Integrated Transcriptomic Analysis of Liver and Muscle Tissues Reveals Candidate Genes and Pathways Regulating Intramuscular Fat Deposition in Beef Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. IMF Measurements
2.3. HE Staining
2.4. RNA Extraction, RNA-Seq Library Construction, and Sequencing
2.5. Transcriptome Data Processing and Differential Gene Expression Analysis
2.6. Functional Enrichment and Network Analysis
2.7. qRT-PCR Validation of RNA-Seq Results
2.8. Statistical Analysis
3. Results
3.1. Differences in Meat Quality of Cattle with Different IMF Contents
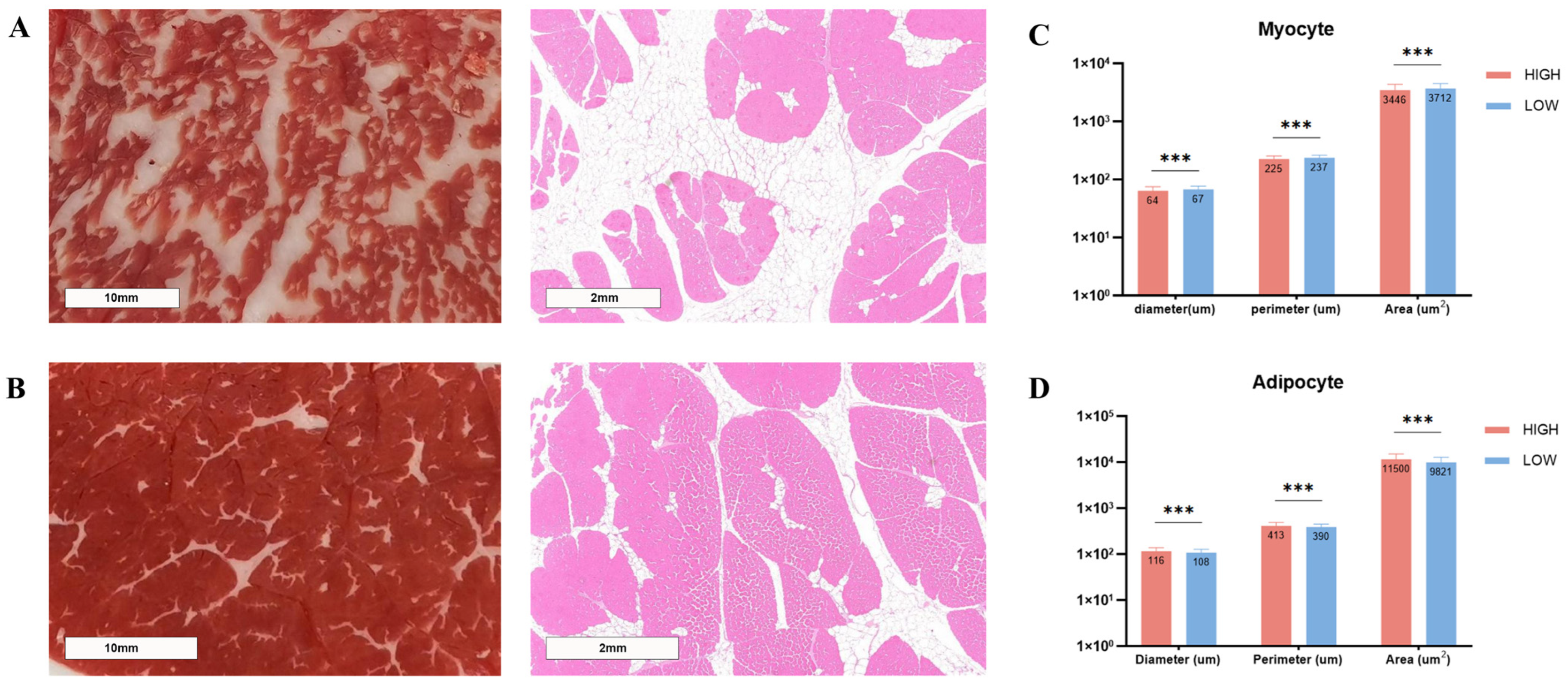
3.2. Morphological Differences Among LD Muscle with Different Levels of IMF Content
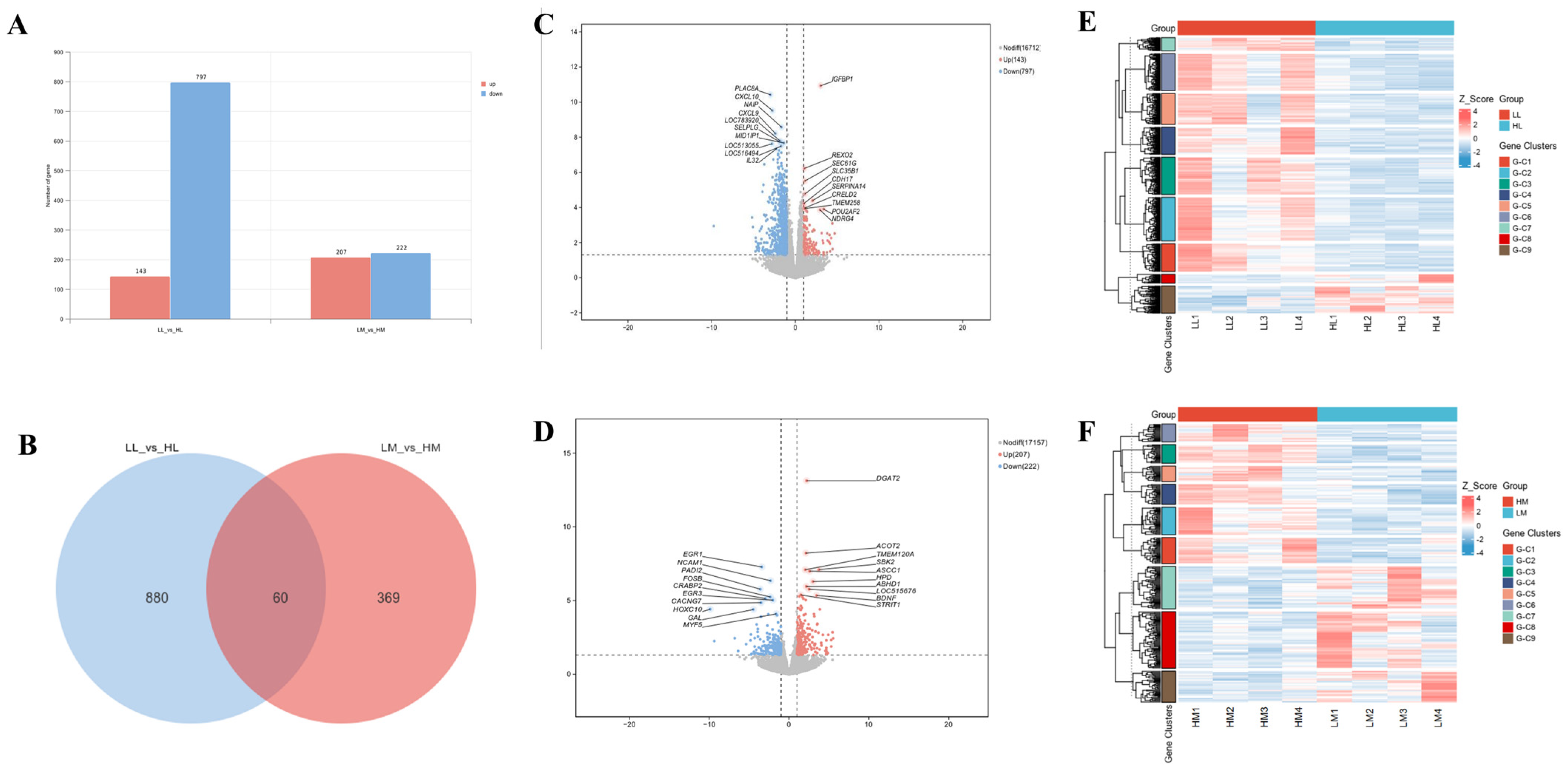
3.3. Summary of RNA-Seq Results and Identification of DEGs
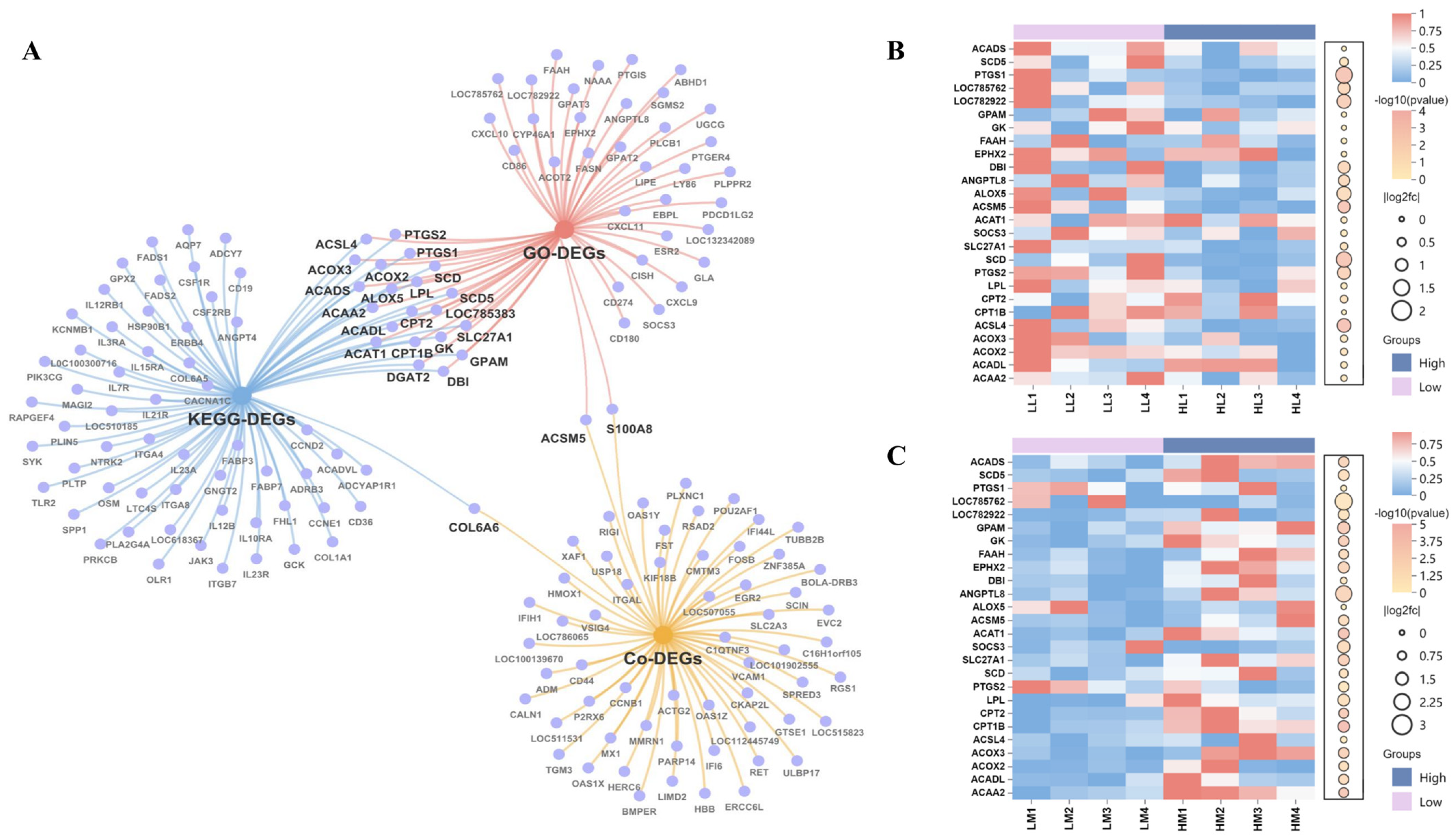
3.4. Functional Enrichment Analysis of DEGs in Liver and Muscle Reveals Tissue-Specific Biological Processes Associated with Intramuscular Fat
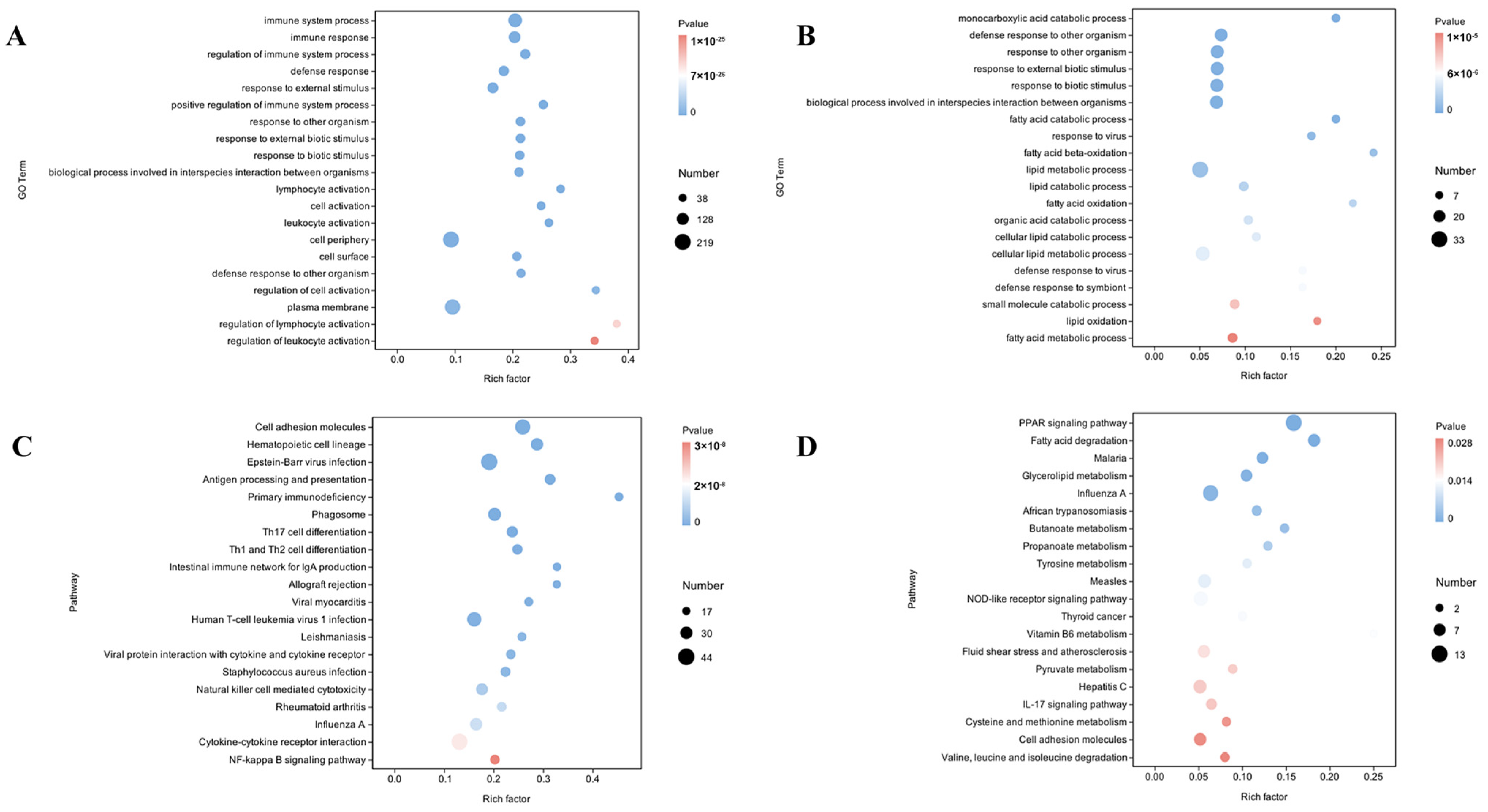
3.4.1. GO Term Enrichment in Liver and Muscle DEGs Related to Intramuscular Fat
3.4.2. KEGG Pathway Enrichment in Liver and Muscle DEGs Related to Intramuscular Fat
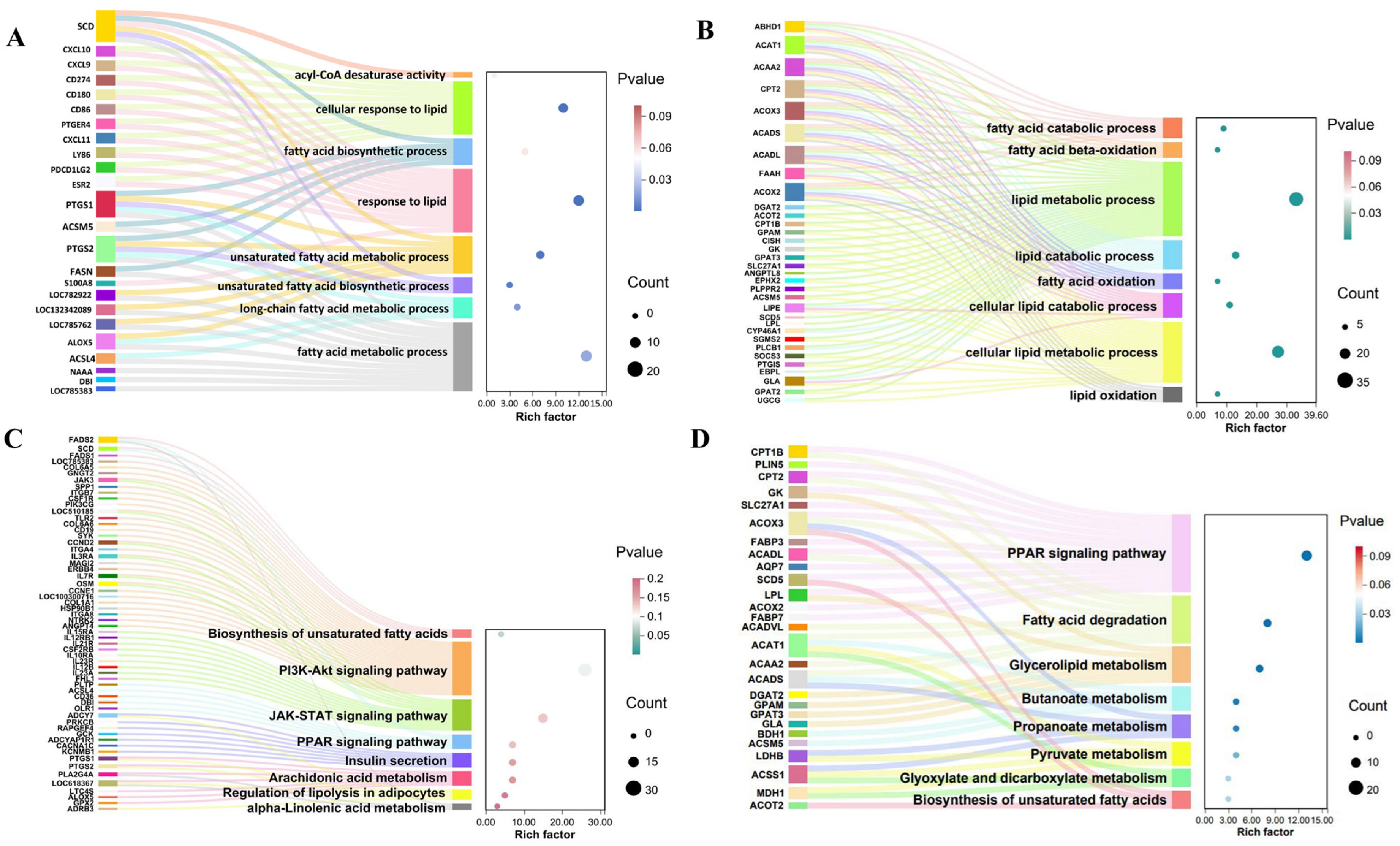
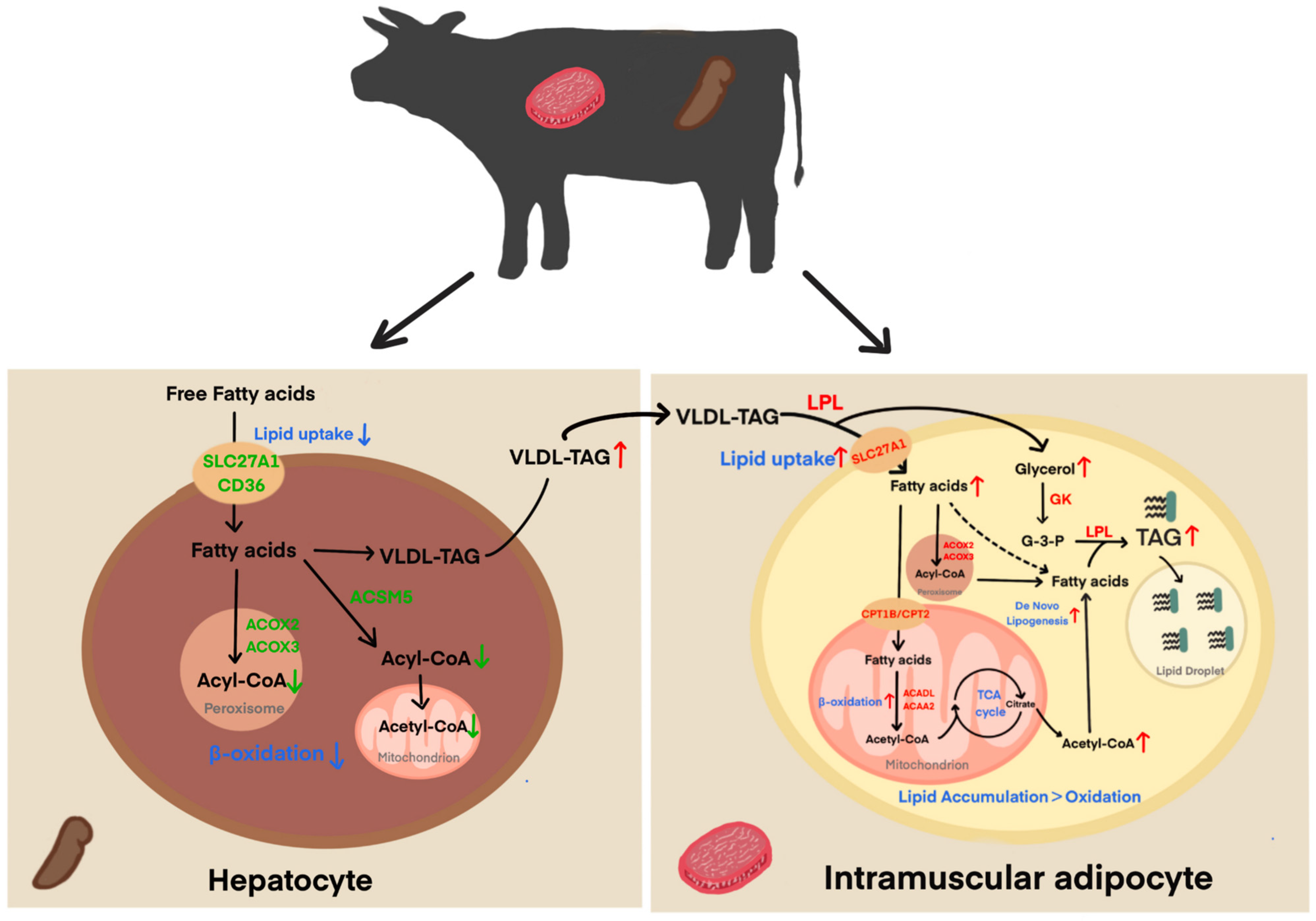
3.5. Candidate Genes in Liver-Muscle Interaction for IMF Deposition
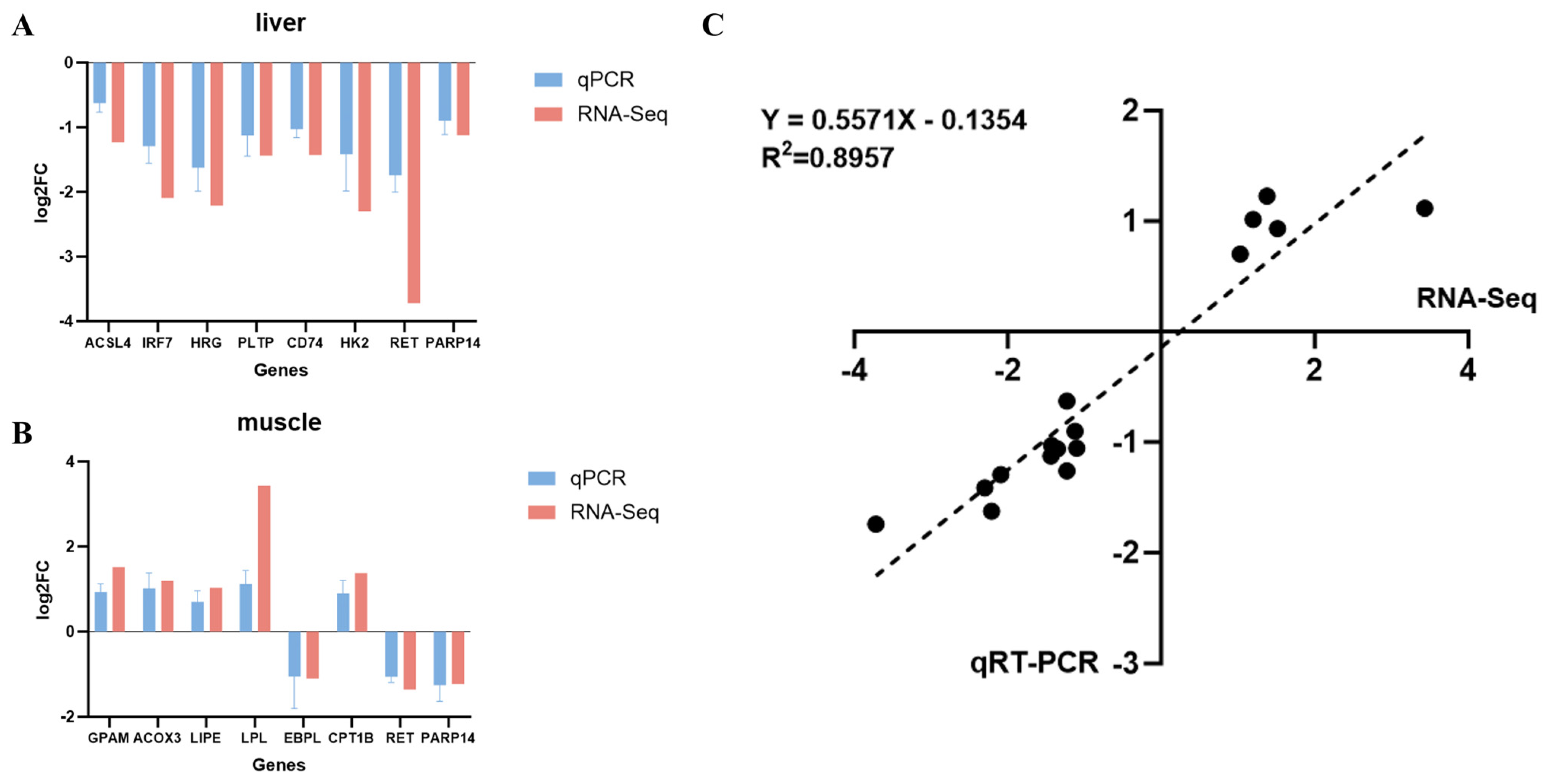
3.6. RNA-Seq Data Validation by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABHD1 | Abhydrolase Domain Containing 1 |
| ACAA2 | Acetyl-CoA Acyltransferase 2 |
| ACADL | Acyl-CoA Dehydrogenase Long Chain |
| ACAT1 | Acetyl-CoA Acetyltransferase 1 |
| ACOT2 | Acyl-CoA Thioesterase 2 |
| ACOX2 | Acyl-CoA Oxidase 2 |
| ACOX3 | Acyl-CoA Oxidase 3 |
| ACSL4 | Acyl-CoA Synthetase Long Chain Family Member 4 |
| ACSM5 | Acyl-CoA Synthetase Medium Chain Family Member 5 |
| ACTB | Actin Beta |
| AdipoQ | Adiponectin |
| C/EBP | CCAAT/Enhancer Binding Protein |
| CD74 | CD74 Molecule |
| CPT1B | Carnitine Palmitoyltransferase 1B |
| CPT2 | Carnitine Palmitoyltransferase 2 |
| CXCL10 | C-X-C Motif Chemokine Ligand 10 |
| DGAT2 | Diacylglycerol O-Acyltransferase 2 |
| EBPL | Emopamil Binding Protein Like |
| EGR1 | Early Growth Response 1 |
| EPHX2 | Epoxide Hydrolase 2 |
| FABPs | Fatty Acid Binding Proteins |
| FASN | Fatty Acid Synthase |
| FATP1 | Fatty Acid Transport Protein 1 |
| GK | Glycerol Kinase |
| GPAM | Glycerol-3-Phosphate Acyltransferase |
