Genomic Characterization and Phylogenetic Relationships of Procypris rabaudi Revealed by Whole-Genome Survey Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Sequencing
2.3. Raw Data Quality Control and Genome Survey
2.4. Draft Genome Assembly and SSR Identification
2.5. Mitochondrial Genome Assembly and Phylogenetic Analysis
2.6. Historical Demography Reconstruction
3. Results
3.1. Size, Heterozygosity Ratio, and Repeat Sequence Ratio of P. rabaudi
3.2. The Draft Genome Assembly and SSR Identification Results
3.3. Mitochondrial Genome Assembly and RSCU Analysis
3.4. Phylogenetic Analysis
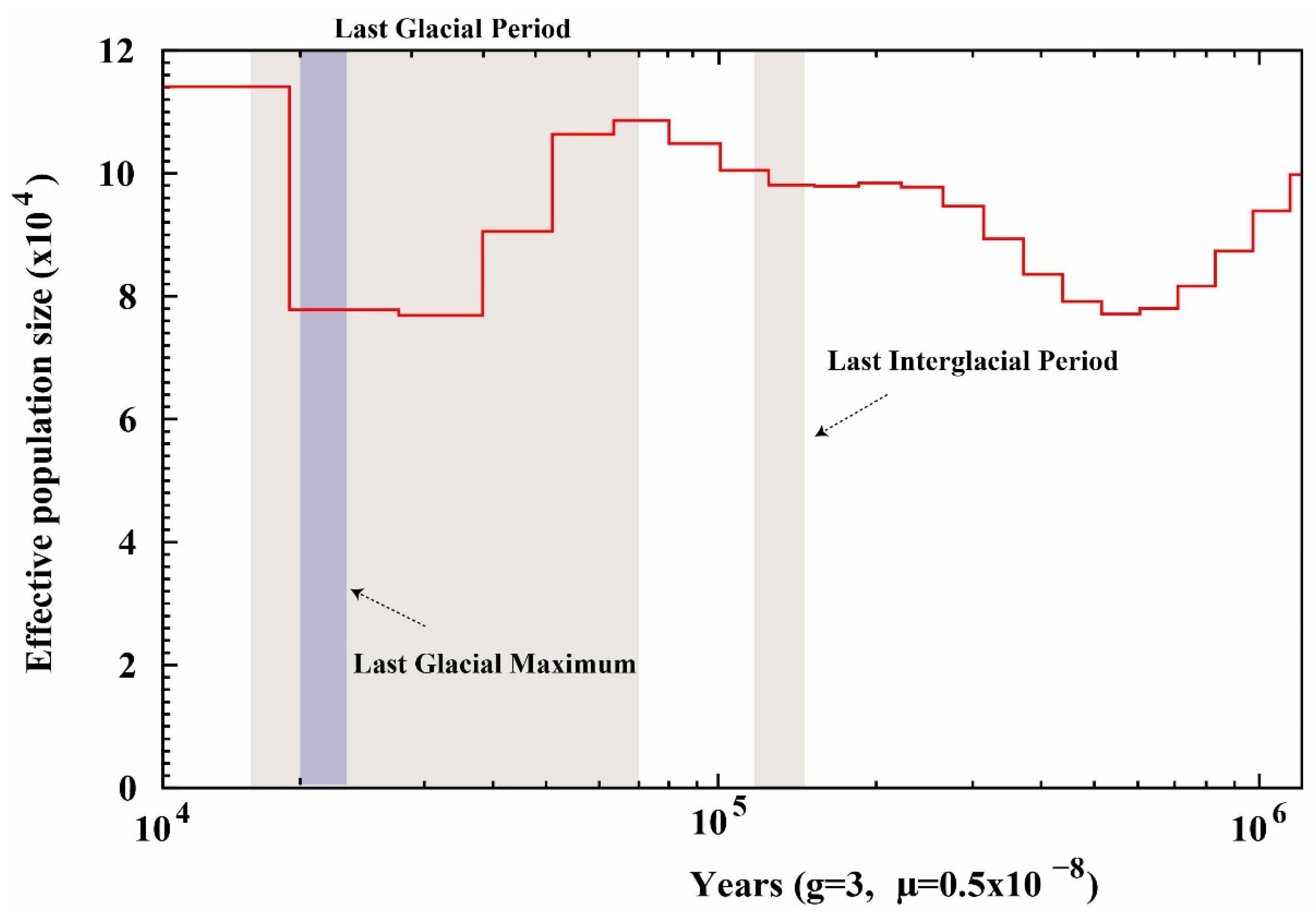
3.5. Population Size Dynamics of P. rabaudi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, T.; Gao, X.; Wang, J.; Jakovlić, I.; Dan, S.-G.; Liu, H.-Z. Life history traits and implications for conservation of rock carp Procypris rabaudi Tchang, an endemic fish in the upper Yangtze River, China. Fish. Sci. 2015, 81, 515–523. [Google Scholar] [CrossRef]
- Probst, W.N.; Tan, D.; Gao, Y.; Drossou, A.; Petereit, C.; Wecker, B.; Xiong, M.; Ueberschär, B.; Chang, J.; Rosenthal, H. Rearing of Procypris rabaudi during early life-history stages. J. Appl. Ichthyol. 2006, 22, 530–535. [Google Scholar] [CrossRef]
- Yang, L.; Mayden, R.L.; Cai, Y. Threatened fishes of the world: Procypris rabaudi (Tchang, 1930) (Cyprinidae). Environ. Biol. Fishes 2009, 84, 275–276. [Google Scholar] [CrossRef]
- He, W.; Dong, Z.; Ma, T.; Yan, H.; Chen, Z.; Yao, W.; Cheng, F. Three Mitochondrial Markers Reveal Genetic Diversity and Structure of Rock Carp (Procypris rabaudi) Endemic to the Upper Yangtze: Implications for Pre-release Genetic Assessment. Fron. Mar. Sci. 2022, 9, 939745. [Google Scholar] [CrossRef]
- Cheng, F.; Wu, Q.; Liu, M.; Radhakrishnan, K.V.; Murphy, B.R.; Xie, S. Impacts of hatchery release on genetic structure of rock carp Procypris rabaudi in the upper Yangtze River, China. Fish. Sci. 2011, 77, 765–771. [Google Scholar] [CrossRef]
- Zhang, X.; Ouyang, M.; Zhang, F.; Wang, J. Study on the genetic structure of wild and hatchery populations of Procypris rabaudi Tchang, an endemic fish in the upper Yangtze River. Fish. Res. 2022, 245, 106134. [Google Scholar] [CrossRef]
- da Fonseca, R.R.; Albrechtsen, A.; Themudo, G.E.; Ramos-Madrigal, J.; Sibbesen, J.A.; Maretty, L.; Zepeda-Mendoza, M.L.; Campos, P.F.; Heller, R.; Pereira, R.J. Next-generation biology: Sequencing and data analysis approaches for non-model organisms. Mar. Genom. 2016, 30, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-generation sequencing technology: Current trends and advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, X.; Qu, Y.; Wang, Y.; Guo, X.; Li, W.; Gao, T.; Qiao, Y. A comprehensive genome survey study unveils the genomic characteristics and phylogenetic evolution of fishes in the Uranoscopidae family. Front. Mar. Sci. 2024, 11, 1383635. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, T.; Song, N.; Qu, Y.; Gao, T. Whole-genome survey reveals interspecific differences in genomic characteristics and evolution of Pampus fish. Front. Mar. Sci. 2024, 10, 1332250. [Google Scholar] [CrossRef]
