Analysis of the Primary Pathogenic Bacteria in Abscess Disease of Musk Deer Using Metagenomic Approaches
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Pathological Section
2.3. DNA Extraction, Library Construction, and Sequencing
2.4. Bioinformatics Analyses
3. Results
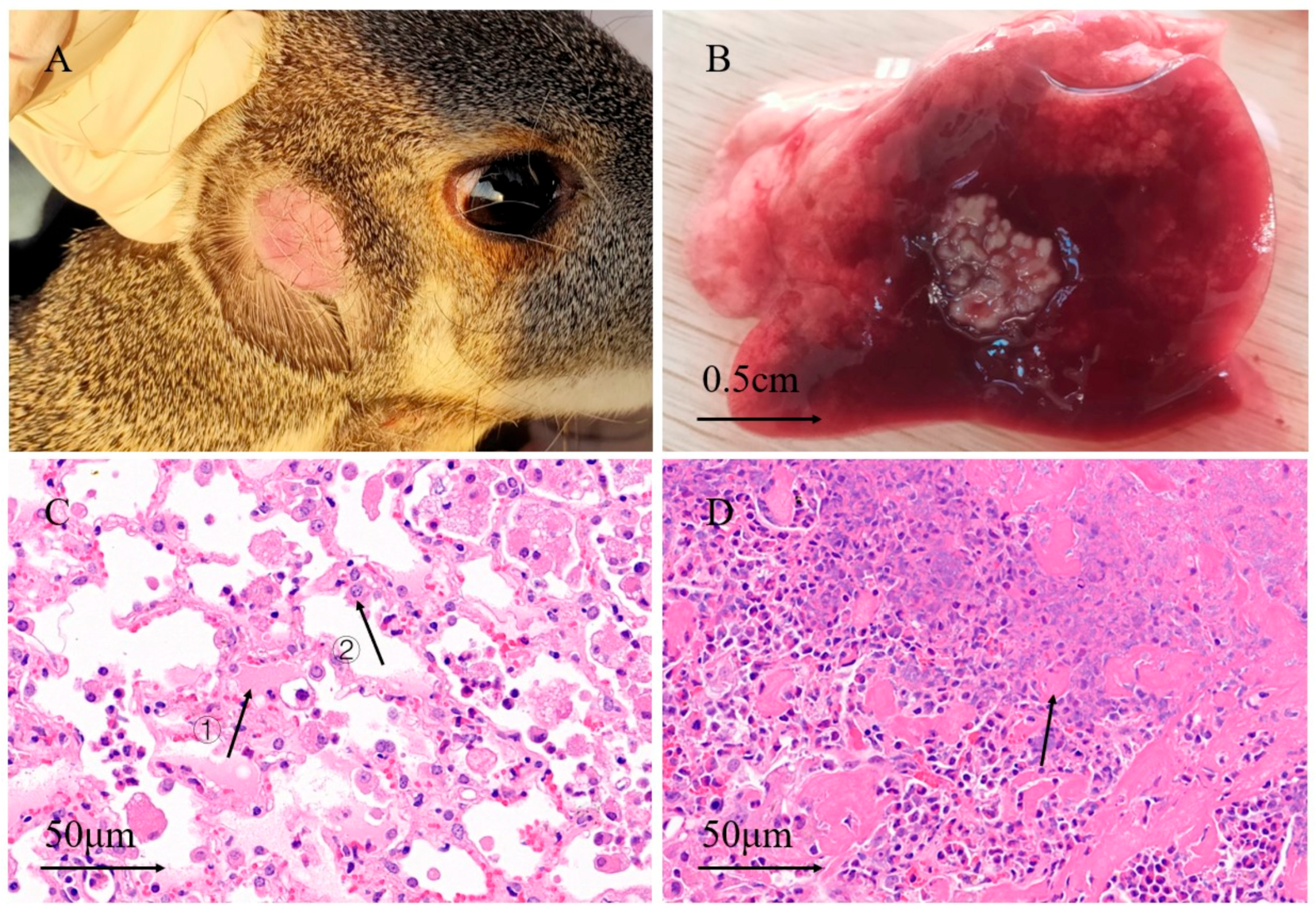
3.1. Anatomy and Pathological Sections
3.2. Raw Data Quality Control
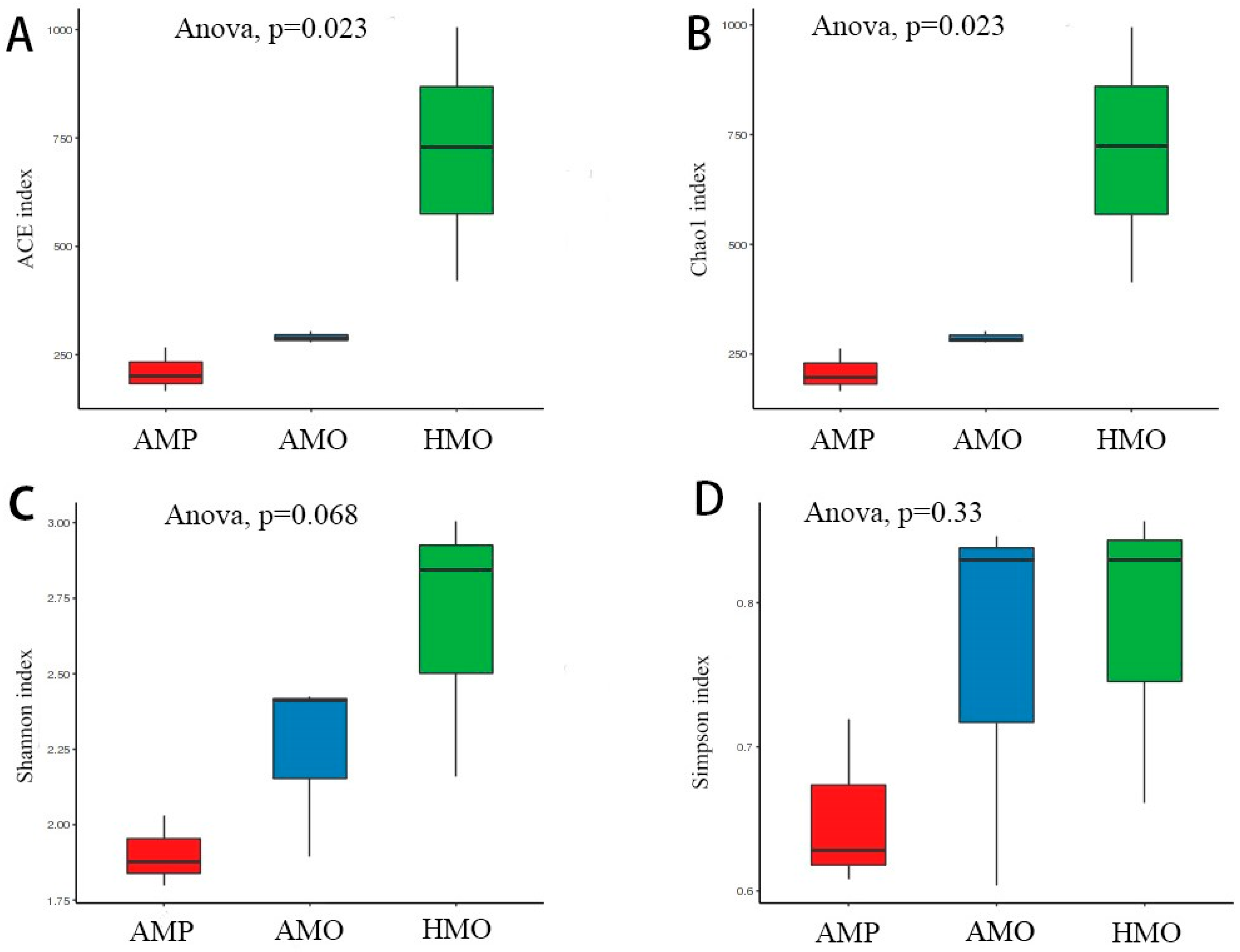
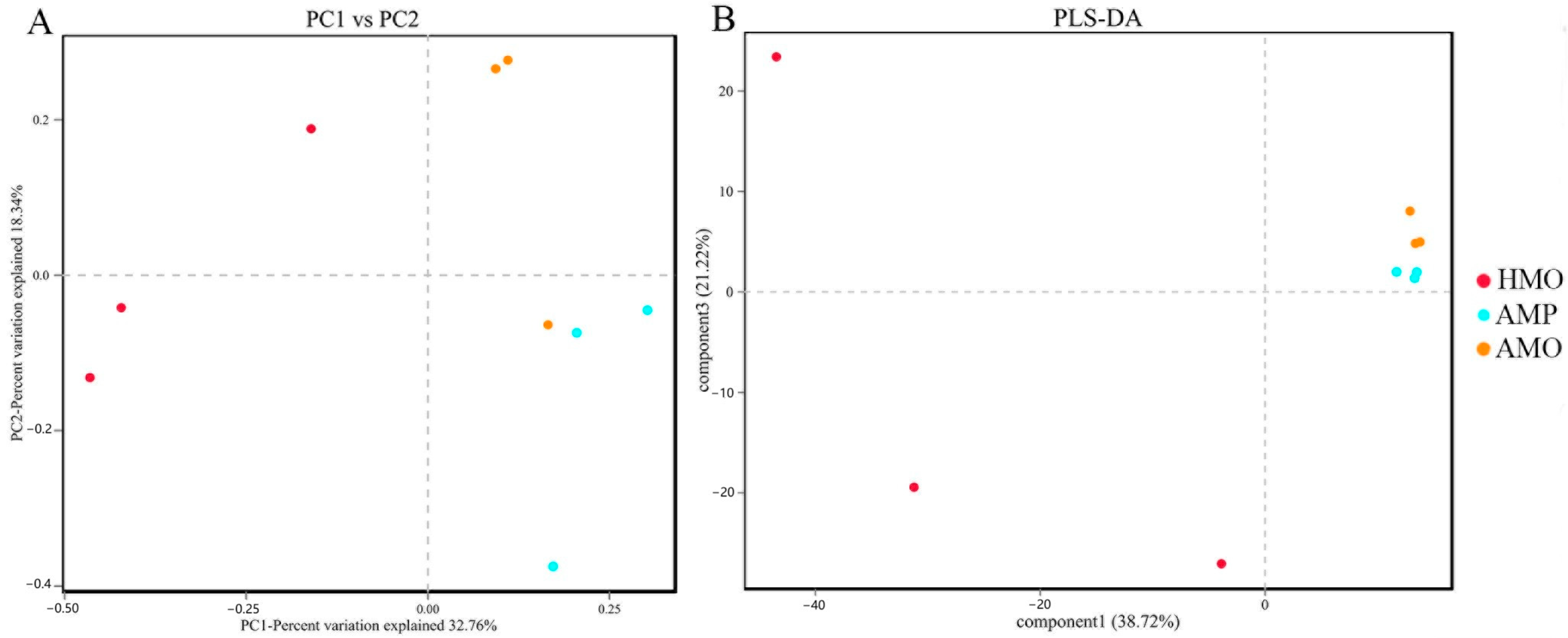
3.3. Biodiversity Composition Analysis Between Groups of Healthy and Abscessed Musk Deer
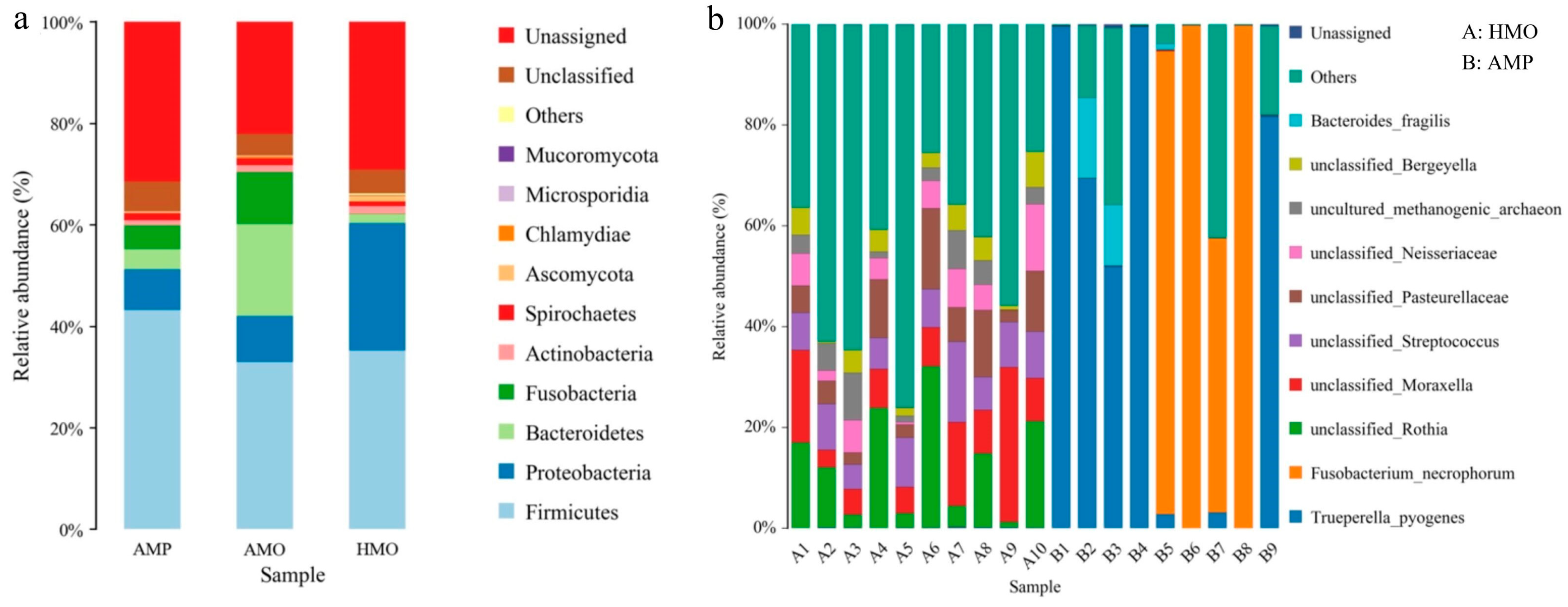
3.4. Species Annotation and Taxonomic Analysis
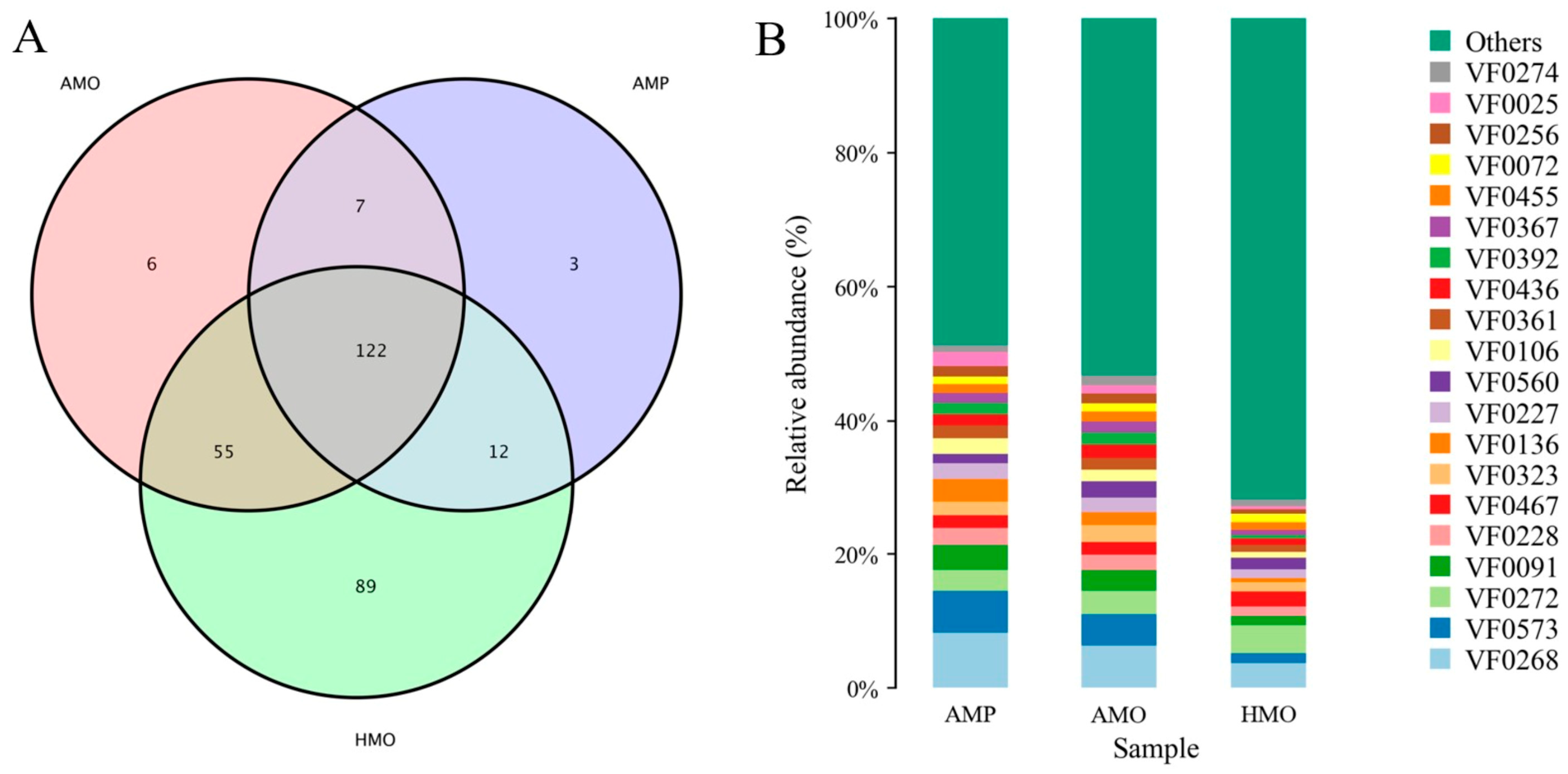
3.5. Annotation and Analysis of Virulence Factors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, J.Y.; Wang, W. The Musk Deer of China; Chinese Forestry Publishing House: Beijing, China, 2006. [Google Scholar]