| HK2 | Hexokinase 2 |
| HRG | Heregulin |
| IGFBP1 | Insulin Like Growth Factor Binding Protein 1 |
| IRF7 | Interferon Regulatory Factor 7 |
| ITGB1 | Integrin Subunit Beta 1 |
| LIPE | Lipase E, Hormone Sensitive Type |
| LPL | Lipoprotein Lipase |
| MC4R | Melanocortin 4 Receptor |
| MYF5 | Myogenic Factor 5 |
| NAIP | NLR Family Apoptosis Inhibitory Protein |
| NCAM1 | Neural Cell Adhesion Molecule 1 |
| PARP14 | Poly(ADP-Ribose) Polymerase 14 |
| PLAC8A | Placenta Specific 8A |
| PLTP | Phospholipid Transfer Protein |
| PPAR | Peroxisome Proliferator Activated Receptor |
| PTGS1 | Prostaglandin-Endoperoxide Synthase 1 |
| PTGS2 | Prostaglandin-Endoperoxide Synthase 2 |
| RAR | Retinoic Acid Receptor |
| RET | Ret Proto-Oncogene |
| REXO2 | RNA exonuclease 2 |
| SCD | Stearoyl-CoA Desaturase |
| SLC27A1 | Solute Carrier Family 27 Member 1 |
| SREBP | Sterol Regulatory Element Binding Protein |
References
- Fu, R.; Li, C.; Wang, L.; Gao, Z. Chinese consumer preference for beef with geographical indications and other attributes. Meat Sci. 2024, 212, 109475. [Google Scholar] [CrossRef]
- Halli, K.; Bohlouli, M.; Schulz, L.; Sundrum, A.; König, S. Estimation of direct and maternal genetic effects and annotation of potential candidate genes for weight and meat quality traits in a genotyped outdoor dual-purpose cattle breed. Transl. Anim. Sci. 2022, 6, txac022. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huang, J.; Wang, X.; Ma, Y. Transcription factors regulate adipocyte differentiation in beef cattle. Anim. Genet. 2020, 51, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.; Kaczmarska, K.; Paterson, J.; Piyasiri, U.; Warner, R. Effect of marbling on volatile generation, oral breakdown and in mouth flavor release of grilled beef. Meat Sci. 2017, 133, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.H.A.; Khan, R.; Abdelnour, S.A.; Abd El-Hack, M.E.; Khafaga, A.F.; Taha, A.; Ohran, H.; Mei, C.; Schreurs, N.M.; Zan, L. Advances of molecular markers and their application for body variables and carcass traits in Qinchuan cattle. Genes 2019, 10, 717. [Google Scholar] [CrossRef]
- Wheeler, T.L.; Cundiff, L.V.; Koch, R.M. Effect of marbling degree on beef palatability in Bos taurus and Bos indicus cattle. J. Anim. Sci. 1994, 72, 3145–3151. [Google Scholar] [CrossRef]