- Xiong, Y.; Lei, X.; Bai, S.; Xiong, Y.; Liu, W.; Wu, W.; Yu, Q.; Dong, Z.; Yang, J.; Ma, X. Genomic survey sequencing, development and characterization of single- and multi-locus genomic SSR markers of Elymus sibiricus L. BMC Plant Biol. 2021, 21, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-j.; Liu, M.-x.; Lu, X.-y.; Sun, S.-s.; Cheng, Y.; Ya, H.-y. Genome survey sequencing and identification of genomic SSR markers for Rhododendron micranthum. Biosci. Rep. 2020, 40, BSR20200988. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiao, W.; Wang, F.; Wang, Y.; Dong, Y.; Nie, W.; Tan, C.; An, S.; Chang, E.; Jiang, Z.; et al. Adaptive divergence, historical population dynamics, and simulation of suitable distributions for Picea Meyeri and P. Mongolica at the whole-genome level. BMC Plant Biol. 2024, 24, 479. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Spence, J.P.; Feng, Y.; Hansen, M.E.B.; Terhorst, J.; Beltrame, M.H.; Ranciaro, A.; Hirbo, J.; Beggs, W.; Thomas, N.; et al. Whole-genome sequencing reveals a complex African population demographic history and signatures of local adaptation. Cell 2023, 186, 923–939.e914. [Google Scholar] [CrossRef]
- Ma, S.; Zhao, X.; Song, N. Whole-genome survey analyses of five goby species provide insights into their genetic evolution and invasion-related genes. Int. J. Mol. Sci. 2024, 25, 3293. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Du, X.; Ma, S.; Song, N.; Zhao, L. Whole-genome survey analyses provide a new perspective for the evolutionary biology of Shimofuri goby, Tridentiger bifasciatus. Animal 2022, 12, 1914. [Google Scholar] [CrossRef]
- Mao, W.; Xu, Z.; Liu, Q.; Li, N.; Liu, L.; Ren, B.; Gao, T.; Liu, C. A Whole-genome survey and the mitochondrial genome of Acanthocepola indica provide insights into its phylogenetic relationships in Priacanthiformes. Animals 2024, 14, 3257. [Google Scholar] [CrossRef]
- Liu, B.; Shi, Y.; Yuan, J.; Hu, X.; Zhang, H.; Li, N.; Li, Z.; Chen, Y.; Mu, D.; Fan, W. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv 2013, arXiv:1308.2012. [Google Scholar] [CrossRef]
- Ranallo-Benavidez, T.R.; Jaron, K.S.; Schatz, M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. Imeta 2023, 2, e87. [Google Scholar] [CrossRef] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Wang, Z.; Ran, M.; Liu, X.; Zhou, C.; Song, Z. Chromosome-level genome assembly of rock carp (Procypris rabaudi). Sci. Data 2025, 12, 914. [Google Scholar] [CrossRef]
- Jung, Y.; Han, D. BWA-MEME: BWA-MEM emulated with a machine learning approach. Bioinformatics 2022, 38, 2404–2413. [Google Scholar] [CrossRef]
- Yue, G.H.; David, L.; Orban, L. Mutation rate and pattern of microsatellites in common carp (Cyprinus carpio L.). Genetica 2007, 129, 329–331. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, X.; Wang, X.; Li, J.; Liu, G.; Kuang, Y.; Xu, J.; Zheng, X.; Ren, L.; Wang, G.; et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Q.; Tang, W.; Huang, Z.; Wang, G.; Wang, Y.; Shi, J.; Xu, H.; Lin, L.; Li, Z.; et al. The evolutionary origin and domestication history of goldfish (Carassius auratus). Proc. Natl. Acad. Sci. USA 2020, 117, 29775–29785. [Google Scholar] [CrossRef]
