Characterization of the Complete Mitochondrial Genome of Angulyagra polyzonata and Its Phylogenetic Status in Viviparidae
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials and DNA Extraction
2.2. Mitochondrial Genome Sequencing and Assembly
2.3. Mitochondrial Genome Structure Annotation and Analysis
2.4. Analysis of Taxonomic Family Development
3. Results and Discussion
3.1. Mitochondrial Structural Characteristics
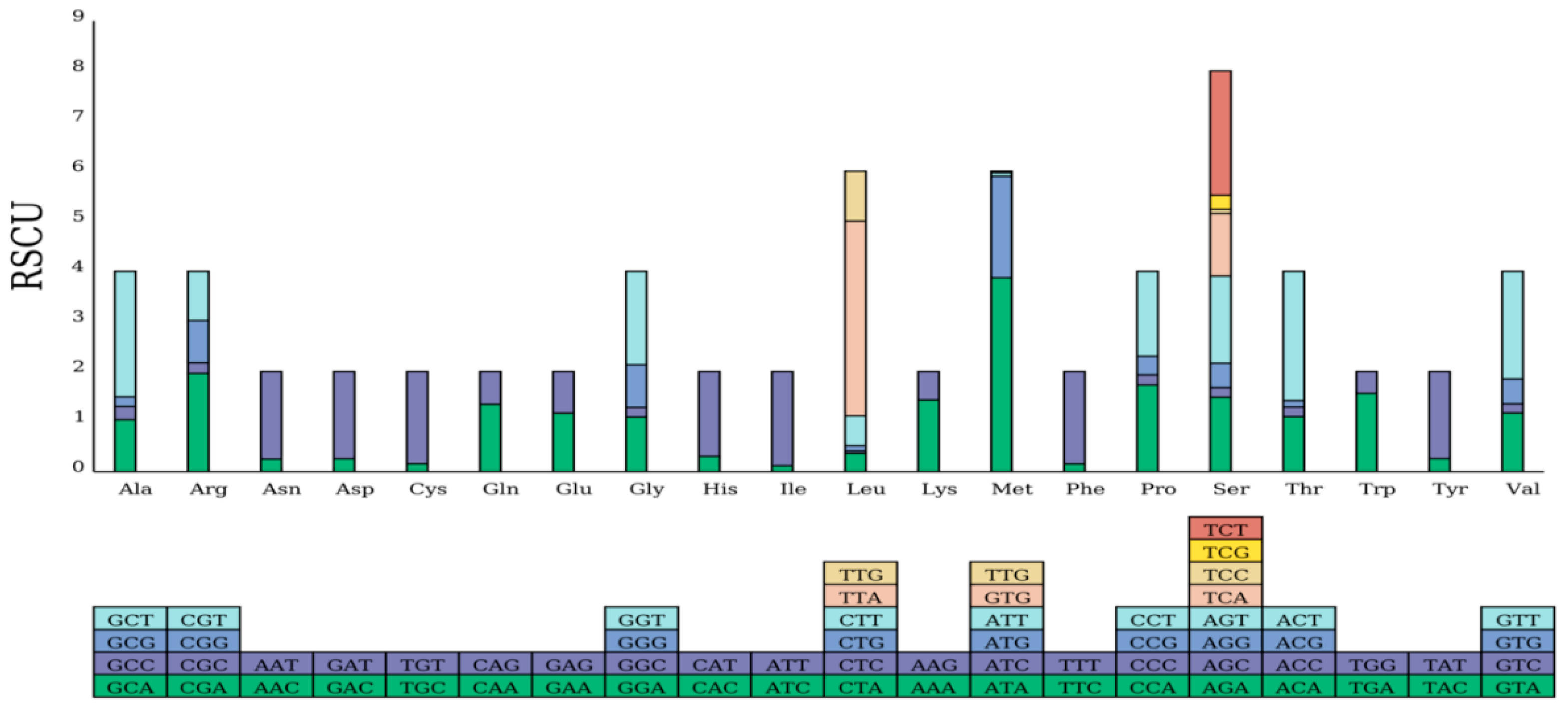
3.2. PCGs in the A. polyzonata Mitochondrial Genome
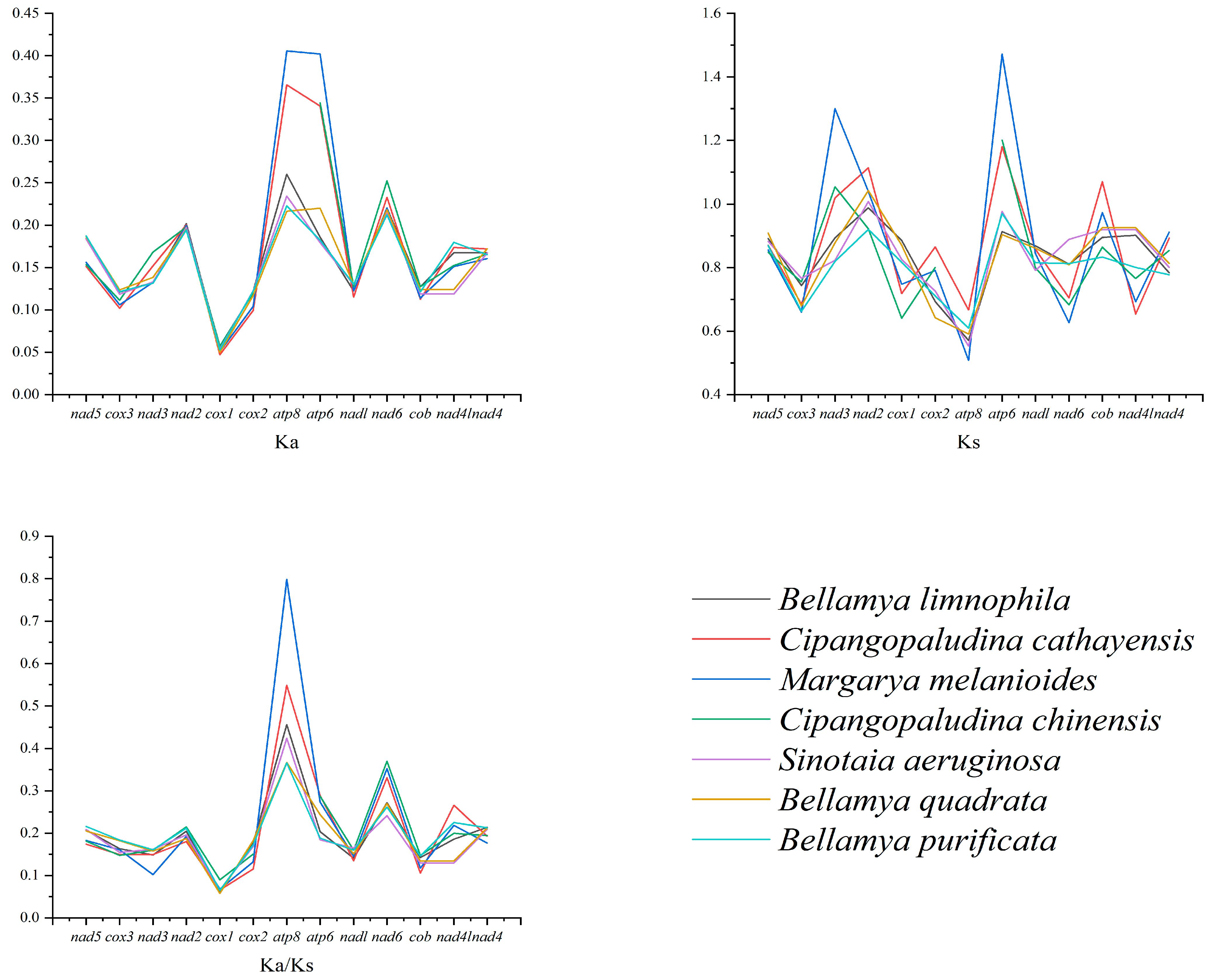
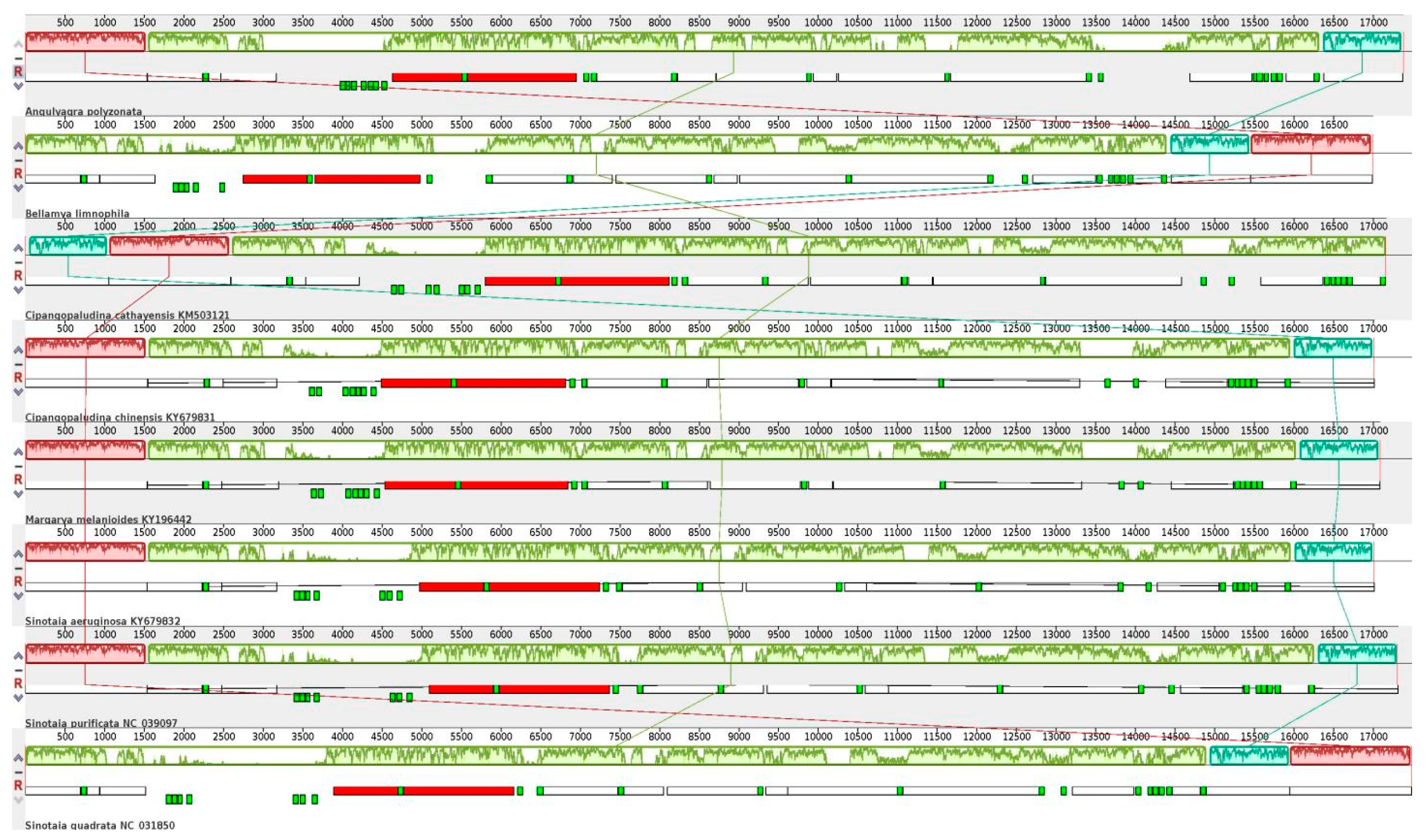
3.3. Comparative Analysis of Viviparidae Mitochondrial Genomes
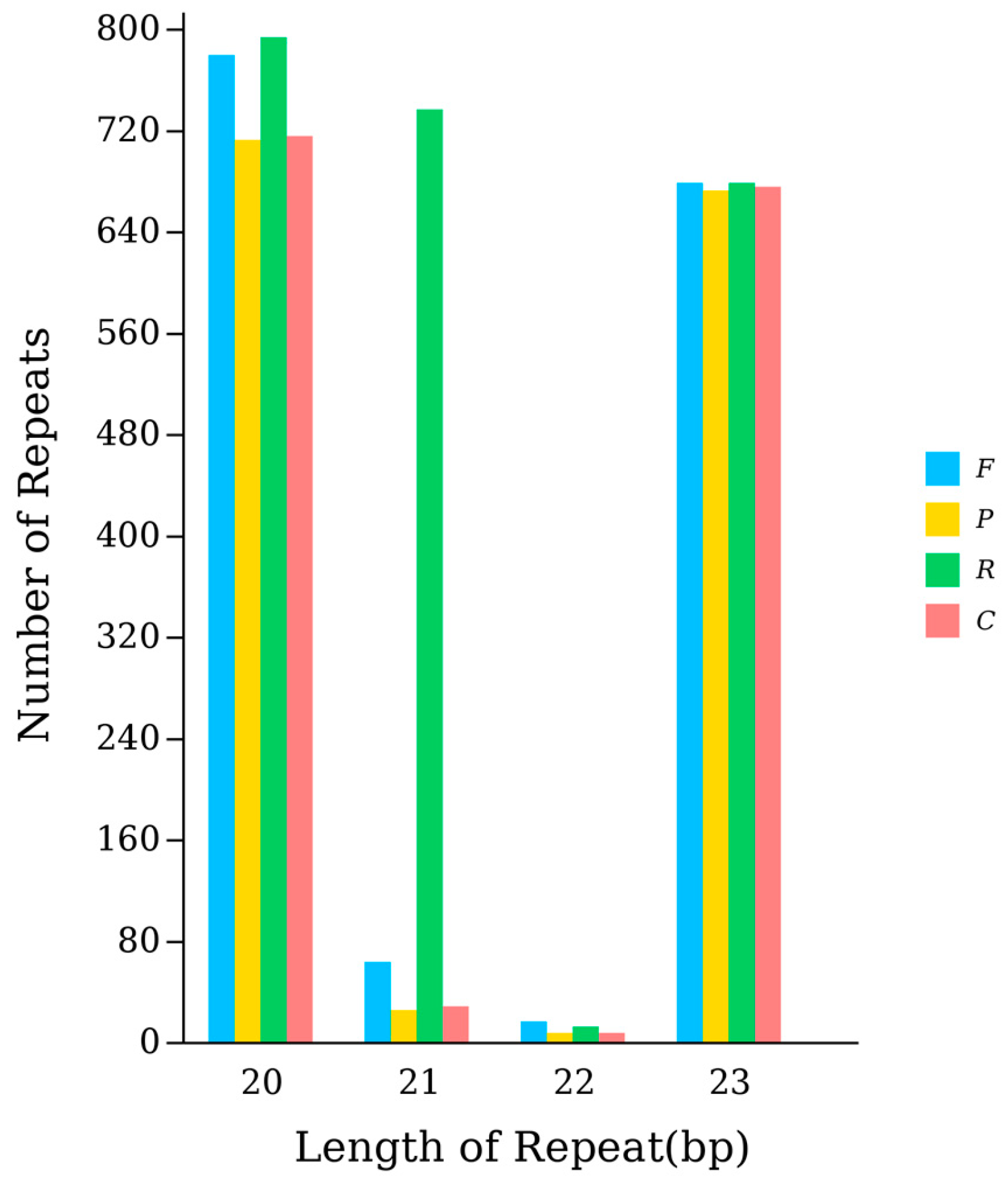
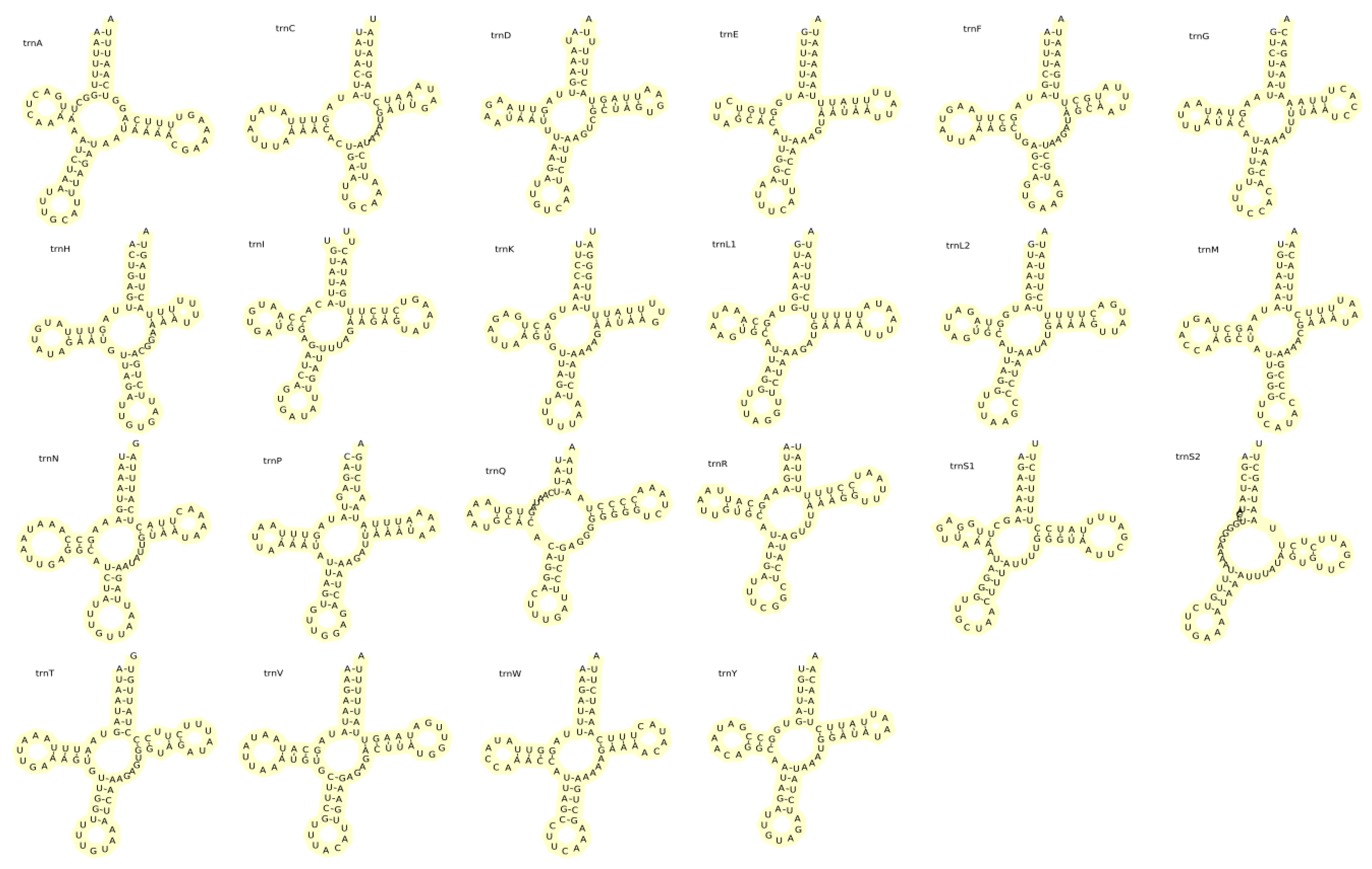
3.4. rRNA and tRNA Genes in the A. polyzonata Mitochondrial Genome
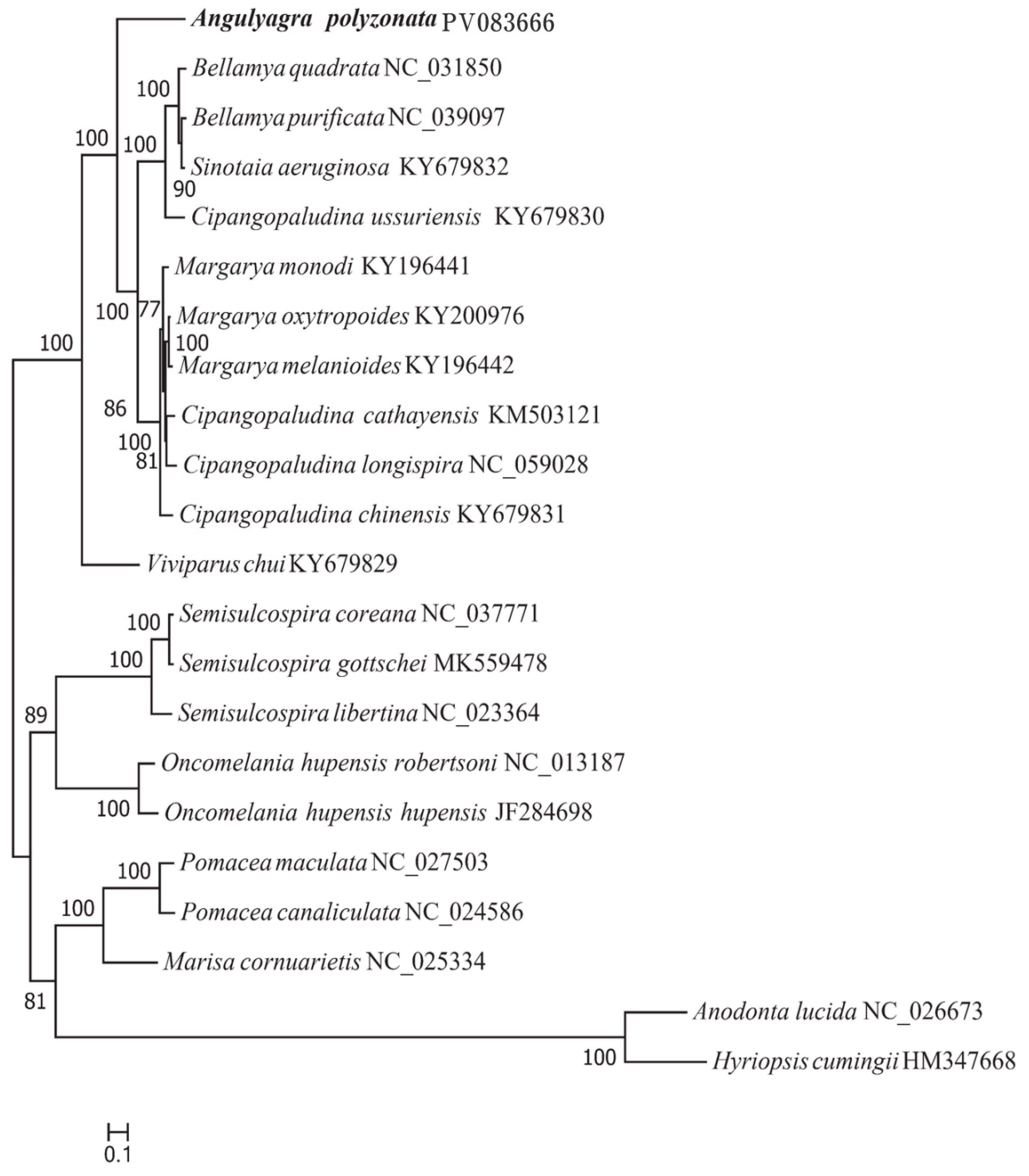
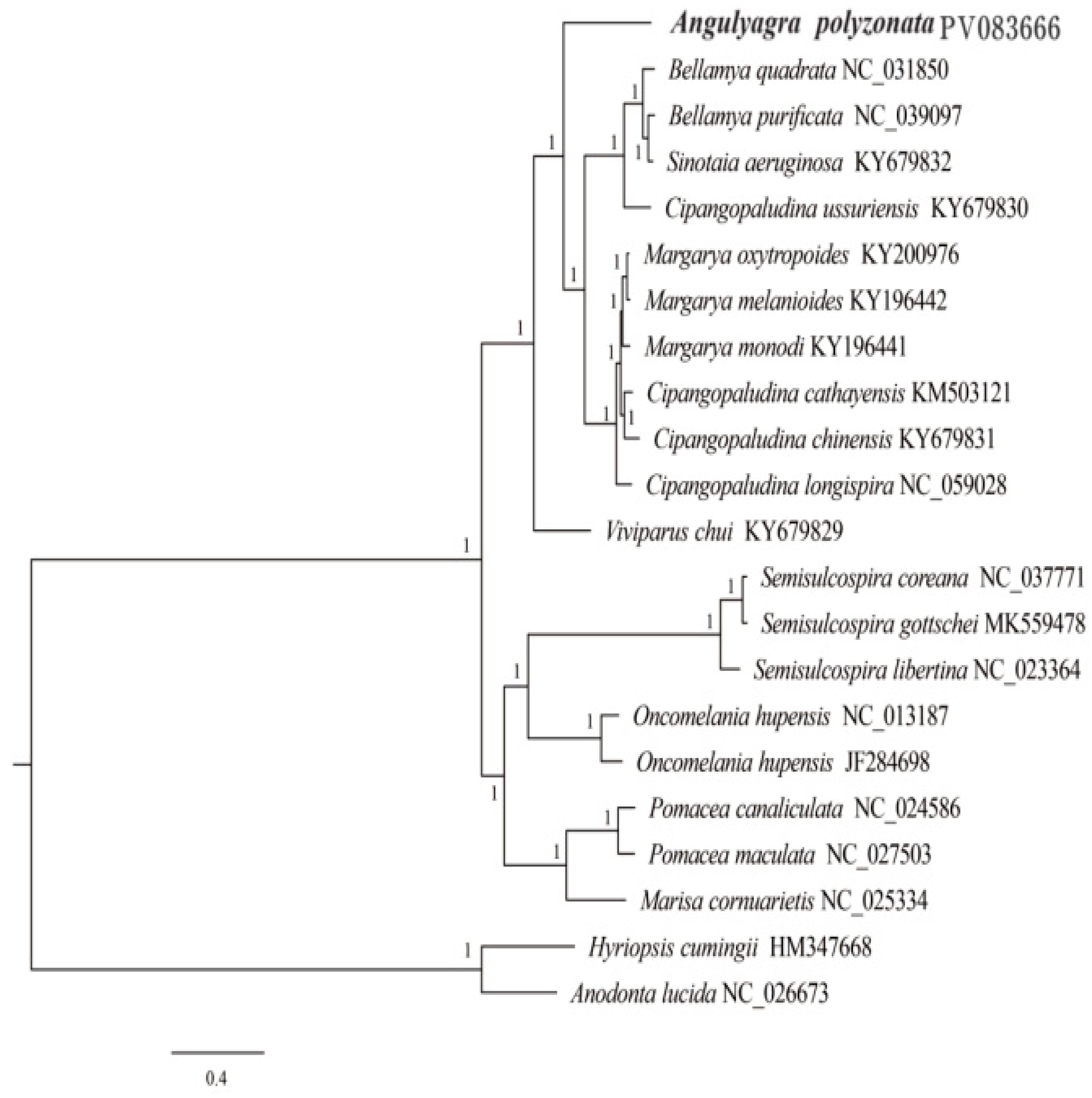
3.5. Phylogenetic Relationship
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liao, Y.; Qin, J. Development status and countermeasures of snail aquaculture in Guangxi. China Fish. 2023, 10, 32–34. [Google Scholar]
- Cheng, L. Research on Inheritance and Development of Luosifen in Liuzhou from the Perspective of Intangible Cultural Heritage. Master’s Thesis, Guangxi University for Nationalities, Nanning, China, 2023. [Google Scholar]
- Sil, M.; Aravind, N.; Karanth, K.P. Role of geography and climatic oscillations in governing into-India dispersal of freshwater snails of the family: Viviparidae. Mol. Phylogenet. Evol. 2019, 138, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Strong, E.E.; Gargominy, O.; Ponder, W.F.; Bouchet, P. Global diversity of gastropods (Gastropoda; Mollusca) in freshwater. Freshw. Anim. Divers. Assess. 2008, 595, 149–166. [Google Scholar]
- Liu, Y.; Zhang, W.; Wang, Y. Distribution of the family Viviparidae from China (Mollusca: Gastropoda). Trans. Chin. Soc. Malacol. 1995, 5–6, 8–16. [Google Scholar]
- Wang, J.; Zhang, D.; Jakovlić, I.; Wang, W. Sequencing of the complete mitochondrial genomes of eight freshwater snail species exposes pervasive paraphyly within the Viviparidae family (Caenogastropoda). PLoS ONE 2017, 12, e0181699. [Google Scholar] [CrossRef]
- Du, L.; Yang, J.; Rintelen, T.v.; Chen, X.; Aldridge, D. Molecular phylogenetic evidence that the Chinese viviparid genus Margarya (Gastropoda: Viviparidae) is polyphyletic. Chin. Sci. Bull. 2013, 58, 2154–2162. [Google Scholar] [CrossRef]
- Hirano, T.; Saito, T.; Chiba, S. Phylogeny of freshwater viviparid snails in Japan. J. Molluscan Stud. 2015, 81, 435–441. [Google Scholar] [CrossRef]
- Huang, X.; Wang, L. Analysis of Cytochrome COxidaseI genes of Five Spesies of Margarya and Cipangopaludina chinensis. J. Hydroecol. 2008, 29, 106–108. [Google Scholar]