- Li, L.H.; Huang, X.Y.; Liu, G.; Wang, W.X.; Wei, N.; Liu, Y.; Hu, D.F.; Meng, M. The Status of Captive Population of Musk Deer and Analysis of its Farming Development in China. J. Sichuan J. Zool. 2012, 31, 492–496. [Google Scholar]
- He, L.; Li, L.H.; Wang, W.X.; Liu, G.; Liu, S.Q.; Liu, W.H.; Meng, M.; Hu, D.F. Discussion about relationship between biological characters and farming development of musk deer. For. Resour. Manage 2014, 02, 26–29. [Google Scholar]
- Li, L.; Wang, B.B.; Ge, Y.F.; Wan, Q.H. Major histocompatibility complex class II polymorphisms in forest musk deer (Moschus berezovskii) and their probable association with purulent disease. Int. J. Immunogenetics 2014, 41, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Yan, Q.G.; Yang, G.Y. The mass diseases of captive musk deer. J. Econ. Anim. 2016, 20, 112–117. [Google Scholar]
- Li, Y.M.; Hu, X.L.; Yang, S.; Zhou, J.T.; Qi, L.; Sun, X.; Fan, M.; Xu, S.; Cha, M.; Zhang, M.; et al. Comparison Between the Fecal Bacterial Microbiota of Healthy and Diarrheic Captive Musk Deer. Front. Microbiol. 2018, 9, 300. [Google Scholar] [CrossRef]
- Wang, J.L.; Zhang, T.Q.; Zhu, C.H.S. Preliminary analysis on the death of forest musk deer. J. Anim. Husb. Vet. Med. 1990, 9, 36–39. [Google Scholar]
- Zhao, K.L.; Li, X.X.; Pa, P.; Zeng, B.; Zhang, X.Y.; Yue, B.S. Isolation and identification on Pathogens of Musk Deer Abscess Disease and Antibiotic Susceptibility Assay. Sichuan Anim. 2011, 30, 522–526. [Google Scholar]