- Zheng, J.; Jiang, J.; Rui, Q.; Li, F.; Liu, S.; Cheng, S.; Chi, M.; Jiang, W. Chromosome-level genome assembly of Acrossocheilus fasciatus using PacBio sequencing and Hi-C technology. Sci. Data 2024, 11, 166. [Google Scholar] [CrossRef]
- Gu, H.; Zhang, X.; Xu, W.; Yang, Z.; Xu, Y.; Miao, X.; Feng, Y. Chromosome-level assemblies of the White bream Parabramis pekinensis. Sci. Data 2025, 12, 871. [Google Scholar] [CrossRef]
- Amos, W.; Harwood, J. Factors affecting levels of genetic diversity in natural populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998, 1366, 177–186. [Google Scholar] [CrossRef]
- Trygve, G.; Nick, R.; Morten, R. The importance of selective breeding in aquaculture to meet future demands for animal protein: A review. Aquaculture 2012, 350, 117–129. [Google Scholar] [CrossRef]
- Shen, Q.; Wu, J.; Yang, C.; Sheng, Q.; Xu, M.; Yi, S.; Song, J. Chromosome-level assembly and annotation of the yellow-shelled fish (Barbodes wynaadensis). Sci. Data 2025, 12, 1196. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Wang, W.; Huang, J.; Luo, M.; Wang, N.; Sun, C.; Lu, J. A chromosome-level genome assembly of big-barbel schizothorcin, Schizothorax macropogon. Sci. Data 2024, 11, 1402. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Liu, H.; Lu, W.; Li, J.; Fang, T.; Gao, N.; Chen, C.; Zhao, X.; Yang, K.; Liu, H. Chromosome-level genome assembly of the smallscale yellowfin (Plagiognathops microlepis). Sci. Data 2024, 11, 1234. [Google Scholar] [CrossRef]
- Shapiro, J.; von Sternberg, R. Why repetitive DNA is essential to genome function. Biol. Rev. Camb. Philos. Soc. 2005, 80, 227–250. [Google Scholar] [CrossRef]
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and diversity of transposable elements in vertebrate genomes. Genome Bio. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015, 7, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Xu, X.; Li, J.; Shen, Y. Primary mapping of QTL for growth-related traits in grass carp (Ctenopharyngodon idellus). Aquacult. Int. 2020, 28, 2275–2285. [Google Scholar] [CrossRef]
- Cheng, J.; Yanagimoto, T.; Song, N.; Gao, T.-x. Population genetic structure of chub mackerel Scomber japonicus in the Northwestern Pacific inferred from microsatellite analysis. Mol. Biol. Rep. 2015, 42, 373–382. [Google Scholar] [CrossRef]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef]
- Lei, Y.; Zhou, Y.; Price, M.; Song, Z. Genome-wide characterization of microsatellite DNA in fishes: Survey and analysis of their abundance and frequency in genome-specific regions. BMC Genom. 2021, 22, 421. [Google Scholar] [CrossRef]
- Wang, Y.; Sha, H.; Li, X.; Zhou, T.; Luo, X.; Zou, G.; Chai, Y.; Liang, H. Microsatellite characteristics of silver carp (Hypophthalmichthysmolitrix) genome and genetic diversity analysis in four cultured populations. Genes 2022, 13, 1267. [Google Scholar] [CrossRef] [PubMed]
- Schlötterer, C.; Tautz, D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992, 20, 211–215. [Google Scholar] [CrossRef]
