Machine Learning Approach for Early Lactation Mastitis Diagnosis Using Total and Differential Somatic Cell Counts
Simple Summary
Abstract
1. Introduction
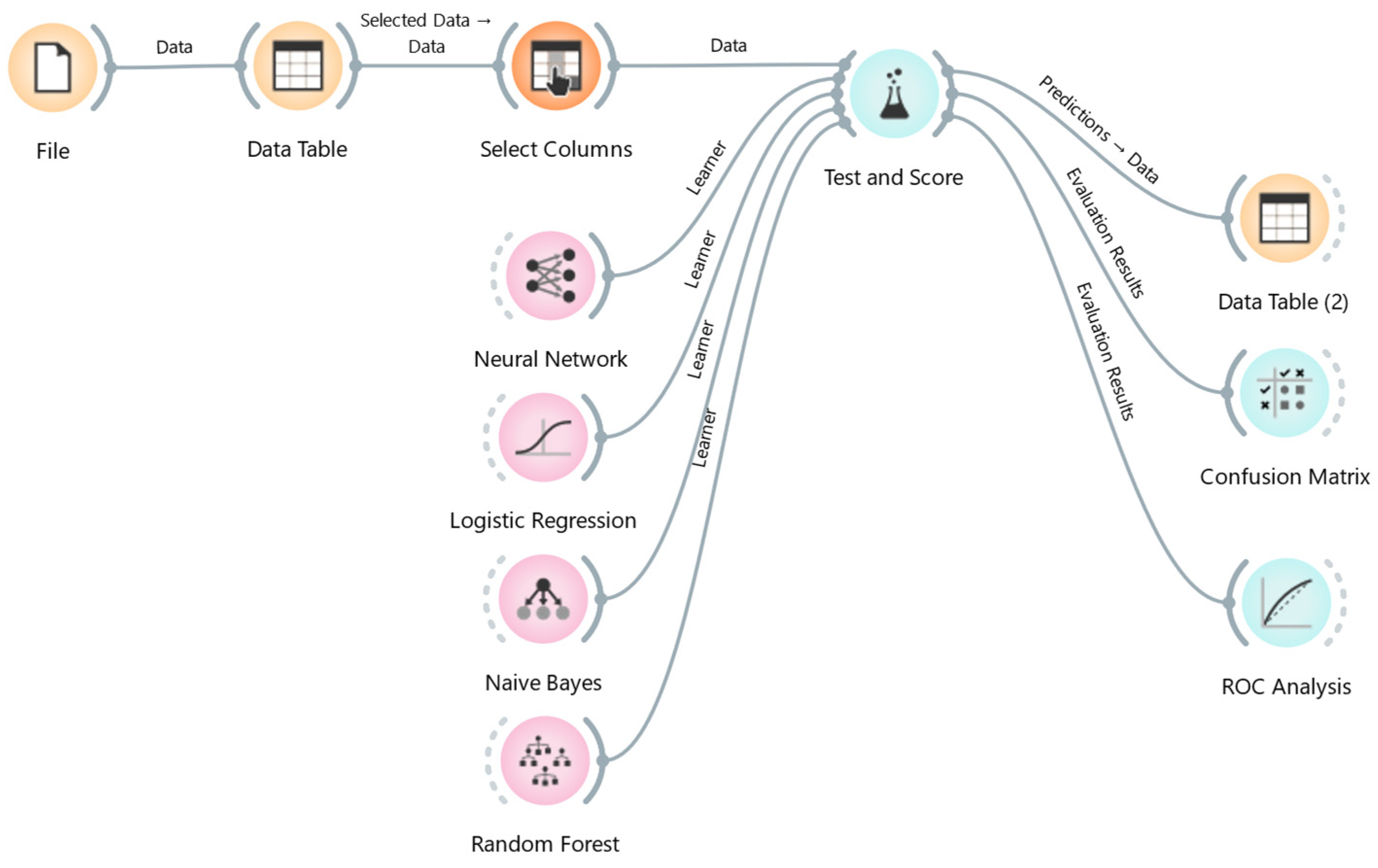
2. Materials and Methods
2.1. Herd and Cow Selection
2.2. Sample Collection
2.3. Cellular Marker Analyses
2.4. Conventional Microbiological Analysis
2.5. Real-Time PCR Analysis
2.6. Statistical Analysis
2.7. Diagnostic Parameters
- -
- Area under the curve (AUC) of the ROC curve: it represents the degree or measure of separability; the higher the AUC, the better the model is at predicting the true status of the sample (positive/negative).