- Sengupta, M.E.; Kristensen, T.K.; Madsen, H.; Jørgensen, A. Molecular phylogenetic investigations of the Viviparidae (Gastropoda: Caenogastropoda) in the lakes of the Rift Valley area of Africa. Mol. Phylogenet. Evol. 2009, 52, 797–805. [Google Scholar] [CrossRef]
- Tian, M.; Fan, B.; Yang, Y.; Fang, F.; Wang, W.; Chen, Y. Studies On Molecular Phylogeny of Margarya melanioides (Gastropoda:Viviparidae). J. Kunming Med. Univ. 2015, 36, 4–8. [Google Scholar]
- Dzung, D.T.; i anh Hoa, N.; Phu, N.T.; iu Ha, N. Response of Freshwater Snail (Angulyagra polyzonata) as Water Biomarker by Heavy Metals (Cd, Cu, Zn, Pb). Vietnam. Acad. Agric. Sci. 2016, 1, 126–130. [Google Scholar]
- Hung, N.M.; The, D.T.; Stauffer, J.R., Jr.; Madsen, H. Feeding behavior of black carp Mylopharyngodon piceus (Pisces: Cyprinidae) on fry of other fish species and trematode transmitting snail species. Biol. Control 2014, 72, 118–124. [Google Scholar] [CrossRef]
- Dung, B.T.; Madsen, H.; The, D.T. Distribution of freshwater snails in family-based VAC ponds and associated waterbodies with special reference to intermediate hosts of fish-borne zoonotic trematodes in Nam Dinh Province, Vietnam. Acta Trop. 2010, 116, 15–23. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, Y.; Tang, J.; Ji, Q.; Wang, J.; Tang, B.; Wang, G. Genome investigation and SSR characteristics analysis of 4 representative shellfish of Viviparidae. Jiangsu Agric. Sci. 2023, 51, 40–47. [Google Scholar]
- Chen, Q.; He, W.; Liu, Y.; Xu, J.; Huang, J. Characteristics of macrozoobenthic community structure in typical wetlands of Macao. South China Fish. Sci. 2015, 11, 1–10. [Google Scholar]
- Nass, M.M.; Nass, S. Intramitochondrial fibers with DNA characteristics I. Fixation and electron staining reactions. J. Cell Biol. 1963, 19, 593–611. [Google Scholar] [CrossRef]
- Brown, W.M.; Vinograd, J. Restriction endonuclease cleavage maps of animal mitochondrial DNAs. Proc. Natl. Acad. Sci. USA 1974, 71, 4617–4621. [Google Scholar] [CrossRef]
- HsuChen, C.-C.; Cleaves, G.R.; Dubin, D.T. A major lysine tRNA with a CUU anticodon in insect mitochondria. Nucleic Acids Res. 1983, 11, 8659–8662. [Google Scholar] [CrossRef]
- Tatarenkov, A.; Avise, J.C. Rapid concerted evolution in animal mitochondrial DNA. Proc. R. Soc. B Biol. Sci. 2007, 274, 1795–1798. [Google Scholar] [CrossRef]
- Pan, B.; Bu, W. Research progress on inheritance and evolution of the mitochondrial genome. Chin. J. Biol. 2005, 40, 1–3. [Google Scholar]
- Lo Presti, R.; Lisa, C.; Di Stasio, L. Molecular genetics in aquaculture. Ital. J. Anim. Sci. 2009, 8, 299–313. [Google Scholar] [CrossRef]