- Park, S.J.; Beak, S.H.; Jung, D.J.S.; Kim, S.Y.; Jeong, I.H.; Piao, M.Y.; Kang, H.J.; Fassah, D.M.; Na, S.W.; Yoo, S.P.; et al. Genetic, management, an5d nutritional factors affecting intramuscular fat deposition in beef cattle—A review. Asian-Australas. J. Anim. Sci. 2018, 31, 1043–1061. [Google Scholar] [CrossRef]
- Michal, J.J.; Zhang, Z.W.; Gaskins, C.T.; Jiang, Z. The bovine fatty acid binding protein 4 gene is significantly associated with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Anim. Genet. 2006, 37, 400–402. [Google Scholar] [CrossRef]
- Chen, J.N.; Jiang, Y.Z.; Cen, W.M.; Xing, S.H.; Zhu, L.; Tang, G.Q.; Li, M.Z.; Jiang, A.A.; Lou, P.E.; Wen, A.X.; et al. Distribution of H-FABP and ACSL4 gene polymorphisms and their associations with intramuscular fat content and backfat thickness in different pig populations. Genet. Mol. Res. 2014, 13, 6759–6772. [Google Scholar] [CrossRef]
- Morsci, N.S.; Schnabel, R.D.; Taylor, J.F. Association analysis of adiponectin and somatostatin polymorphisms on BTA1 with growth and carcass traits in Angus cattle. Anim. Genet. 2006, 37, 554–562. [Google Scholar] [CrossRef]
- Choi, Y.; Davis, M.E.; Chung, H. Effects of genetic variants in the promoter region of the bovine adiponectin (ADIPOQ) gene on marbling of Hanwoo beef cattle. Meat Sci. 2015, 105, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Albrecht, E.; Schering, L.; Kuehn, C.; Yang, R.; Zhao, Z.; Maak, S. Agouti signaling protein and its receptors as potential molecular markers for intramuscular and body fat deposition in cattle. Front. Physiol. 2018, 9, 172. [Google Scholar] [CrossRef]
- Wu, S.A.; Kersten, S.; Qi, L. Lipoprotein lipase and its regulators: An unfolding story. Trends Endocrinol. Metab. 2021, 32, 48–61. [Google Scholar] [CrossRef]
- Gao, Y.Y.; Cheng, G.; Cheng, Z.X.; Bao, C.; Yamada, T.; Cao, G.F.; Bao, S.Q.; Schreurs, N.M.; Zan, L.S.; Tong, B. Association of variants in FABP4, FASN, SCD, SREBP1 and TCAP genes with intramuscular fat, carcass traits and body size in Chinese Qinchuan cattle. Meat Sci. 2022, 192, 108882. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Yang, R.; Lu, C.; Qiu, Z.; Yan, C.; Zhao, Z. Differential expression of PPARγ, FASN, and ACADM genes in various adipose tissues and longissimus dorsi muscle from Yanbian yellow cattle and Yan yellow cattle. Asian-Australas. J. Anim. Sci. 2014, 27, 10–18. [Google Scholar] [CrossRef]