- Thomas, E.K. Repetitive DNA regulates gene expression. Science 2023, 381, 1289–1290. [Google Scholar] [CrossRef]
- Tobar-Tosse, F.; Veléz, P.; Ocampo-Toro, E.; Moreno, P. Structure, clustering and functional insights of repeats configurations in the upstream promoter region of the human coding genes. BMC Genom. 2018, 19, 862. [Google Scholar] [CrossRef]
- Zhang, X.; Yue, B.; Jiang, W.; Song, Z. The complete mitochondrial genome of rock carp Procypris rabaudi (Cypriniformes: Cyprinidae) and phylogenetic implications. Mol. Biol. Rep. 2009, 36, 981–991. [Google Scholar] [CrossRef]
- Li, Z.; Han, Y.; Li, Y.; Wu, W.; Lei, J.; Wang, D.; Lin, Y.; Wang, X. Whole mitochondrial genome sequencing and phylogenetic tree construction for Procypris mera (Lin 1933). Animals 2024, 14, 2672. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, T.; Li, H.; Deng, L. The mitochondrial genome of Linichthys laticeps (Cypriniformes: Cyprinidae): Characterization and phylogeny. Genes 2023, 14, 1938. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Tan, C.; Zhao, L.; Hu, Z.; Su, C.; Li, F.; Ma, Y.; Zhang, W.; Hao, X.; Zou, W.; et al. The complete mitochondrial genome of the Luciocyprinus langsoni (Cypriniformes: Cyprinidae): Characterization, phylogeny, and genetic diversity analysis. Genes 2024, 15, 1621. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Morio, A.; Tsutsumi, R.; Kondo, T.; Miyoshi, H.; Kato, T.; Narasaki, S.; Satomi, S.; Nakaya, E.; Kuroda, M.; Sakaue, H.; et al. Leucine induces cardioprotection in vitro by promoting mitochondrial function via mTOR and Opa-1 signaling. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 2979–2986. [Google Scholar] [CrossRef]
- Wu, X.; Wang, L.; Chen, S.; Zan, R.; Xiao, H.; Zhang, Y.P. The complete mitochondrial genomes of two species from Sinocyclocheilus (Cypriniformes: Cyprinidae) and a phylogenetic analysis within Cyprininae. Mol. Biol. Rep. 2010, 37, 2163–2171. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Y.; Yang, F.; Han, S.; Wang, X.; Xie, R.; He, S. Identification analysis of Procypris rabaudi by RAPD. J. Wuhan Univ. (Nat. Sci. Ed.) 2004, 50, 477–481. (In Chinese) [Google Scholar]
- Karamanlidis, A.A.; Skrbinšek, T.; Amato, G.; Dendrinos, P.; Gaughran, S.; Kasapidis, P.; Kopatz, A.; Stronen, A.V. Genetic and demographic history define a conservation strategy for earth’s most endangered pinniped, the Mediterranean monk seal Monachus monachus. Sci. Rep. 2021, 11, 373. [Google Scholar] [CrossRef]
- Nielsen, E.E.; Hemmer-Hansen, J.; Larsen, P.F.; Bekkevold, D. Population genomics of marine fishes: Identifying adaptive variation in space and time. Mol. Ecol. 2009, 18, 3128–3150. [Google Scholar] [CrossRef]
- Obreht, I.; De Vleeschouwer, D.; Wörmer, L.; Kucera, M.; Varma, D.; Prange, M.; Laepple, T.; Wendt, J.; Nandini-Weiss, S.D.; Schulz, H.; et al. Last Interglacial decadal sea surface temperature variability in the eastern Mediterranean. Nat. Geosci. 2022, 15, 812–818. [Google Scholar] [CrossRef]
- Hu, A.; Meehl, G.A.; Otto-Bliesner, B.L.; Waelbroeck, C.; Han, W.; Loutre, M.-F.; Lambeck, K.; Mitrovica, J.X.; Rosenbloom, N. Influence of Bering Strait flow and North Atlantic circulation on glacial sea-level changes. Nat. Geosci. 2010, 3, 118–121. [Google Scholar] [CrossRef]
