Pathogenomic Insights into Piscirickettsia salmonis with a Focus on Virulence Factors, Single-Nucleotide Polymorphism Identification, and Resistance Dynamics
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomic Dataset Selection
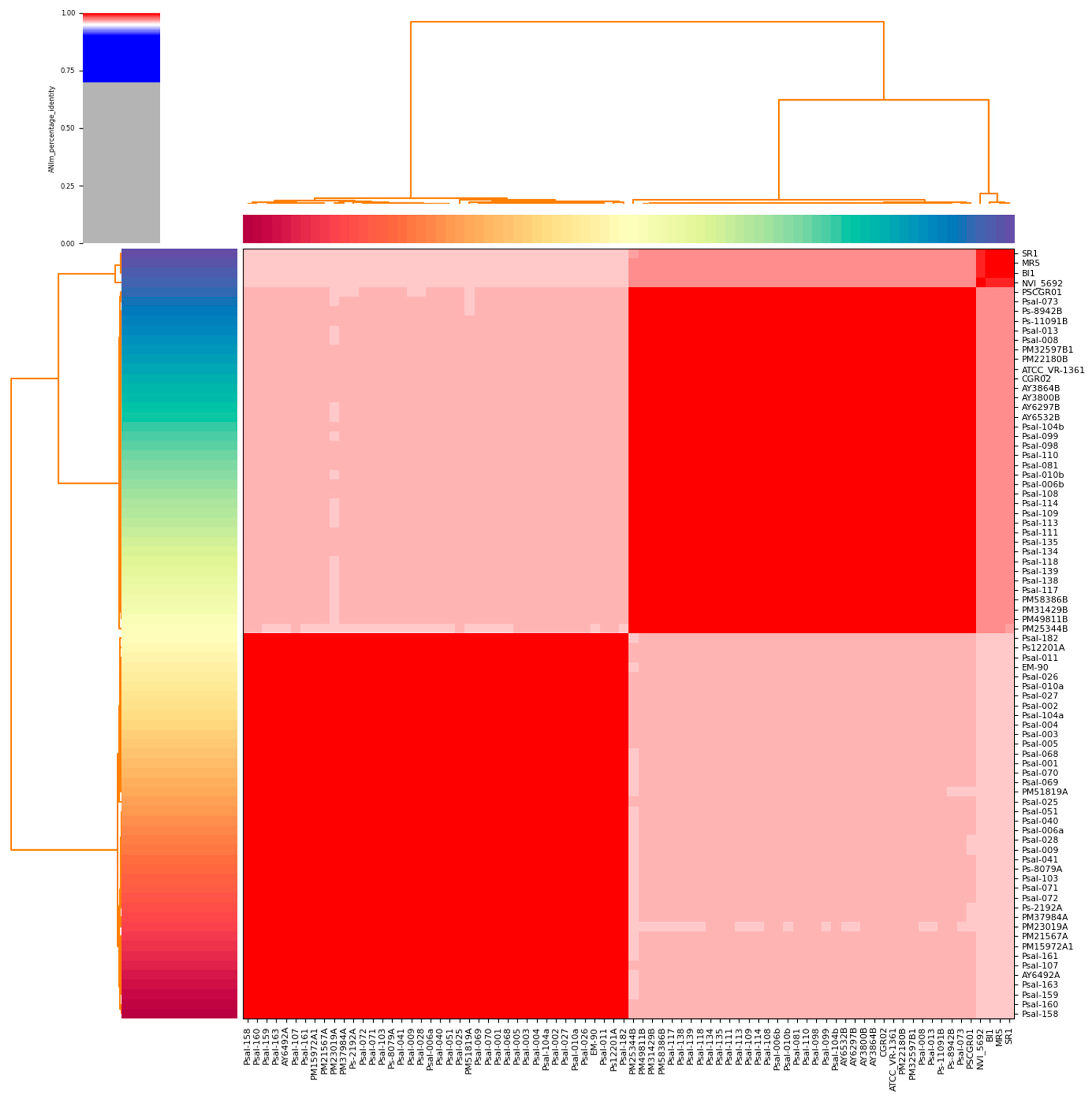
2.2. Species Validation Using Average Nucleotide Identity
2.3. Pan-Genome Analysis
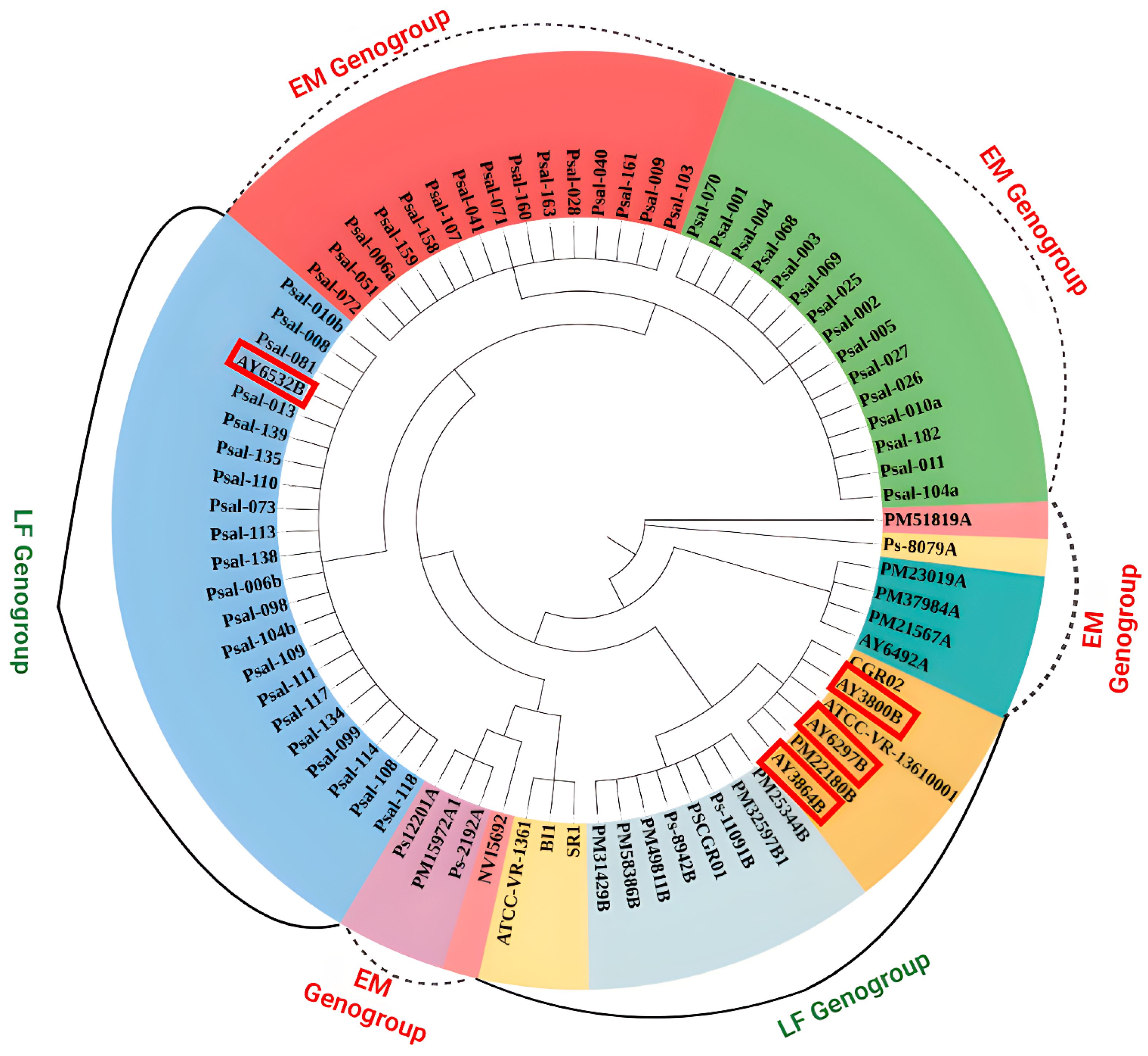
2.4. Phylogenetic Analysis and SNP Identification
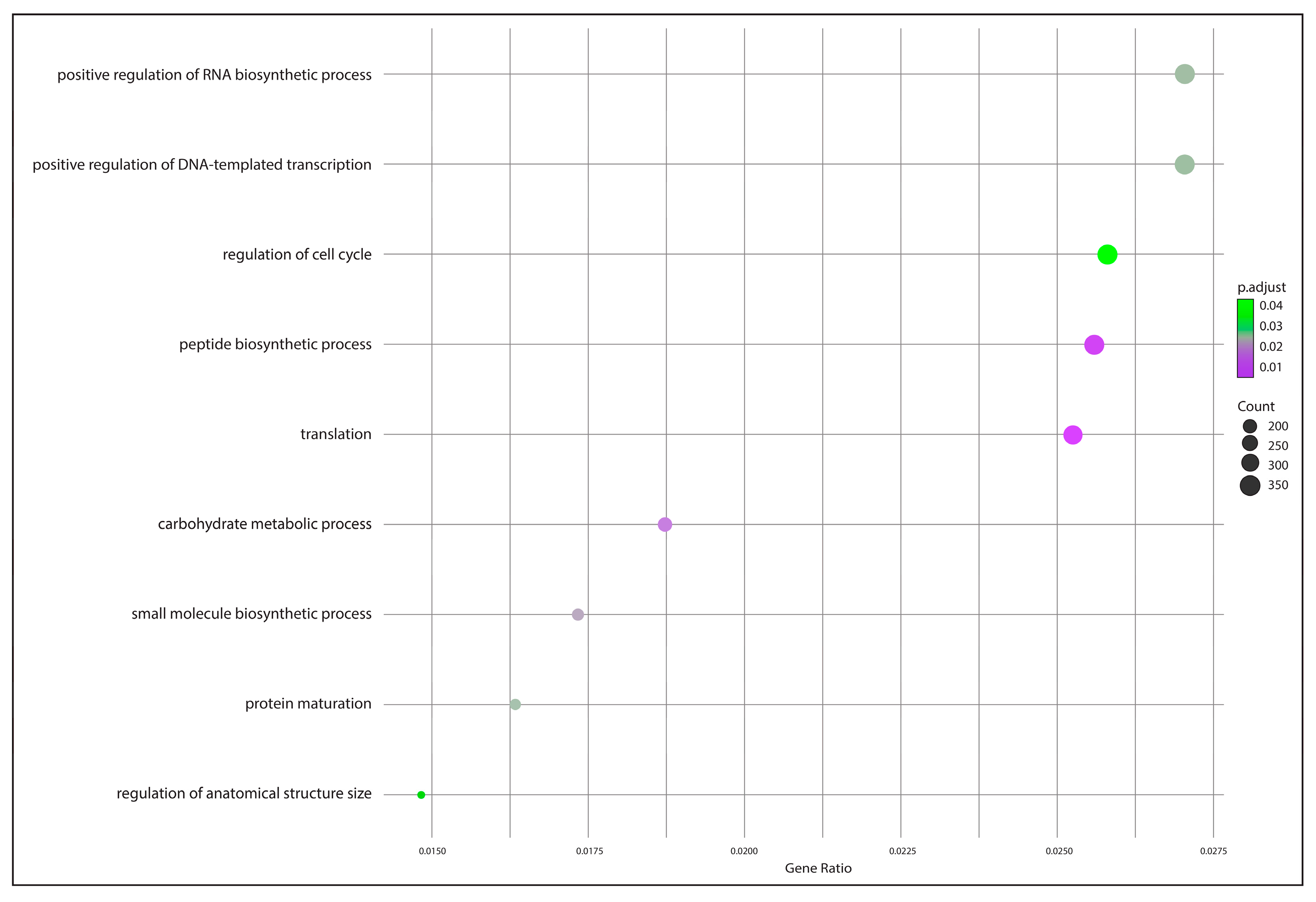
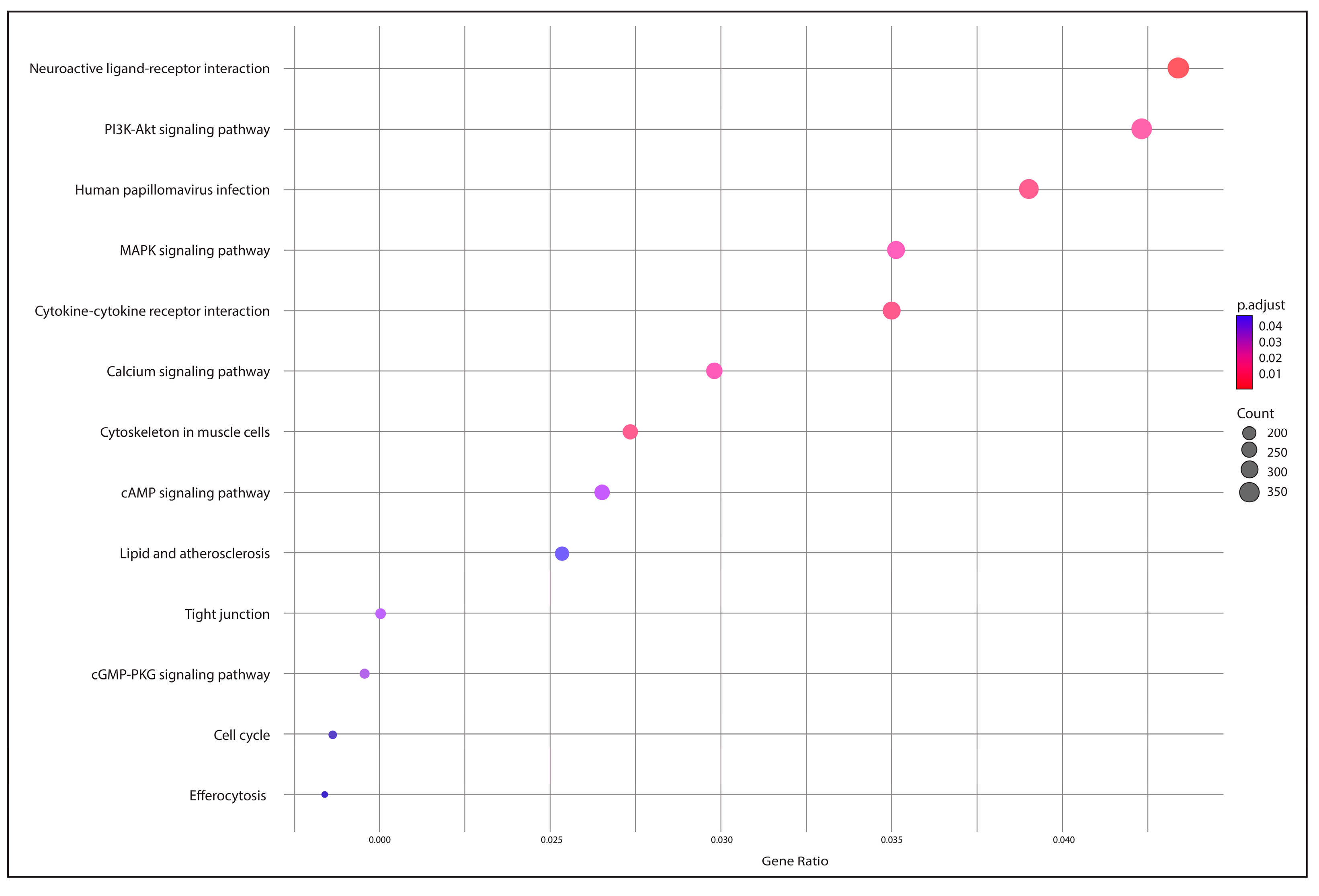
2.5. Functional Annotation and Identification of Gene Encoders
3. Results
3.1. Genomic Features and ANI
3.2. Pan-Genome Characteristics
3.3. Phylogenetic Relationship and SNP Identification
3.4. Functional Annotation and Gene Encoding Factor Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). The State of World Fisheries and Aquaculture 2022; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Marinho-Neto, F.A.; Claudiano, G.S.; Yunis-Aguinaga, J.; Cueva-Quiroz, V.A.; Kobashigawa, K.K.; Cruz, N.R.; Moraes, F.R.; Moraes, J.R. Morphological, microbiological and ultrastructural aspects of sepsis by Aeromonas hydrophila in Piaractus mesopotamicus. PLoS ONE 2019, 14, e0222626. [Google Scholar] [CrossRef]
- Shirajum Monir, M.; Yusoff, S.M.; Mohamad, A.; Ina-Salwany, M. Vaccination of tilapia against motile Aeromonas septicemia: A review. J. Aquat. Anim. Health 2020, 32, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Iversen, A.; Asche, F.; Hermansen, Ø.; Nystøyl, R. Production cost and competitiveness in major salmon farming countries 2003–2018. Aquaculture 2020, 522, 735089. [Google Scholar] [CrossRef]
- Hegde, A.; Kabra, S.; Basawa, R.M.; Khile, D.A.; Abbu, R.U.F.; Thomas, N.A.; Manickam, N.B.; Raval, R. Bacterial diseases in marine fish species: Current trends and future prospects in disease management. World J. Microbiol. Biotechnol. 2023, 39, 317. [Google Scholar] [CrossRef] [PubMed]