- Li, K.; Zhang, S.; Gu, Y.; Wang, J.; Yang, Y.; Mao, W. Transcriptomic data of BT549 triple negative breast cancer cells treated with 20 µM NU7441, a DNA-dependent kinase inhibitor. Data Brief 2024, 53, 110183. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Salmela, L.; Rivals, E. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 2014, 30, 3506–3514. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Y.; Yin, C.; Wang, J.; Chen, Y. Characterization of the complete mitochondrial genome of Neolissochilus hendersoni (Herre, 1940) (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B 2023, 8, 133–135. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 1–7. [Google Scholar] [CrossRef]
- Orešič, M.; Shalloway, D. Specific correlations between relative synonymous codon usage and protein secondary structure. J. Mol. Biol. 1998, 281, 31–48. [Google Scholar] [CrossRef]
- Ke, Z.; Zhou, K.; Hou, M.; Luo, H.; Li, Z.; Pan, X.; Zhou, J.; Jing, T.; Ye, H. Characterization of the complete mitochondrial genome of the elongate loach and its phylogenetic implications in Cobitidae. Animals 2023, 13, 3841. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, A.; Ambrosio, L.; Di Martino, A.; Knijn, A.; De Sabato, L.; Vaccari, G.; Di Bartolo, I.; Morabito, S.; Palamara, A.T.; Stefanelli, P.; et al. Selective Pressure and Evolution of SARS-CoV-2 Lineages BF. 7 and BQ. 1.1 Circulating in Italy from July to December 2022. Microorganisms 2024, 12, 908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, J.; Long, M.; Fan, C. gKaKs: The pipeline for genome-level Ka/Ks calculation. Bioinformatics 2013, 29, 645–646. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Aberer, A.J.; Goll, C.; Smith, S.A.; Berger, S.A.; Izquierdo-Carrasco, F. RAxML-Light: A tool for computing terabyte phylogenies. Bioinformatics 2012, 28, 2064–2066. [Google Scholar] [CrossRef]
- Rempel, E.M.; Marcus, J.M.; Detwiler, J.T. The complete mitochondrial genome of the file ramshorn snail Planorbella pilsbryi (Mollusca: Gastropoda: Hygrophila: Planorbidae). Mitochondrial DNA Part B 2021, 6, 3181–3183. [Google Scholar] [CrossRef]
- Dong, H.; Yuan, H.; Yang, X.; Shan, W.; Zhou, Q.; Tao, F.; Zhao, C.; Bai, J.; Li, X.; Ma, Y.; et al. Phylogenetic analysis of some species of the Anopheles Hyrcanus Group (Diptera: Culicidae) in China based on complete mitochondrial genomes. Genes 2023, 14, 1453. [Google Scholar] [CrossRef]
- Xu, Y.; Zeng, S.; Meng, Y.; Yang, D.; Yang, S. The mitochondrial genome of Huaaristarchorum (Heude, 1889)(Gastropoda, Cerithioidea, Semisulcospiridae) and its phylogenetic implications. ZooKeys 2024, 1192, 237. [Google Scholar] [CrossRef]
- Wu, M.-M.; Cheng, H.-Z.; Li, L.-L.; Xie, G.-L. The complete mitochondrial genome of the freshwater snail Cipangopaludina ampullacea (Küster, 1852)(Gastropoda: Viviparidae). Mitochondrial DNA Part B 2022, 7, 1599–1601. [Google Scholar] [CrossRef]