- Moisá, S.J.; Shike, D.W.; Faulkner, D.B.; Meteer, W.T.; Keisler, D.; Loor, J.J. Central role of the PPARγ gene network in coordinating beef cattle intramuscular adipogenesis in response to weaning age and nutrition. Gene Regul. Syst. Biol. 2014, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Choy, L.; Derynck, R. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J. Biol. Chem. 2003, 278, 9609–9619. [Google Scholar] [CrossRef]
- Abebe, B.K.; Wang, H.; Li, A.; Zan, L. A review of the role of transcription factors in regulating adipogenesis and lipogenesis in beef cattle. J. Anim. Breed. Genet. 2024, 141, 235–256. [Google Scholar] [CrossRef]
- Carobbio, S.; Hagen, R.M.; Lelliott, C.J.; Slawik, M.; Medina-Gomez, G.; Tan, C.-Y.; Sicard, A.; Atherton, H.J.; Barbarroja, N.; Bjursell, M.; et al. Adaptive changes of the Insig1/SREBP1/SCD1 set point help adipose tissue to cope with increased storage demands of obesity. Diabetes 2013, 62, 3697–3708. [Google Scholar] [CrossRef]
- Xu, H.Y.; Shao, J.; Yin, B.Z.; Zhang, L.M.; Fang, J.C.; Zhang, J.S.; Xia, G.J. Bovine bta-microRNA-1271 promotes preadipocyte differentiation by targeting activation transcription factor 3. Biochemistry 2020, 85, 749–757. [Google Scholar] [CrossRef]
- Ma, X.; Wei, D.; Cheng, G.; Li, S.; Wang, L.; Wang, Y.; Wang, X.; Zhang, S.; Wang, H.; Zan, L. Bta-miR-130a/b regulates preadipocyte differentiation by targeting PPARG and CYP2U1 in beef cattle. Mol. Cell. Probes 2018, 42, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Raza, S.H.A.; Cheng, G.; Ma, X.; Wang, J.; Zan, L. Bta-miR-376a targeting KLF15 interferes with adipogenesis signaling pathway to promote differentiation of Qinchuan beef cattle preadipocytes. Animals 2020, 10, 2362. [Google Scholar] [CrossRef] [PubMed]
- Mir, B.A.; Albrecht, E.; Ali, A.; Hansson, O.; Maak, S. MicroRNA-100 reduced fetal bovine muscle satellite cell myogenesis and augmented intramuscular lipid deposition by modulating IGF1R. Cells 2022, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yu, S.; Guo, J.; Wang, J.; Mei, C.; Abbas Raza, S.H.; Cheng, G.; Zan, L. Comprehensive analysis of transcriptome and metabolome reveals regulatory mechanism of intramuscular fat content in beef cattle. J. Agric. Food Chem. 2024, 72, 2911–2924. [Google Scholar] [CrossRef]
- Wang, S.; Pan, C.; Sheng, H.; Yang, M.; Yang, C.; Feng, X.; Hu, C.; Ma, Y. Construction of a molecular regulatory network related to fat deposition by multi-tissue transcriptome sequencing of Jiaxian red cattle. iScience 2023, 26, 108346. [Google Scholar] [CrossRef]