- Schober, I.; Bunk, B.; Carril, G.; Freese, H.M.; Ojeda, N.; Riedel, T.; Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Flores-Herrera, P.A.; et al. Ongoing diversification of the global fish pathogen Piscirickettsia salmonis through genetic isolation and transposition bursts. ISME J. 2023, 17, 2247–2258. [Google Scholar] [CrossRef]
- Toranzo, A.E.; Magariños, B.; Romalde, J.L. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 2005, 246, 37–61. [Google Scholar] [CrossRef]
- Gomez, F.A.; Tobar, J.A.; Henríquez, V.; Sola, M.; Altamirano, C.; Marshall, S.H. Evidence of the presence of a functional Dot/Icm type IV-B secretion system in the fish bacterial pathogen Piscirickettsia salmonis. PLoS ONE 2013, 8, e54934. [Google Scholar] [CrossRef]
- Rozas, M.; Enríquez, R. Piscirickettsiosis and Piscirickettsia salmonis in fish: A review. J. Fish Dis. 2013, 37, 163–188. [Google Scholar] [CrossRef]
- Rozas-Serri, M. Why Does Piscirickettsia salmonis Break the Immunological Paradigm in Farmed Salmon? Biological Context to Understand the Relative Control of Piscirickettsiosis. Front. Immunol. 2022, 13, 856896. [Google Scholar] [CrossRef]
- Evensen, Ø. Immunization strategies against Piscirickettsia salmonis infections: Review of vaccination approaches and modalities and their associated immune response profiles. Front. Immunol. 2016, 7, 482. [Google Scholar] [CrossRef]
- Maisey, K.; Montero, R.; Christodoulides, M. Vaccines for piscirickettsiosis (salmonid rickettsial septicaemia, SRS): The Chile perspective. Expert Rev. Vaccines 2017, 16, 215–228. [Google Scholar] [CrossRef] [PubMed]
- López-Cortés, X.A.; Ávila-Salas, F.; Orellana, C.; Santos, L.S. Strategy based on data mining and maldi-mass spectrometry for control disease of srs in salmo salar. In Proceedings of the 2018 IEEE International Conference on Automation/XXIII Congress of the Chilean Association of Automatic Control (ICA-ACCA), Concepcion, Chile, 17–19 October 2018; pp. 1–6. [Google Scholar]
- Aravena, P.; Pulgar, R.; Ortiz-Severín, J.; Maza, F.; Gaete, A.; Martínez, S.; Serón, E.; González, M.; Cambiazo, V. PCR-RFLP Detection and Genogroup Identification of Piscirickettsia salmonis in Field Samples. Pathogens 2020, 9, 358. [Google Scholar] [CrossRef] [PubMed]