- Yin, N.; Zhao, S.; Huang, X.-C.; Ouyang, S.; Wu, X.-P. Complete mitochondrial genome of the freshwater snail Tarebia granifera (Lamarck, 1816)(Gastropoda: Cerithioidea: Thiaridae). Mitochondrial DNA Part B 2022, 7, 259–261. [Google Scholar] [CrossRef]
- Nasu, K.; Yokoyama, Y.; Sun, Y.; Suzuki-Matsubara, M.; Teramoto, T.; Moriyama, A.; Kawase, M.; Kumazawa, Y. Mitochondrial genome of Cipangopaludina japonica (Gastropoda: Viviparidae) with a tRNA gene rearrangement. Mitochondrial DNA Part B 2020, 5, 1340–1341. [Google Scholar] [CrossRef]
- Hui, M.; Zhang, Y.; Wang, A.; Sha, Z. The First Genome Survey of the Snail Provanna glabra Inhabiting Deep-Sea Hydrothermal Vents. Animals 2023, 13, 3313. [Google Scholar] [CrossRef] [PubMed]
- Kartavtsev, Y.P.; Masalkova, N.A. Structure, Evolution, and Mitochondrial Genome Analysis of Mussel Species (Bivalvia, Mytilidae). Int. J. Mol. Sci. 2024, 25, 6902. [Google Scholar] [CrossRef] [PubMed]
- Kartavtsev, Y.P. Some examples of the use of molecular markers for needs of basic biology and modern society. Animals 2021, 11, 1473. [Google Scholar] [CrossRef] [PubMed]
- Redin, A.D.; Kartavtsev, Y.P. The Mitogenome Structure of Righteye Flounders (Pleuronectidae): Molecular Phylogeny and Systematics of the Family in East Asia. Diversity 2022, 14, 805. [Google Scholar] [CrossRef]
- Majumdar, C.; Demir, M.; Merrill, S.R.; Hashemian, M.; David, S.S. FSHing for DNA Damage: Key Features of MutY Detection of 8-Oxoguanine: Adenine Mismatches. Acc. Chem. Res. 2024, 57, 1019–1031. [Google Scholar] [CrossRef]
- Guo, F.-B.; Yu, X.-J. Separate base usages of genes located on the leading and lagging strands in Chlamydia muridarum revealed by the Z curve method. BMC Genom. 2007, 8, 1–8. [Google Scholar] [CrossRef]
- Ouyang, B.; Li, Y.; Wang, J.; Wei, Z.; Shi, A. The Complete Mitochondrial Genomes of Penthe kochi (Coleoptera: Tetratomidae) with Its Phylogenetic Implications. Curr. Issues Mol. Biol. 2024, 46, 10795–10805. [Google Scholar] [CrossRef]
- Consuegra, S.; John, E.; Verspoor, E.; de Leaniz, C.G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. 2015, 47, 1–10. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Kundu, S.; Kim, H.-W.; Lee, J.; Chung, S.; Lee, S.R.; Gietbong, F.Z.; Wibowo, A.; Kang, K. Mitogenomic architecture and phylogenetic relationship of European barracuda, Sphyraena sphyraena (Teleostei: Sphyraenidae) from the Atlantic Ocean. Fishes 2023, 8, 573. [Google Scholar] [CrossRef]
- Patil, M.P.; Kim, J.-O.; Yoo, S.H.; Seo, Y.B.; Lee, Y.-J.; Kim, J.-K.; Kitamura, S.-I.; Kim, G.-D. Complete mitogenome and phylogenetic analysis of a marine ray-finned fish, Alcichthys elongatus (Perciformes: Cottidae). Fishes 2023, 8, 513. [Google Scholar] [CrossRef]