- Hoareau, T.B.; Boissin, E.; Berrebi, P. Evolutionary history of a widespread Indo-Pacific goby: The role of Pleistocene sea-level changes on demographic contraction/expansion dynamics. Mol. Phylogenet. Evol. 2012, 62, 566–572. [Google Scholar] [CrossRef]
- He, D.; Chen, Y. Biogeography and molecular phylogeny of the genus Schizothorax (Teleostei: Cyprinidae) in China inferred from cytochrome b sequences. J. Biogeogr. 2006, 33, 1448–1460. [Google Scholar] [CrossRef]
- He, D.; Chen, Y. Phylogeographic structure of Schizothorax o’connori (Cyprinidae: Schizothoracinae) in the Nujiang River inferred from mtDNA sequences. Biochem. Syst. Ecol. 2009, 635, 251–262. [Google Scholar] [CrossRef]
- Gu, L.; Liu, Y.; Que, P.; Zhang, Z. Quaternary climate and environmental changes have shaped genetic differentiation in a Chinese pheasant endemic to the eastern margin of the Qinghai-Tibetan Plateau. Mol. Phylogenet. Evol. 2013, 67, 129–139. [Google Scholar] [CrossRef] [PubMed]




| Type | Reads Number | Base Count (Gb) | Read Length (bp) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| raw | 1,112,770,540 | 166.92 | 150 | 99.61 | 98.50 | 38.91 |
| dedup | 1,069,509,006 | 157.79 | 147 | 99.61 | 98.49 | 38.65 |
| K-mer Number | K-mer Depth | Genome Size (bp) | Revised Genome Size (bp) | Heterozygous Ratio (%) | Repeat (%) |
|---|---|---|---|---|---|
| 140,489,127,949 | 92 | 1,495,420,000 | 1,490,725,446 | 0.44 | 61.47 |
| Assembly Level | Total Number | Total Base (bp) | Max Length (bp) | N50 Length (bp) | N90 Length (bp) |
|---|---|---|---|---|---|
| Scaffold | 1,017,551 | 1,881,216,104 | 77,646 | 7176 | 1292 |
| Contig | 4,385,755 | 1,368,591,351 | 10,405 | 469 | 129 |
| Assembly Level | N90 Length (bp) |
|---|---|
| Total number of sequences examined | 1,017,551 |
| Total size of examined sequences (bp) | 1,881,216,104 |
| Total number of identified SSRs | 1,151,980 |
| Number of SSR containing sequences | 398,918 |
| Number of sequences containing more than 1 SSR | 227,348 |
| Number of SSRs present in compound formation | 156,448 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Han, X.; Luo, R.; Liu, Q.; Yuan, Z.; He, W. Genomic Characterization and Phylogenetic Relationships of Procypris rabaudi Revealed by Whole-Genome Survey Analysis. Animals 2026, 16, 246. https://doi.org/10.3390/ani16020246
Han X, Luo R, Liu Q, Yuan Z, He W. Genomic Characterization and Phylogenetic Relationships of Procypris rabaudi Revealed by Whole-Genome Survey Analysis. Animals. 2026; 16(2):246. https://doi.org/10.3390/ani16020246
Chicago/Turabian StyleHan, Xiaolu, Renhui Luo, Qi Liu, Zengbao Yuan, and Wenping He. 2026. "Genomic Characterization and Phylogenetic Relationships of Procypris rabaudi Revealed by Whole-Genome Survey Analysis" Animals 16, no. 2: 246. https://doi.org/10.3390/ani16020246
APA StyleHan, X., Luo, R., Liu, Q., Yuan, Z., & He, W. (2026). Genomic Characterization and Phylogenetic Relationships of Procypris rabaudi Revealed by Whole-Genome Survey Analysis. Animals, 16(2), 246. https://doi.org/10.3390/ani16020246