- Mandakovic, D.; Glasner, B.; Maldonado, J.; Aravena, P.; González, M.; Cambiazo, V.; Pulgar, R. Genomic-Based Restriction Enzyme Selection for Specific Detection of Piscirickettsia salmonis by 16S rDNA PCR-RFLP. Front. Microbiol. 2016, 7, 643. [Google Scholar] [CrossRef][Green Version]
- Rozas-Serri, M.; Ildefonso, R.; Peña, A.; Enríquez, R.; Barrientos, S.; Maldonado, L. Comparative pathogenesis of piscirickettsiosis in Atlantic salmon (Salmo salar L.) post-smolt experimentally challenged with LF-89-like and EM-90-like Piscirickettsia salmonis isolates. J. Fish Dis. 2017, 40, 1451–1472. [Google Scholar] [CrossRef] [PubMed]
- Mauel, M.J.; Ware, C.; Smith, P.A. Culture of Piscirickettsia salmonis on enriched blood agar. J. Vet. Diagn. Investig. 2008, 20, 213–214. [Google Scholar] [CrossRef]
- Mauel, M.J.; Giovannoni, S.J.; Fryer, J.L. Development of polymerase chain reaction assays for detection, identification, and differentiation of Piscirickettsia salmonis. Dis. Aquat. Org. 1996, 26, 189–195. [Google Scholar] [CrossRef]
- Karatas, S.; Mikalsen, J.; Steinum, T.M.; Taksdal, T.; Bordevik, M.; Colquhoun, D.J. Real time PCR detection of Piscirickettsia salmonis from formalin-fixed paraffin-embedded tissues. J. Fish Dis. 2008, 31, 747–753. [Google Scholar] [CrossRef]
- Lannan, C.; Ewing, S.; Fryer, J. A fluorescent antibody test for detection of the rickettsia causing disease in Chilean salmonids. J. Aquat. Anim. Health 1991, 3, 229–234. [Google Scholar] [CrossRef]
- Sousa, P.S.; Silva, I.N.; Moreira, L.M.; Veríssimo, A.; Costa, J. Differences in Virulence Between Legionella pneumophila Isolates From Human and Non-human Sources Determined in Galleria mellonella Infection Model. Front. Cell. Infect. Microbiol. 2018, 8, 97. [Google Scholar] [CrossRef]
- Fernández-Bravo, A.; Figueras, M.J. An update on the genus Aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef]