- Greenwood, P.L.; Siddell, J.P.; Walmsley, B.J.; Geesink, G.H.; Pethick, D.W.; McPhee, M.J. Postweaning substitution of grazed forage with a high-energy concentrate has variable long-term effects on subcutaneous fat and marbling in Bos taurus genotypes. J. Anim. Sci. 2015, 93, 4132–4143. [Google Scholar] [CrossRef]
- Heeren, J.; Scheja, L. Metabolic-associated fatty liver disease and lipoprotein metabolism. Mol. Metab. 2021, 50, 101238. [Google Scholar] [CrossRef]
- Nguyen, P.; Leray, V.; Diez, M.; Serisier, S.; Bloc’h, J.L.; Siliart, B.; Dumon, H. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. 2008, 92, 272–283. [Google Scholar] [CrossRef]
- NRC. Nutrient Requirements of Beef Cattle, 8th rev. ed.; National Academies Press: Washington, DC, USA, 2016. [Google Scholar]
- Matsunari, H.; Watanabe, M.; Nakano, K.; Enosawa, S.; Umeyama, K.; Uchikura, A.; Yashima, S.; Fukuda, T.; Klymiuk, N.; Kurome, M.; et al. Modeling lethal X-linked genetic disorders in pigs with ensured fertility. Proc. Natl. Acad. Sci. USA 2018, 115, 708–713. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Zhang, X.; Wang, D.; Jin, G.; Li, B.; Xu, F.; Cheng, J.; Zhang, F.; Wu, S.; et al. The comprehensive liver transcriptome of two cattle breeds with different intramuscular fat content. Biochem. Biophys. Res. Commun. 2017, 490, 1018–1025. [Google Scholar] [CrossRef]
- Dehghanian Reyhan, V.; Ghafouri, F.; Sadeghi, M.; Miraei-Ashtiani, S.R.; Kastelic, J.P.; Barkema, H.W.; Shirali, M. Integrated Comparative transcriptome and circRNA-lncRNA-miRNA-mRNA ceRNA regulatory network analyses identify molecular mechanisms associated with intramuscular fat content in beef cattle. Animals 2023, 13, 2598. [Google Scholar] [CrossRef]
- Gao, K.; Han, S.; Li, Z.; Luo, Z.; Lv, S.; Choe, H.M.; Paek, H.J.; Quan, B.; Kang, J.; Yin, X. Analysis of metabolome and transcriptome of longissimus thoracis and subcutaneous adipose tissues reveals the regulatory mechanism of meat quality in MSTN mutant castrated male finishing pigs. Meat Sci. 2024, 207, 109370. [Google Scholar] [CrossRef]
- Costa, T.C.; Gionbelli, M.P.; Duarte, M.S. Fetal programming in ruminant animals: Understanding the skeletal muscle development to improve meat quality. Anim. Front. 2021, 11, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Owens, F.N.; Dubeski, P.; Hanson, C.F. Factors that alter the growth and development of ruminants. J. Anim. Sci. 1993, 71, 3138–3150. [Google Scholar] [CrossRef]