- -
- Accuracy: expressed as a proportion of correctly classified subjects [true positive (TP) + true negative (TN)] among all subjects.
- -
- Sensitivity (Se): the proportion of TP/[TP + false positive (FP)].
- -
- Specificity (Sp): the proportion of TN/[false negative (FN) + TP].
- -
- Positive predictive value (PPV): TP/(TP + FN).
- -
- Negative predictive value (NPV): TN/(TN + FP).
3. Results
3.1. Data Description
3.2. Machine Learning Analysis
4. Discussion
4.1. Intramammary Infections
4.2. Machine Learning Approach
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Dairy Market Review: Overview of Global Market Developments in 2024; FAO: Rome, Italy, 2024. [Google Scholar]
- Martin, T.; Gasselin, P.; Hostiou, N.; Feron, G.; Laurens, L.; Purseigle, F.; Ollivier, G. Robots and transformations of work in farm: A systematic review of the literature and a research agenda. Agron. Sustain. Dev. 2022, 42, 66. [Google Scholar] [CrossRef]
- Cogato, A.; Brscic, M.; Guo, H.; Marinello, F.; Pezzuolo, A. Challenges and Tendencies of Automatic Milking Systems (AMS): A 20-Years Systematic Review of Literature and Patents. Animals 2021, 11, 356. [Google Scholar] [CrossRef]
- Marino, R.; Petrera, F.; Abeni, F. Scientific Productions on Precision Livestock Farming: An Overview of the Evolution and Current State of Research Based on a Bibliometric Analysis. Animals 2023, 13, 2280. [Google Scholar] [CrossRef]
- Garcia, S.N.; Osburn, B.I.; Cullor, J.S. A one health perspective on dairy production and dairy food safety. One Health 2019, 7, 100086. [Google Scholar] [CrossRef] [PubMed]
- Bramer, M. Principles of Data Mining, 4th ed.; Srpinger: London, UK, 2020; p. 576. [Google Scholar]
- Qamar, U.; Raza, M.S. Data Science Concepts and Techniques with Applications; Springer: Singapore, 2020; p. 196. [Google Scholar]
- Zhou, X.J.; Xu, C.; Wang, H.; Xu, W.; Zhao, Z.X.; Chen, M.X.; Jia, B.; Huang, B.Y. The Early Prediction of Common Disorders in Dairy Cows Monitored by Automatic Systems with Machine Learning Algorithms. Animals 2022, 12, 1251. [Google Scholar] [CrossRef] [PubMed]
- Lasser, J.; Matzhold, C.; Egger-Danner, C.; Fuerst-Waltl, B.; Steininger, F.; Wittek, T.; Klimek, P. Integrating diverse data sources to predict disease risk in dairy cattle-a machine learning approach. J. Anim. Sci. 2021, 99, skab294. [Google Scholar] [CrossRef]
- Bobbo, T.; Biffani, S.; Taccioli, C.; Penasa, M.; Cassandro, M. Comparison of machine learning methods to predict udder health status based on somatic cell counts in dairy cows. Sci. Rep. 2021, 11, 13642. [Google Scholar] [CrossRef]
- Bobbo, T.; Matera, R.; Pedota, G.; Manunza, A.; Cotticelli, A.; Neglia, G.; Biffani, S. Exploiting machine learning methods with monthly routine milk recording data and climatic information to predict subclinical mastitis in Italian Mediterranean buffaloes. J. Dairy Sci. 2023, 106, 1942–1952. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Zhou, X.J.; Wang, H.; Xu, C.; Zhao, Z.X.; Xu, W.; Deng, Z.J. The Prediction of Clinical Mastitis in Dairy Cows Based on Milk Yield, Rumination Time, and Milk Electrical Conductivity Using Machine Learning Algorithms. Animals 2024, 14, 427. [Google Scholar] [CrossRef]