- Happold, J.; Sadler, R.; Meyer, A.; Hillman, A.; Cowled, B.; Mackenzie, C.; Gallardo Lagno, A.L.; Cameron, A. Effectiveness of vaccination for the control of salmonid rickettsial septicaemia in commercial salmon and trout farms in Chile. Aquaculture 2020, 520, 734968. [Google Scholar] [CrossRef]
- Dinh-Hung, N.; Mwamburi, S.M.; Dong, H.T.; Rodkhum, C.; Meemetta, W.; Linh, N.V.; Mai, H.N.; Dhar, A.K.; Hirono, I.; Senapin, S.; et al. Unveiling Insights into the Whole Genome Sequencing of Mycobacterium spp. Isolated from Siamese Fighting Fish (Betta splendens). Animals 2024, 14, 2833. [Google Scholar] [CrossRef] [PubMed]
- Mwamburi, S.M.; Islam, S.I.; Dinh-Hung, N.; Dangsawat, O.; Sowanpreecha, R.; Khang, L.T.P.; Montha, N.; Therdtatha, P.; Dwinanti, S.H.; Permpoonpattana, P.; et al. Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential. Microorganisms 2024, 12, 2476. [Google Scholar] [CrossRef]
- Nourdin-Galindo, G.; Sánchez, P.; Molina, C.F.; Espinoza-Rojas, D.A.; Oliver, C.; Ruiz, P.; Vargas-Chacoff, L.; Cárcamo, J.G.; Figueroa, J.E.; Mancilla, M.; et al. Comparative Pan-Genome Analysis of Piscirickettsia salmonis Reveals Genomic Divergences within Genogroups. Front. Cell. Infect. Microbiol. 2017, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, N.H.; Elsherbiny, B.A.; Moffit, S.M.; Mohamed, J.H.; Abouelkheir, S.S.; Abdella, B. Cell biology and microbial interactions in algal cells. In Handbook of Research on Algae as a Sustainable Solution for Food, Energy, and the Environment; IGI Global: Hershey, PA, USA, 2022; pp. 84–108. [Google Scholar]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.; Humphris, S.; Elphinstone, J.; Toth, I. Genomics and Taxonomy in Diagnostics for Food Security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Abdella, B.; Abozahra, N.A.; Shokrak, N.M.; Mohamed, R.A.; El-Helow, E.R. Whole spectrum of Aeromonas hydrophila virulence determinants and the identification of novel SNPs using comparative pathogenomics. Sci. Rep. 2023, 13, 7712. [Google Scholar] [CrossRef]
