–2509. [Google Scholar] [CrossRef] [PubMed]
- Frey, N.; Barrientos, T.; Shelton, J.M.; Frank, D.; Rütten, H.; Gehring, D.; Kuhn, C.; Lutz, M.; Rothermel, B.; Basselduby, R. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 2004, 10, 1336–1343. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Ma, J.; Wang, N.; Wang, D.; Xu, G. Molecular cloning and characterization of different expression of MYOZ2 and MYOZ3 in tianfu goat. PLoS ONE 2013, 8, e82550. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shen, T.; Randall, W.R.; Schneider, M.F. Signaling pathways in activity-dependent fiber type plasticity in adult skeletal muscle. J. Muscle Res. Cell Motil. 2005, 26, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Osio, A.; Tan, L.; Chen, S.N.; Lombardi, R.; Nagueh, S.F.; Shete, S.; Roberts, R.; Willerson, J.T.; Marian, A.J. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ. Res. 2007, 100, 766–768. [Google Scholar] [CrossRef] [PubMed]
- Alessandra, R.; Suet Nee, C.; Raffaella, L.; Gabriela, R.; Marian, A.J. Pathogenesis of hypertrophic cardiomyopathy caused by myozenin 2 mutations is independent of calcineurin activity. Cardiovasc. Res. 2013, 97, 44–54. [Google Scholar]
- Posch, M.G.; Thiemann, L.; Tomasov, P.; Veselka, J.; Cardim, N.; Garcia-Castro, M.; Coto, E.; Perrot, A.; Geier, C.; Dietz, R. Sequence analysis of myozenin 2 in 438 european patients with familial hypertrophic cardiomyopathy. Med. Sci. Monit. 2008, 14, CR372–CR374. [Google Scholar] [PubMed]
- Shaulian, E.; Karin, M. Ap-1 in cell proliferation and survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M. Ap-1 transcription factor in cell differentiation and survival. Am. J. Hum. Genet. 2005, 85, 737–744. [Google Scholar]
- Angel, P.; Karin, M. The role of jun, fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1991, 1072, 129–157. [Google Scholar] [CrossRef]
- Dony, C.; Gruss, P. Proto-oncogene c-fos expression in growth regions of fetal bone and mesodermal web tissue. Nature 1987, 328, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Mccabe, L.R.; Kockx, M.; Lian, J.; Stein, J.; Stein, G. Selective expression of fos- and jun-related genes during osteoblast proliferation and differentiation. Exp. Cell Res. 1995, 218, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Dollenmeier, P.; Turner, D.C.; Eppenberger, H.M. Proliferation and differentiation of chick skeletal muscle cells cultured in a chemically defined medium. Exp. Cell Res. 1981, 135, 47–61. [Google Scholar] [CrossRef]
- Liu, X.; Mcfarland, D.C.; Nestor, K.E.; Velleman, S.G. Expression of fibroblast growth factor 2 and its receptor during skeletal muscle development from turkeys with different growth rates. Domest. Anim. Endocrinol. 2003, 25, 215–229. [Google Scholar] [CrossRef]
- Brunetti, A.; Goldfine, I.D. Role of myogenin in myoblast differentiation and its regulation by fibroblast growth factor. J. Biol. Chem. 1990, 265, 5960–5963. [Google Scholar] [PubMed]
- Olwin, B.B.; Arthur, K.; Hannon, K.; Hein, P.; Zhou, Z.; Zuber, M.E.; Kudla, A.J.; Mcfall, A.; Rapraeger, A.C.; Riley, B. Role of fgfs in skeletal muscle and limb development. Mol. Reprod. Dev. 1994, 39, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Barclay, C.; Li, A.W.; Geldenhuys, L.; Bagumanibasheka, M.; Porter, G.A.; Veugelers, P.J.; Murphy, P.R.; Casson, A.G. Basic fibroblast growth factor (FGF-2) overexpression is a risk factor for esophageal cancer recurrence and reduced survival, which is ameliorated by coexpression of the FGF-2 antisense gene. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 7683–7691. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Güth, R.; Jonsson, C.B.; Unguez, G.A.S. Macrurus myogenic regulatory factors induce mammalian skeletal muscle differentiation: Evidence for functional conservation of MRFs. Int. J. Dev. Biol. 2009, 53, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Potthoff, M.J.; Arnold, M.A.; Mcanally, J.; Richardson, J.A.; Basselduby, R.; Olson, E.N. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell. Biol. 2007, 27, 8143–8151. [Google Scholar] [CrossRef] [PubMed]
- Fakhfakh, R.; Michaud, A.; Tremblay, J.P. Blocking the myostatin signal with a dominant negative receptor improves the success of human myoblast transplantation in dystrophic mice. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar] [PubMed]
- Ren, H. Profiles of Gene Expression and Histologigical Analysis in Fetal Skeletal Muscle between Texel and Ujumqin Sheep (Ovis Aries) during the Second Half of Gestation; Chinese Academy of Agricultural Sciences: Beijing, China, 2010. [Google Scholar]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Liu, R.; Zhao, H.; Di, R.; Lu, Z.; Liu, E.; Wang, Y.; Chu, M.; Wei, C. Identification and Characterization of Long Noncoding RNAs in Ovine Skeletal Muscle. Animals 2018, 8, 127. https://doi.org/10.3390/ani8070127
Li Q, Liu R, Zhao H, Di R, Lu Z, Liu E, Wang Y, Chu M, Wei C. Identification and Characterization of Long Noncoding RNAs in Ovine Skeletal Muscle. Animals. 2018; 8(7):127. https://doi.org/10.3390/ani8070127
Chicago/Turabian StyleLi, Qing, Ruizao Liu, Huijing Zhao, Ran Di, Zengkui Lu, Enmin Liu, Yuqin Wang, Mingxing Chu, and Caihong Wei. 2018. "Identification and Characterization of Long Noncoding RNAs in Ovine Skeletal Muscle" Animals 8, no. 7: 127. https://doi.org/10.3390/ani8070127
APA StyleLi, Q., Liu, R., Zhao, H., Di, R., Lu, Z., Liu, E., Wang, Y., Chu, M., & Wei, C. (2018). Identification and Characterization of Long Noncoding RNAs in Ovine Skeletal Muscle. Animals, 8(7), 127. https://doi.org/10.3390/ani8070127