- Fadul-Pacheco, L.; Delgado, H.; Cabrera, V.E. Exploring machine learning algorithms for early prediction of clinical mastitis. Int. Dairy J. 2021, 119, 105051. [Google Scholar] [CrossRef]
- Satola, A.; Satola, K. Performance comparison of machine learning models used for predicting subclinical mastitis in dairy cows: Bagging, boosting, stacking, and super-learner ensembles versus single machine learning models. J. Dairy Sci. 2024, 107, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- Mitsunaga, T.M.; Garcia, B.L.N.; Pereira, L.B.R.; Costa, Y.C.B.; da Silva, R.F.; Delbem, A.C.B.; dos Santos, M.V. Current Trends in Artificial Intelligence and Bovine Mastitis Research: A Bibliometric Review Approach. Animals 2024, 14, 2023. [Google Scholar] [CrossRef]
- Kiouvrekis, Y.; Vasileiou, N.G.C.; Katsarou, E.I.; Lianou, D.T.; Michael, C.K.; Zikas, S.; Katsafadou, A.I.; Bourganou, M.V.; Liagka, D.V.; Chatzopoulos, D.C.; et al. The Use of Machine Learning to Predict Prevalence of Subclinical Mastitis in Dairy Sheep Farms. Animals 2024, 14, 2295. [Google Scholar] [CrossRef]
- N.M.C. Laboratory Handbook on Bovine Mastitis; N.M.C.: New Prague, MN, USA, 2017. [Google Scholar]
- Damm, M.; Holm, C.; Blaabjerg, M.; Bro, M.N.; Schwarz, D. Differential somatic cell count-A novel method for routine mastitis screening in the frame of Dairy Herd Improvement testing programs. J. Dairy Sci. 2017, 100, 4926–4940. [Google Scholar] [CrossRef] [PubMed]
- Zecconi, A.; Meroni, G.; Sora, V.; Mattina, R.; Cipolla, M.; Zanini, L. Total and differential cell counts as a tool to identify intramammary infections in cows after calving. Animals 2021, 11, 727. [Google Scholar] [CrossRef]
- Zecconi, A.; Zanini, L.; Cipolla, M.; Stefanon, B. Factors Affecting the Patterns of Total Amount and Proportions of Leukocytes in Bovine Milk. Animals 2020, 10, 992. [Google Scholar] [CrossRef] [PubMed]
- Demsar, J.; Curk, T.; Erjavec, A.; Gorup, C.; Hocevar, T.; Milutinovic, M.; Mozina, M.; Polajnar, M.; Toplak, M.; Staric, A.; et al. Orange data mining toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Hosseini, S.; Sardo, S.R. Data mining tools—A case study for network intrusion detection. Multimed. Tools Appl. 2021, 80, 4999–5019. [Google Scholar] [CrossRef]
- Antas, J.; Rocha Silva, R.; Bernardino, J. Assessment of SQL and NoSQL Systems to Store and Mine COVID-19 Data. Computers 2022, 11, 29. [Google Scholar] [CrossRef]
- Ozella, L.; Rebuli, K.B.; Forte, C.; Giacobini, M. A Literature Review of Modeling Approaches Applied to Data Collected in Automatic Milking Systems. Animals 2023, 13, 1916. [Google Scholar] [CrossRef]