- Podrzaj, L.; Burtscher, J.; Domig, K.J. Comparative genomics provides insights into genetic diversity of Clostridium tyrobutyricum and potential implications for late blowing defects in cheese. Front. Microbiol. 2022, 13, 889551. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Rajput, A.; Chauhan, S.M.; Mohite, O.S.; Hyun, J.C.; Ardalani, O.; Jahn, L.J.; Sommer, M.O.A.; Palsson, B.O. Pangenome analysis reveals the genetic basis for taxonomic classification of the Lactobacillaceae family. Food Microbiol. 2023, 115, 104334. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Horsfield, S.T.; Croucher, N.J.; Lees, J.A. Accurate and fast graph-based pangenome annotation and clustering with ggCaller. bioRxiv 2023. [Google Scholar] [CrossRef]
- Soares, S.C.; Silva, A.; Trost, E.; Blom, J.; Ramos, R.; Carneiro, A.; Ali, A.; Santos, A.R.; Pinto, A.C.; Diniz, C.; et al. The pan-genome of the animal pathogen Corynebacterium pseudotuberculosis reveals differences in genome plasticity between the biovar ovis and equi strains. PLoS ONE 2013, 8, e53818. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Xu, S.; Hu, E.; Cai, Y.; Xie, Z.; Luo, X.; Zhan, L.; Tang, W.; Wang, Q.; Liu, B.; Wang, R.; et al. Using clusterProfiler to characterize multiomics data. Nat. Protoc. 2024, 19, 3292–3320. [Google Scholar] [CrossRef] [PubMed]
- Man, T.J.B.d.; Limbago, B.M. SSTAR, a Stand-Alone Easy-To-Use Antimicrobial Resistance Gene Predictor. mSphere 2016, 1, e00050-15. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Khang, L.T.P.; Dinh-Hung, N.; Islam, S.I.; Dwinanti, S.H.; Mwamburi, S.M.; Permpoonpattana, P.; Linh, N.V. A Review of In Silico Approaches for Discovering Natural Viral Protein Inhibitors in Aquaculture Disease Control. J. Fish Dis. 2025, e14129. [Google Scholar] [CrossRef] [PubMed]
- Kaas, R.S.; Friis, C.; Ussery, D.W.; Aarestrup, F.M. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genom. 2012, 13, 577. [Google Scholar] [CrossRef]