- Zhang, K.; Zhu, K.; Liu, Y.; Zhang, H.; Gong, L.; Jiang, L.; Liu, L.; Lü, Z.; Liu, B. Novel gene rearrangement in the mitochondrial genome of Muraenesox cinereus and the phylogenetic relationship of Anguilliformes. Sci. Rep. 2021, 11, 2411. [Google Scholar] [CrossRef] [PubMed]
- Garey, J.R.; Wolstenholme, D.R. Platyhelminth mitochondrial DNA: Evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J. Mol. Evol. 1989, 28, 374–387. [Google Scholar] [CrossRef]
- Iriarte, A.; Lamolle, G.; Musto, H. Codon usage bias: An endless tale. J. Mol. Evol. 2021, 89, 589–593. [Google Scholar] [CrossRef]
- Novembre, J.A. Accounting for background nucleotide composition when measuring codon usage bias. Mol. Biol. Evol. 2002, 19, 1390–1394. [Google Scholar] [CrossRef]
- Rao, Y.; Wu, G.; Wang, Z.; Chai, X.; Nie, Q.; Zhang, X. Mutation bias is the driving force of codon usage in the Gallus gallus genome. DNA Res. 2011, 18, 499–512. [Google Scholar] [CrossRef]
- Salim, H.M.; Cavalcanti, A.R. Factors influencing codon usage bias in genomes. J. Braz. Chem. Soc. 2008, 19, 257–262. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.-S.; Wang, J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef]
- Cvijović, I.; Good, B.H.; Desai, M.M. The effect of strong purifying selection on genetic diversity. Genetics 2018, 209, 1235–1278. [Google Scholar] [CrossRef]
- Shah, P.; McCandlish, D.M.; Plotkin, J.B. Contingency and entrenchment in protein evolution under purifying selection. Proc. Natl. Acad. Sci. USA 2015, 112, E3226–E3235. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, W.; Li, Y.; Wang, J.; Bai, Y.; Liu, H. The Complete Mitogenomes of Two Species of Snakehead Fish (Perciformes: Channidae): Genome Characterization and Phylogenetic Analysis. Diversity 2024, 16, 346. [Google Scholar] [CrossRef]
- Baeza, J.A. The complete mitochondrial genome of the Caribbean spiny lobster Panulirus argus. Sci. Rep. 2018, 8, 17690. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, T.A.; MacInnis, M.J.; Bieler, R.; Boore, J.L.; Collins, T.M. Sessile snails, dynamic genomes: Gene rearrangements within the mitochondrial genome of a family of caenogastropod molluscs. BMC Genom. 2010, 11, 1–24. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, J.-E.; Xia, J.; Yang, J.; Guo, J.; Deng, Z.; Luo, M. Comparative characterization of the complete mitochondrial genomes of the three apple snails (Gastropoda: Ampullariidae) and the phylogenetic analyses. Int. J. Mol. Sci. 2018, 19, 3646. [Google Scholar] [CrossRef]