- Afifi, M.; Kabera, F.; Stryhn, H.; Roy, J.P.; Heider, L.C.; Godden, S.; Montelpare, W.; Sanchez, J.; Dufour, S. Antimicrobial-based dry cow therapy approaches for cure and prevention of intramammary infections: A protocol for a systematic review and meta-analysis. Anim. Health Res. Rev. 2018, 19, 74–78. [Google Scholar] [CrossRef]
- Koskinen, M.T.; Wellenberg, G.J.; Sampimon, O.C.; Holopainen, J.; Rothkamp, A.; Salmikivi, L.; van Haeringen, W.A.; Lam, T.; Pyorala, S. Field comparison of real-time polymerase chain reaction and bacterial culture for identification of bovine mastitis bacteria. J. Dairy Sci. 2010, 93, 5707–5715. [Google Scholar] [CrossRef] [PubMed]
- Taponen, S.; Salmikivi, L.; Simojoki, H.; Koskinen, M.T.; Pyorala, S. Real-time polymerase chain reaction-based identification of bacteria in milk samples from bovine clinical mastitis with no growth in conventional culturing. J. Dairy Sci. 2009, 92, 2610–2617. [Google Scholar] [CrossRef]
- Schwarz, D.; Lipkens, Z.; Piepers, S.; De Vliegher, S. Investigation of differential somatic cell count as a potential new supplementary indicator to somatic cell count for identification of intramammary infection in dairy cows at the end of the lactation period. Prev. Vet. Med. 2019, 172, 104803. [Google Scholar] [CrossRef] [PubMed]
- Kirkeby, C.; Toft, N.; Schwarz, D.; Farre, M.; Nielsen, S.S.; Zervens, L.; Hechinger, S.; Halasa, T. Differential somatic cell count as an additional indicator for intramammary infections in dairy cows. J. Dairy Sci. 2020, 103, 1759–1775. [Google Scholar] [CrossRef] [PubMed]
- Zecconi, A.; Zaghen, F.; Meroni, G.; Sora, V.; Martino, P.A.; Laterza, G.; Zanini, L. Early Milk Total and Differential Cell Counts as a Diagnostic Tool to Improve Antimicrobial Therapy Protocols. Animals 2023, 13, 1143. [Google Scholar] [CrossRef]
- Luo, W.K.; Dong, Q.; Feng, Y. Risk prediction model of clinical mastitis in lactating dairy cows based on machine learning algorithms. Prev. Vet. Med. 2023, 221, 106059. [Google Scholar] [CrossRef]
- Goyache, F.; Díez, J.; López, S.; Pajares, G.; Santos, B.; Fernández, I.; Prieto, M. Machine Learning as an aid to management decisions on high somatic cell counts in dairy farms. Arch. Tierz.—Arch. Anim. Breed. 2005, 48, 138–148. [Google Scholar] [CrossRef]
- De Campos, M.A.; Oppermann, M.L.R.; Genro, V.K.; Leitao, C.B.; Hirakata, V.N.; Reichelt, A.J. 170-LB: Predictors of Diabetes First Diagnosed in Pregnancy: A Machine-Learning Model. Diabetes 2020, 69, 170-LB. [Google Scholar] [CrossRef]
- Tougui, I.; Jilbab, A.; El Mhamdi, J. Heart disease classification using data mining tools and machine learning techniques. Health Technol. 2020, 10, 1137–1144. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E.; Petrovski, K.R. Comprehensive analysis of machine learning models for prediction of sub-clinical mastitis: Deep Learning and Gradient-Boosted Trees outperform other models. Comput. Biol. Med. 2019, 114, 103456. [Google Scholar] [CrossRef] [PubMed]