- Herrera, V.; Olavarría, N.; Saavedra, J.; Yuivar, Y.; Bustos, P.; Almarza, O.; Mancilla, M. Complete Lipopolysaccharide of Piscirickettsia salmonis Is Required for Full Virulence in the Intraperitoneally Challenged Atlantic Salmon, Salmo salar, Model. Front. Cell. Infect. Microbiol. 2022, 12, 845661. [Google Scholar] [CrossRef]
- Saavedra, J.; Hernandez, N.; Osses, A.; Castillo, A.; Cancino, A.; Grothusen, H.; Navas, E.; Henriquez, P.; Bohle, H.; Bustamante, F.; et al. Prevalence, geographic distribution and phenotypic differences of Piscirickettsia salmonis EM-90-like isolates. J. Fish Dis. 2017, 40, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Beaz-Hidalgo, R.; Alperi, A.; Buján, N.; Romalde, J.L.; Figueras, M.J. Comparison of phenotypical and genetic identification of Aeromonas strains isolated from diseased fish. Syst. Appl. Microbiol. 2010, 33, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Fernández-No, I.C.; Böhme, K.; Caamaño-Antelo, S.; Barros-Velázquez, J.; Calo-Mata, P. Identification of single nucleotide polymorphisms (SNPs) in the 16S rRNA gene of foodborne Bacillus spp. Food Microbiol. 2015, 46, 239–245. [Google Scholar] [CrossRef]
- Medini, D.; Donati, C.; Tettelin, H.; Masignani, V.; Rappuoli, R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005, 15, 589–594. [Google Scholar] [CrossRef]
- Diene, S.M.; Merhej, V.; Henry, M.; El Filali, A.; Roux, V.; Robert, C.; Azza, S.; Gavory, F.; Barbe, V.; La Scola, B.; et al. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Mol. Biol. Evol. 2013, 30, 369–383. [Google Scholar] [CrossRef]
- Saleh, A.; Elkenany, R.; Younis, G. Virulent and Multiple Antimicrobial Resistance Aeromonas hydrophila Isolated from Diseased Nile Tilapia Fish (Oreochromis niloticus) in Egypt with Sequencing of Some Virulence-Associated Genes. Biocontrol Sci. 2021, 26, 167–176. [Google Scholar] [CrossRef]
- Wang, G.; Clark, C.G.; Liu, C.; Pucknell, C.; Munro, C.K.; Kruk, T.M.; Caldeira, R.; Woodward, D.L.; Rodgers, F.G. Detection and characterization of the hemolysin genes in Aeromonas hydrophila and Aeromonas sobria by multiplex PCR. J. Clin. Microbiol. 2003, 41, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Poot-Hernandez, A.C.; Rodriguez-Vazquez, K.; Perez-Rueda, E. The alignment of enzymatic steps reveals similar metabolic pathways and probable recruitment events in Gammaproteobacteria. BMC Genom. 2015, 16, 957. [Google Scholar] [CrossRef] [PubMed]