- Tang, Y.; Ma, W.; Chen, X.; Nie, G.; Zhou, C. Four new complete mitochondrial genomes of Gobioninae fishes (Teleostei: Cyprinidae) and their phylogenetic implications. PeerJ 2024, 12, e16632. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, T.; Yu, F. The New Mitochondrial Genome of Hemiculterella wui (Cypriniformes, Xenocyprididae): Sequence, Structure, and Phylogenetic Analyses. Genes 2023, 14, 2110. [Google Scholar] [CrossRef]
- Bravo, H.; Baeza, J.A.; van der Meij, S.E. Genomic survey sequencing and complete mitochondrial genome of the elkhorn coral crab Domecia acanthophora (Desbonne in Desbonne & Schramm, 1867)(Decapoda: Brachyura: Domeciidae). J. Crustac. Biol. 2023, 43, ruad046. [Google Scholar]
- Kilpert, F.; Podsiadlowski, L. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genom. 2006, 7, 1–18. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, X.; Li, Y.; Han, Z.; Xu, W.; Dong, J.; Wei, H.; Li, X. Mitochondrial genome of Chinese grass shrimp, Palaemonetes sinensis and comparison with other Palaemoninae species. Sci. Rep. 2019, 9, 17301. [Google Scholar] [CrossRef]
- Chen, H.; Yang, P. Progress of the Nuclear Acid with the Mismatched Base Pairs. Prog. Chem. 2002, 14, 133. [Google Scholar]
- Chen, L.; Lin, Y.; Xiao, Q.; Lin, Y.; Du, Y.; Lin, C.; Ward-Fear, G.; Hu, C.; Qu, Y.; Li, H. Characterization of the complete mitochondrial genome of the many-lined sun skink (Eutropis multifasciata) and comparison with other Scincomorpha species. Genomics 2021, 113, 2526–2536. [Google Scholar] [CrossRef]
- Lu, H.-F.; Du, L.-N.; Li, Z.-Q.; Chen, X.-Y.; Yang, J.-X. Morphological analysis of the Chinese Cipangopaludina species (Gastropoda; Caenogastropoda: Viviparidae). Zool. Res. 2014, 35, 510. [Google Scholar]


| Family | Genus | Species | Accession Number |
|---|---|---|---|
| Unionidae | Anodonta | Anodonta lucida | NC_026673 |
| Hyriopsis | Hyriopsis cumingii | HM347668 | |
| Ampullariidae | Pomacea | Pomacea maculata | NC_027503 |
| Pomacea canaliculata | NC_024586 | ||
| Marisa | Marisa cornuarietis | NC_025334 | |
| Semisulcospiridae | Semisulcospira | Semisulcospira coreana | NC_037771 |
| Semisulcospira gottschei | MK559478 | ||
| Semisulcospira libertina | NC 023364 | ||
| Hydrobiidae | Oncomelania Gredler | Oncomelania hupensis robertsoni | NC_013187 |
| Oncomelania hupensis hupensis | JF284698 | ||
| Viviparidae | Bellamya | Bellamya quadrata | NC_031850 |
| Bellamya purificata | NC _039097 | ||
| Sinotaia aeruginosa | KY679832 | ||
| Cipangopaludina | Cipangopaludina ussuriensis | KY679830 | |
| Cipangopaludina cathayensis | KM503121 | ||
| Cipangopaludina longispira | NC_059028 | ||
| Cipangopaludina chinensis | KY679831 | ||
| Viviparus chui | KY679829 | ||
| Margarya | Margarya monodi | KY196441 | |
| Margarya oxytropoides | KY200976 | ||
| Margarya melanioides | KY196442 | ||