- Wang, Z.; An, X.; Yang, Y.; Zhang, L.; Jiao, T.; Zhao, S. Comprehensive analysis of the longissimus dorsi transcriptome and metabolome reveals the regulatory mechanism of different varieties of meat quality. J. Agric. Food Chem. 2023, 71, 1234–1245. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, F.; Li, R.; Li, L.; Ren, M.; Jin, M.; Zhou, J.; Wang, C.; Li, S. Integrated 4D analysis of intramuscular fat deposition: Quantitative proteomic and transcriptomic studies in Wannanhua pig longissimus dorsi muscle. Animals 2024, 14, 167. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wang, L.; Wang, S.; Chen, R.; Zhang, T.; Ma, H.; Lu, H.; Yuan, G. Transcriptomic analysis of thigh muscle of Lueyang black-bone chicken in free-range and caged feeding. Anim. Biotechnol. 2023, 34, 785–795. [Google Scholar] [CrossRef]
- Lim, D.; Chai, H.-H.; Lee, S.-H.; Cho, Y.-M.; Choi, J.-W.; Kim, N.-K. Gene expression patterns associated with peroxisome proliferator-activated receptor (PPAR) signaling in the longissimus dorsi of Hanwoo (Korean cattle). Asian-Australas. J. Anim. Sci. 2015, 28, 1075–1083. [Google Scholar] [CrossRef]
- Liu, X.; Li, S.; Wang, L.; Zhang, W.; Wang, Y.; Gui, L.; Zan, L.; Zhao, C. The effect of FATP1 on adipocyte differentiation in Qinchuan beef cattle. Animals 2021, 11, 2789. [Google Scholar] [CrossRef]
- Zhao, Z.D.; Li, A.N.; Wei, S.J.; Wang, M.M.; Li, S.J.; Zan, L.S. Genetic polymorphisms of the FATP1 gene and their associations with meat quality traits in Chinese Qinchuan cattle. Genet. Mol. Res. 2015, 14, 17439–17446. [Google Scholar] [CrossRef]
- Yang, X.; Ma, X.; Mei, C.; Zan, L. A genome-wide landscape of mRNAs, lncRNAs, circRNAs and miRNAs during intramuscular adipogenesis in cattle. BMC Genom. 2022, 23, 691. [Google Scholar] [CrossRef]
- Wanders, R.J.; Vreken, P.; Ferdinandusse, S.; Jansen, G.A.; Waterham, H.R.; van Roermund, C.W.; Van Grunsven, E.G. Peroxisomal fatty acid alpha- and beta-oxidation in humans: Enzymology, peroxisomal metabolite transporters and peroxisomal diseases. Biochem. Soc. Trans. 2001, 29 Pt 2, 250–267. [Google Scholar] [CrossRef]
- Zhang, M.; Zha, X.; Ma, X.; La, Y.; Guo, X.; Chu, M.; Bao, P.; Yan, P.; Wu, X.; Liang, C. Genome-wide transcriptome profiling reveals the mechanisms underlying hepatic metabolism under different raising systems in yak. Animals 2024, 14, 695. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, J.; Zhang, K.; Cheng, G.; Mei, C.; Zan, L. Integrated multi-omics analysis reveals variation in intramuscular fat among muscle locations of Qinchuan cattle. BMC Genom. 