- Halasa, T.; Kirkeby, C. Differential Somatic Cell Count: Value for Udder Health Management. Front. Vet. Sci. 2020, 7, 609055. [Google Scholar] [CrossRef] [PubMed]
- Zecconi, A.; Vairani, D.; Cipolla, M.; Rizzi, N.; Zanini, L. Assessment of Subclinical Mastitis Diagnostic Accuracy by Differential Cell Count in Individual Cow Milk. Ital. J. Anim. Sci. 2018, 18, 460–465. [Google Scholar] [CrossRef]


| N | SCC 1 ± Std. Dev (Log10/mL) | DSCC 2 ± Std. Dev (%) | PLCC 3 ± Std. Dev (Log10/mL) | |
|---|---|---|---|---|
| Lactation period | ||||
| A (5–15 d) | 88 | 4.98 ± 0.22 a,4 | 63.1 ± 17.7 a | 4.76 ± 0.90 a |
| B (16–45 d) | 111 | 4.78 ± 0.83 a | 61.9 ± 18.1 a | 4.55 ± 0.93 a |
| C (46–90 d) | 225 | 5.00 ± 0.82 b | 64.4 ± 18.3 a | 4.79 ± 0.92 b |
| Parturition | ||||
| Primiparous | 133 | 4.81 ± 0.66 a | 63.8 ± 19.5 a | 4.60 ± 0.73 a |
| Pluriparous | 291 | 5.01 ± 0.92 b | 64.0 ± 15.2 a | 4.80 ± 1.02 b |
| Lactation Period | S. aureus | S. agalactiae | S. uberis | S. dysgalactiae | Negative |
|---|---|---|---|---|---|
| A (5–15 d) | 2.3 | 2.3 | 17.0 | 4.5 | 73.9 |
| B (16–45 d) | 5.4 | 0.0 | 8.9 | 0.9 | 84.8 |
| C (46–90 d) | 14.5 | 0.0 | 19.3 | 3.1 | 63.1 |
| Total | 9.6 | 0.5 | 16.1 | 2.8 | 71.0 |
| Parturition | S. aureus | S. agalactiae | S. uberis | S. dysgalactiae | Negative |
|---|---|---|---|---|---|
| Primiparous | 6.8 | 0.0 | 10.5 | 3.0 | 79.7 |
| Pluriparous | 11.0 | 0.7 | 18.6 | 2.7 | 67.0 |
| Total | 9.6 | 0.5 | 16.1 | 2.8 | 71.0 |
| Lactation Period | Quarter (N) | MajP 1 | Other Pathogens | Negative |
|---|---|---|---|---|
| A (5–15 d) | 352 | 1.9% | 18.8% | 79.3% |
| B (16–45 d) | 444 | 2.9% | 22.3% | 74.8% |
| C (46–90 d) | 900 | 6.5% | 25.6% | 67.9% |
| Total | 1696 | 4.6% | 23.3% | 72.1% |
| Parturition | Quarter (N) | MajP 1 | Other Pathogens | Negative |
|---|---|---|---|---|
| Primiparous | 648 | 2.6% | 22.8% | 74.6% |
| Pluriparous | 1048 | 5.9% | 23.6% | 70.5% |
| Total | 1696 | 4.6% | 23.3% | 72.1% |
| Model | Parameter | AUC 4 | Accuracy | Sensitivity | PPV 5 | Specificity | NPV 6 |
|---|---|---|---|---|---|---|---|
| Logistic regression | PLCC 1 | 0.740 | 0.774 | 57.6% | 17.6% | 78.9% | 96.0% |
| SCC 2 | 0.743 | 0.774 | 57.6% | 17.6% | 78.9% | 96.0% | |
| DSCC 3 | 0.665 | 0.763 | 0.0% | 0.0% | 100% | 76.3% | |
| Neural network | PLCC | 0.733 | 0.760 | 42.3% | 20.4% | 78.7% | 91.4% |
| SCC | 0.739 | 0.765 | 51.2% | 19.4% | 79.0% | 93.7% | |
| DSCC | 0.651 | 0.760 | 33.3% | 0.9% | 76.3% | 99.4% | |
| Naïve Bayes | PLCC | 0.711 | 0.758 | 48.1% | 24.1% | 79.6% | 91.9% |
| SCC | 0.717 | 0.758 | 48.1% | 24.1% | 79.6% | 91.9% | |
| DSCC | 0.662 | 0.763 | 0.0% | 0.0% | 76.3% | 100.0% | |
| Random forest | PLCC | 0.684 | 0.745 | 45.6% | 38.0% | 81.6% | 85.9% |
| SCC | 0.656 | 0.727 | 40.9% | 33.3% | 80.4% | 85.0% | |