| Genes | Annotation | Position, SNPs Unique for P. salmonis |
|---|---|---|
| dnaK | Chaperone protein | 5 C > T/A, 408 A > T/-/G, 717 A > T/C, 724 G > T/A, 807 A > T/-/C, 1325 G > T/A, 1327 G > A/C, 1445 T > G/C, 1452 T > G/C, 1546 A > T/G, 1555 G > A/C, 1560 T > A/G, 1565 C > T/A, 1596 T > -/G/C, 1608 C > T/A/G, 1617 G > A/C, 1627 C > A/G, 1630 G > T/A,1690 A > T/G, 1697 A > T/G, 1717 G > A/C, 1761 T > -/A/G, |
| ftsZ | Filamenting temperature-sensitive mutant Z | 297 T > G/C, 423 T > A/C, 459 A > G/T, 537 A > T/C, 612 C > CA/CG, 633 T > A/G, 634 G > T/C, 864 T > A/G, 876 C > G/T, 906 A > G/T, 975 T > A/C, 993 G > T/C, 1050 G > T/C, 1077 G > A/T, 1116 G > T/C, |
| gyrA | DNA gyrase A | 36 A > /AT, 57 G > C/A/-, 192 T > G/C/-, 216 C > G/A/-, 373 A > G/C, 376 G > T/C, 380 T > G/C, 381 A > T/C, 382 T > G/C, 388 C > G/T, 390 G > T/C, 392 T > C/A, 397 C > G/T, 399 A > T/C, 402 G > T/C, 404 T > G/A, 406 T > C/A, 416 G > T/C, 419 A > G/C, 422 T > C/A, 428 A > G/C, 429 A > T/C, 431 A > G/T, 432 A > G/T, 433 A > T/C, 437 A > G/C, 440 T > G/C, 441 G > C/A, 443 T > C/A, 459 A > T/C, 461 C > G/T, 463 C > T/A, 468 T > G/A, 479 A > G/C, 480 G > C/A, 486 A > G/T, 493 G > T/A, 494 G > C/A, 495 A > G/T, 498 G > T/C, 501 G > C/A, 502 A > G/C, 505 C > T/A, 514 C > G/A/CAA, 518 G > T/A, 2106 A > G/T/-, 2343 C > G/A/-, |
| rpoB | RNA polymerase, beta subunit | 1887 G > T/C, 2655 G > A/C, |
| Gene Functional Category | Virulence Gene | Product | Replicon |
|---|---|---|---|
| Nutritional/Metabolic factor | ggt | gamma-glutamyltranspeptidase | Chromosome |
| Adherence | htpB | Hsp60, 60K heat shock protein | Chromosome |
| Adherence | tufA | Elongation factor Tu | Chromosome |
| Effector delivery system | icmO/dotL | Dot/Icm type IV secretion system coupling protein | Chromosome |