2023, 24, 367. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cui, H.; Fu, R.; Zheng, M.; Liu, R.; Zhao, G.; Wen, J. The regulation of IMF deposition in pectoralis major of fast- and slow-growing chickens at hatching. J. Anim. Sci. Biotechnol. 2017, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- López-Viñas, E.; Bentebibel, A.; Gurunathan, C.; Morillas, M.; de Arriaga, D.; Serra, D.; Asins, G.; Hegardt, F.G.; Gómez-Puertas, P. Definition by functional and structural analysis of two malonyl-CoA sites in carnitine palmitoyltransferase 1A. J. Biol. Chem. 2007, 282, 18212–18224. [Google Scholar] [CrossRef]
- Chen, W.; Xiao, Y.; Yang, F.; Liufu, S.; Gong, Y.; Li, Z.; Zhang, S.; Tang, S.; Li, B.; Ma, H. Integrated analysis of muscle transcriptome, miRNA, and proteome of Chinese indigenous breed Ningxiang pig in three developmental stages. Front. Genet. 2024, 15, 1393834. [Google Scholar] [CrossRef]
- Houten, S.M.; Wanders, R.J. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 2010, 33, 469–477. [Google Scholar] [CrossRef]
- Li, A.; Li, Y.; Wang, Y.; Wang, Y.; Li, X.; Qubi, W.; Xiong, Y.; Zhu, J.; Liu, W.; Lin, Y. ACADL Promotes the differentiation of goat intramuscular adipocytes. Animals 2023, 13, 281. [Google Scholar] [CrossRef]
- Chang, C.C.; Sakashita, N.; Ornvold, K.; Lee, O.; Chang, E.T.; Dong, R.; Lin, S.; Lee, C.Y.; Strom, S.C.; Kashyap, R.; et al. Immunological quantitation and localization of ACAT-1 and ACAT-2 in human liver and small intestine. J. Biol. Chem. 2000, 275, 28083–28092. [Google Scholar] [CrossRef]
- Silva-Vignato, B.; Coutinho, L.L.; Poleti, M.D.; Cesar, A.S.M.; Moncau, C.T.; Regitano, L.C.A.; Balieiro, J.C.C. Gene co-expression networks associated with carcass traits reveal new pathways for muscle and fat deposition in Nelore cattle. BMC Genom. 2019, 20, 32. [Google Scholar] [CrossRef] [PubMed]
- Kiema, T.R.; Harijan, R.K.; Strozyk, M.; Fukao, T.; Alexson, S.E.; Wierenga, R.K. The crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase (T1): Insight into the reaction mechanism of its thiolase and thioesterase activities. Acta Crystallogr. D Biol. Crystallogr. 2014, 70 Pt 12, 3212–3225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Wang, X.; Ji, Y.; Cheng, S.; Wang, M.; Zhang, C.; Yu, X.; Zhao, R.; Zhang, W.; et al. Acetyl-coenzyme A acyltransferase 2 promote the differentiation of sheep precursor adipocytes into adipocytes. J. Cell. Biochem. 