| DSCC | 0.630 | 0.690 | 30.6% | 24.1% | 77.8% | 83.0% |
| Model | Parameter | AUC 4 | Accuracy | Sensitivity | PPV 5 | Specificity | NPV 6 |
|---|---|---|---|---|---|---|---|
| Logistic regression | PLCC 1 | 0.816 | 0.952 | 0.0% | 0.0% | 95.3% | 99.9% |
| SCC 2 | 0.821 | 0.952 | 0.0% | 0.0% | 95.3% | 99.8% | |
| DSCC 3 | 0.686 | 0.953 | n.a. 7 | 0.0% | 95.3% | 100.0% | |
| Neural network | PLCC | 0.806 | 0.952 | 0.0% | 0.0% | 95.3% | 99.9% |
| SCC | 0.811 | 0.951 | 16.7% | 1.2% | 95.4% | 99.7% | |
| DSCC | 0.658 | 0.953 | n.a. | 0.0% | 95.3% | 100.0% | |
| Naïve Bayes | PLCC | 0.770 | 0.953 | n.a. | 0.0% | 95.3% | 100.0% |
| SCC | 0.780 | 0.953 | n.a. | 0.0% | 95.3% | 100.0% | |
| DSCC | 0.639 | 0.953 | n.a. | 0.0% | 95.3% | 100.0% | |
| Random forest | PLCC | 0.723 | 0.933 | 21.5% | 16.5% | 96.0% | 97.1% |
| SCC | 0.711 | 0.943 | 32.7% | 20.0% | 96.2% | 98.0% | |
| DSCC | 0.602 | 0.951 | 0.0% | 0.0% | 95.3% | 99.7% |
| Model | Parameter | AUC 4 | Accuracy | Sensitivity | PPV 5 | Specificity | NPV 6 |
|---|---|---|---|---|---|---|---|
| Logistic regression | PLCC 1 | 0.638 | 0.728 | 61.4% | 16.9% | 73.8% | 95.7% |
| SCC 2 | 0.637 | 0.732 | 64.3% | 17.0% | 73.9% | 96.1% | |
| DSCC 3 | 0.595 | 0.710 | n.a. 7 | 0.0% | 71.0% | 100.0% | |
| Neural network | PLCC | 0.657 | 0.733 | 60.5% | 22.9% | 74.9% | 93.9% |
| SCC | 0.653 | 0.733 | 60.7% | 22.5% | 74.8% | 94.0% | |
| DSCC | 0.621 | 0.716 | 55.8% | 10.0% | 72.5% | 96.7% | |
| Naïve Bayes | PLCC | 0.618 | 0.713 | 51.6% | 18.6% | 73.6% | 92.9% |
| SCC | 0.616 | 0.710 | n.a. | 0.0% | 71.0% | 100.0% | |
| DSCC | 0.587 | 0.710 | n.a. | 0.0% | 71.0% | 100.0% | |
| Random forest | PLCC | 0.586 | 0.657 | 39.6% | 34.8% | 74.6% | 78.3% |
| SCC | 0.595 | 0.683 | 43.1% | 29.2% | 74.4% | 84.3% | |
| DSCC | 0.543 | 0.660 | 35.1% | 20.1% | 72.2% | 84.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zecconi, A.; Zaghen, F.; Meroni, G.; Sommariva, F.; Ferrari, S.; Sora, V. Machine Learning Approach for Early Lactation Mastitis Diagnosis Using Total and Differential Somatic Cell Counts. Animals 2025, 15, 1125. https://doi.org/10.3390/ani15081125
Zecconi A, Zaghen F, Meroni G, Sommariva F, Ferrari S, Sora V. Machine Learning Approach for Early Lactation Mastitis Diagnosis Using Total and Differential Somatic Cell Counts. Animals. 2025; 15(8):1125. https://doi.org/10.3390/ani15081125
Chicago/Turabian StyleZecconi, Alfonso, Francesca Zaghen, Gabriele Meroni, Flavio Sommariva, Silvio Ferrari, and Valerio Sora. 2025. "Machine Learning Approach for Early Lactation Mastitis Diagnosis Using Total and Differential Somatic Cell Counts" Animals 15, no. 8: 1125. https://doi.org/10.3390/ani15081125
APA StyleZecconi, A., Zaghen, F., Meroni, G., Sommariva, F., Ferrari, S., & Sora, V. (2025). Machine Learning Approach for Early Lactation Mastitis Diagnosis Using Total and Differential Somatic Cell Counts. Animals, 15(8), 1125. https://doi.org/10.3390/ani15081125