| Motility | flrA | sigma-54 dependent transcriptional activator | Chromosome I |
| Adherence | rpoS | RNA polymerase sigma factor | Chromosome |
| Regulation | csrA | carbon storage regulator | Chromosome |
| Regulation | rpoS | RNA polymerase sigma factor | Chromosome |
| Motility | fliA | flagellar biosynthesis sigma factor | Chromosome I |
| Motility | flhA | flagellar biosynthesis protein | Chromosome I |
| Nutritional/Metabolic factor | feoB | ferrous iron transporter B | Chromosome |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, S.I.; Shahed, K.; Ahamed, M.I.; Khang, L.T.P.; Jung, W.-K.; Sangsawad, P.; Dinh-Hung, N.; Permpoonpattana, P.; Linh, N.V. Pathogenomic Insights into Piscirickettsia salmonis with a Focus on Virulence Factors, Single-Nucleotide Polymorphism Identification, and Resistance Dynamics. Animals 2025, 15, 1176. https://doi.org/10.3390/ani15081176
Islam SI, Shahed K, Ahamed MI, Khang LTP, Jung W-K, Sangsawad P, Dinh-Hung N, Permpoonpattana P, Linh NV. Pathogenomic Insights into Piscirickettsia salmonis with a Focus on Virulence Factors, Single-Nucleotide Polymorphism Identification, and Resistance Dynamics. Animals. 2025; 15(8):1176. https://doi.org/10.3390/ani15081176
Chicago/Turabian StyleIslam, Sk Injamamul, Khandker Shahed, Md Imtiaz Ahamed, Luu Tang Phuc Khang, Won-Kyo Jung, Papungkorn Sangsawad, Nguyen Dinh-Hung, Patima Permpoonpattana, and Nguyen Vu Linh. 2025. "Pathogenomic Insights into Piscirickettsia salmonis with a Focus on Virulence Factors, Single-Nucleotide Polymorphism Identification, and Resistance Dynamics" Animals 15, no. 8: 1176. https://doi.org/10.3390/ani15081176
APA StyleIslam, S. I., Shahed, K., Ahamed, M. I., Khang, L. T. P., Jung, W.-K., Sangsawad, P., Dinh-Hung, N., Permpoonpattana, P., & Linh, N. V. (2025). Pathogenomic Insights into Piscirickettsia salmonis with a Focus on Virulence Factors, Single-Nucleotide Polymorphism Identification, and Resistance Dynamics. Animals, 15(8), 1176. https://doi.org/10.3390/ani15081176