2019, 120, 8021–8031. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wang, H.; Ma, X.; Zan, L. MicroRNA-224 impairs adipogenic differentiation of bovine preadipocytes by targeting LPL. Mol. Cell. Probes 2019, 44, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Criado-Mesas, L.; Ballester, M.; Crespo-Piazuelo, D.; Castelló, A.; Fernández, A.I.; Folch, J.M. Identification of eQTLs associated with lipid metabolism in Longissimus dorsi muscle of pigs with different genetic backgrounds. Sci. Rep. 2020, 10, 9845. [Google Scholar] [CrossRef]
- Cui, H.; Wang, Y.; Zhu, Y.; Liu, X.; Liu, L.; Wang, J.; Tan, X.; Wang, Y.; Xing, S.; Luo, N.; et al. Genomic insights into the contribution of de novo lipogenesis to intramuscular fat deposition in chicken. J. Adv. Res. 2024, 65, 19–31. [Google Scholar] [CrossRef]

| Traits | High IMF Group | Low IMF Group | p |
|---|---|---|---|
| Carcass weight/kg | 421.50 ± 44.16 | 425.00 ± 55.24 | 0.924 |
| IMF/% | 29.02 ± 6.28 | 11.70 ± 1.10 | 0.010 |
| Fat color | 2.00 ± 0.00 | 2.50 ± 0.58 | 0.134 |
| Meat color | 2.00 ± 0.00 | 3.00 ± 0.00 | - |
| Marbling level | 4.25 ± 0.50 | 1.75 ± 0.50 | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Liu, T.; Peng, P.; Fu, Y.; Shi, S.; Liang, S.; Chen, X.; Wang, K.; Zhou, R. Integrated Transcriptomic Analysis of Liver and Muscle Tissues Reveals Candidate Genes and Pathways Regulating Intramuscular Fat Deposition in Beef Cattle. Animals 2025, 15, 1306. https://doi.org/10.3390/ani15091306
Wang S, Liu T, Peng P, Fu Y, Shi S, Liang S, Chen X, Wang K, Zhou R. Integrated Transcriptomic Analysis of Liver and Muscle Tissues Reveals Candidate Genes and Pathways Regulating Intramuscular Fat Deposition in Beef Cattle. Animals. 2025; 15(9):1306. https://doi.org/10.3390/ani15091306
Chicago/Turabian StyleWang, Siwei, Tingting Liu, Peng Peng, Yurong Fu, Shaoqing Shi, Shuang Liang, Xi Chen, Kun Wang, and Rongyan Zhou. 2025. "Integrated Transcriptomic Analysis of Liver and Muscle Tissues Reveals Candidate Genes and Pathways Regulating Intramuscular Fat Deposition in Beef Cattle" Animals 15, no. 9: 1306. https://doi.org/10.3390/ani15091306
APA StyleWang, S., Liu, T., Peng, P., Fu, Y., Shi, S., Liang, S., Chen, X., Wang, K., & Zhou, R. (2025). Integrated Transcriptomic Analysis of Liver and Muscle Tissues Reveals Candidate Genes and Pathways Regulating Intramuscular Fat Deposition in Beef Cattle. Animals, 15(9), 1306. https://doi.org/10.3390/ani15091306