| Angulyagra | Angulyagra polyzonata | PV083666 |
| Angulyagra_polyzonata | Size (bp) | A% | T% | G% | C% | A + T% | G + C% | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|---|
| Mitogenome | 17,379 | 31.24 | 43.27 | 16.04 | 9.45 | 74.51 | 25.49 | −0.161 | 0.259 |
| PCGs | 11,123 | 26.95 | 44.40 | 17.96 | 10.69 | 71.35 | 28.65 | −0.245 | 0.254 |
| tRNAs | 1484 | 35.85 | 37.74 | 16.11 | 10.30 | 73.59 | 26.41 | −0.026 | 0.219 |
| rRNAs | 2260 | 36.06 | 37.92 | 17.39 | 8.63 | 73.98 | 26.02 | −0.025 | 0.337 |
| Gene | Position | Intergenic Length | Codon | ||||
|---|---|---|---|---|---|---|---|
| Stand | From | To | Size | Start | Stop | ||
| cox1 | H | 1 | 1536 | 1536 | 0 | ATG | TAA |
| cox2 | H | 1545 | 2237 | 693 | 8 | ATG | TAA |
| trnD-GTC | H | 2244 | 2308 | 65 | 6 | / | / |
| atp8 | H | 2311 | 2466 | 156 | 2 | ATG | TAA |
| atp6 | H | 2469 | 3164 | 696 | 2 | ATG | TAA |
| trnM-CAT | L | 3973 | 4038 | 66 | 808 | / | / |
| trnY-GTA | L | 4041 | 4104 | 64 | 2 | / | / |
| trnC-GCA | L | 4114 | 4181 | 68 | 9 | / | / |
| trnG-TCC | L | 4240 | 4305 | 66 | 58 | / | / |
| trnW-TCA | L | 4324 | 4390 | 67 | 18 | / | / |
| trnQ-TTG | L | 4389 | 4454 | 66 | −2 | / | / |
| trnE-TTC | L | 4496 | 4561 | 66 | 41 | / | / |
| rrnS | H | 4631 | 5511 | 881 | 69 | / | / |
| trnV-TAC | H | 5506 | 5577 | 72 | −6 | / | / |
| rrnL | H | 5578 | 6956 | 1379 | 0 | / | / |
| trnL2-TAA | H | 7044 | 7110 | 67 | 87 | / | / |
| trnL1-TAG | H | 7139 | 7206 | 68 | 28 | / | / |
| nad1 | H | 7207 | 8148 | 942 | 0 | ATG | TAA |
| trnP-TGG | H | 8149 | 8214 | 66 | 0 | / | / |
| nad6 | H | 8228 | 8710 | 483 | 13 | ATT | TAG |
| cob | H | 8716 | 9853 | 1138 | 5 | ATG | T-- |
| trnS2-TGA | H | 9854 | 9917 | 64 | 0 | / | / |
| nad4l | H | 9936 | 10,232 | 297 | 18 | ATG | TAA |
| nad4 | H | 10,256 | 11,596 | 1341 | 23 | TGG | TAA |
| trnH-GTG | H | 11,598 | 11,661 | 64 | 1 | / | / |
| nad5 | H | 11,668 | 13,377 | 1710 | 6 | ATT | TAA |
| trnF-GAA | H | 13,383 | 13,450 | 68 | 5 | / | / |
| trnT-TGT | H | 13,531 | 13,601 | 71 | 80 | / | / |
| cox3 | H | 14,687 | 15,466 | 780 | 1085 | ATG | TAA |
| trnK-TTT | H | 15,485 | 15,549 | 65 | 18 | / | / |
| trnA-TGC | H | 15,553 | 15,622 | 70 | 3 | / | / |
| trnR-TCG | H | 15,622 | 15,684 | 63 | −1 | / | / |
| trnI-GAT | H | 15,709 | 15,776 | 68 | 24 | / | / |
| trnN-GTT | H | 15,788 | 15,857 | 70 | 11 | / | / |
| nad3 | H | 15,898 | 16,249 | 352 | 40 | ATG | T-- |
| trnS1-GCT | H | 16,250 | 16,318 | 69 | 0 | / | / |
| nad2 | H | 16,373 | 17,371 | 999 | 54 | ATT | TAA |
| Codon | No. | RSCU | Codon | No. | RSCU | Codon | No. | RSCU |
|---|---|---|---|---|---|---|---|---|
| UAA() | 12 | 1.8462 | AAG(K) | 25 | 0.5682 | CGG(R) | 12 | 0.842 |
| UAG() | 1 | 0.1538 | CUA(L) | 34 | 0.3684 | CGU(R) | 14 | 0.9824 |
| GCA(A) | 43 | 1.036 | CUC(L) | 4 | 0.0432 | AGA(S) | 70 | 1.4856 |
| GCC(A) | 11 | 0.2652 | CUG(L) | 10 | 0.1086 | AGC(S) | 9 | 0.1912 |
| GCG(A) | 8 | 0.1928 | CUU(L) | 55 | 0.5958 | AGG(S) | 23 | 0.488 |
| GCU(A) | 104 | 2.506 | UUA(L) | 359 | 3.888 | AGU(S) | 82 | 1.74 |
| UGC(C) | 5 | 0.1588 | UUG(L) | 92 | 0.9966 | UCA(S) | 59 | 1.252 |
| UGU(C) | 58 | 1.8412 | AUA(M) | 151 | 3.8718 | UCC(S) | 4 | 0.0848 |
| GAC(D) | 10 | 0.2564 | AUC(M) | 0 | 0 | UCG(S) | 13 | 0.276 |
| GAU(D) | 68 | 1.7436 | AUG(M) | 79 | 2.0256 | UCU(S) | 117 | 2.4824 |
| GAA(E) | 52 | 1.1686 | AUU(M) | 3 | 0.0768 | ACA(T) | 34 | 1.0968 |
| GAG(E) | 37 | 0.8314 | GUG(M) | 0 | 0 | ACC(T) | 6 | 0.1936 |
| UUC(F) | 27 | 0.1552 | UUG(M) | 1 | 0.0258 | ACG(T) | 4 | 0.1292 |
| UUU(F) | 321 | 1.8448 | AAC(N) | 16 | 0.25 | ACU(T) | 80 | 2.5808 |
| GGA(G) | 68 | 1.0924 | AAU(N) | 112 | 1.75 | GUA(V) | 90 | 1.1728 |
| GGC(G) | 12 | 0.1928 | CCA(P) | 51 | 1.7288 | GUC(V) | 14 | 0.1824 |
| GGG(G) | 53 | 0.8516 | CCC(P) | 6 | 0.2032 | GUG(V) | 38 | 0.4952 |
| GGU(G) | 116 | 1.8636 | CCG(P) | 11 | 0.3728 | GUU(V) | 165 | 2.15 |
| CAC(H) | 11 | 0.3056 | CCU(P) | 50 | 1.6948 | UGA(W) | 83 | 1.566 |
| CAU(H) | 61 | 1.6944 | CAA(Q) | 41 | 1.3442 | UGG(W) | 23 | 0.434 |
| AUC(I) | 19 | 0.1172 | CAG(Q) | 20 | 0.6558 | UAC(Y) | 20 | 0.2614 |
| AUU(I) | 305 | 1.8828 | CGA(R) | 28 | 1.9648 | UAU(Y) | 133 | 1.7386 |
| AAA(K) | 63 | 1.4318 | CGC(R) | 3 | 0.2104 | / | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Zhou, K.; Pan, X.; Lin, Y.; Peng, J.; Qin, J.; Ke, Z.; Han, Y.; Chen, Z.; Du, X.; et al. Characterization of the Complete Mitochondrial Genome of Angulyagra polyzonata and Its Phylogenetic Status in Viviparidae. Animals 2025, 15, 1284. https://doi.org/10.3390/ani15091284
Zhang S, Zhou K, Pan X, Lin Y, Peng J, Qin J, Ke Z, Han Y, Chen Z, Du X, et al. Characterization of the Complete Mitochondrial Genome of Angulyagra polyzonata and Its Phylogenetic Status in Viviparidae. Animals. 2025; 15(9):1284. https://doi.org/10.3390/ani15091284
Chicago/Turabian StyleZhang, Shengjie, Kangqi Zhou, Xianhui Pan, Yong Lin, Jinxia Peng, Junqi Qin, Zhenlin Ke, Yaoquan Han, Zhong Chen, Xuesong Du, and et al. 2025. "Characterization of the Complete Mitochondrial Genome of Angulyagra polyzonata and Its Phylogenetic Status in Viviparidae" Animals 15, no. 9: 1284. https://doi.org/10.3390/ani15091284
APA StyleZhang, S., Zhou, K., Pan, X., Lin, Y., Peng, J., Qin, J., Ke, Z., Han, Y., Chen, Z., Du, X., Li, W., Wei, P., & Wang, D. (2025). Characterization of the Complete Mitochondrial Genome of Angulyagra polyzonata and Its Phylogenetic Status in Viviparidae. Animals, 15(9), 1284. https://doi.org/10.3390/ani15091284