- Zhao, K.L.; Liu, Y.; Zhang, X.Y.; Paha’erding Palahati Wang, H.N.; Yue, B.S. Detection and characterization of antibiotic-resistance genes in Arcanobacterium pyogenes strains from abscesses of forest musk deer. J. Med. Microbiol. 2011, 60, 1820–1826. [Google Scholar] [CrossRef]
- Heimesaat, M.M.; Escher, U.; Grunau, A.; Kühl, A.A.; Bereswill, S. Multidrug-Resistant Pseudomonas aeruginosa Accelerate Intestinal, Extra-Intestinal, and Systemic Inflammatory Responses in Human Microbiota-Associated Mice with Subacute Ileitis. Front. Immunol. 2019, 10, 49. [Google Scholar] [CrossRef]
- Cohen, B.S.; Belser, E.H.; Killmaster, C.H.; Bowers, J.W.; Irwin, B.J.; Yabsley, M.J.; Miller, K.V. Epizootiology of cranial abscess disease in white-tailed deer. (Odocoileus virginianus) of Georgia, USA. J. Wildl. Dis. 2015, 51, 609–618. [Google Scholar] [CrossRef]
- Joob, B.; Wiwanitkit, V. Klebsiella pneumoniae Invasive Liver Abscess Syndrome and Endophthalmitis. J. Emerg. Med. 2017, 53, 917. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Cheng, J.G.; Li, Q.B.; Cai, Y.H.; Jia, L.; Zhao, C. Isolation and identification of the pathogen of Escherichia coli suppurosis in musk deer. J. Heilongjiang Anim. Sci. Vet. Med. 2006, 11, 81–83. [Google Scholar]
- Li, Q.B.; Yan, Q.G.; Kang, J.P.; Li, P.; Xiong, W.P. Isolation and Identification of Purulent Bacteria from Forest Musk Deer (Moschus berezovskii). J. BBC Wildl. Mag. 2012, 30, 211–213. [Google Scholar]
- Tang, J.; Hu, H.; Wang, Y.Q.; Li, F.R.; Liu, W.H. Isolation and Identification of Corynebacterium Pyogenes from Forest Musk Deer. J. China Herbiv. 2011, 31, 71–73. [Google Scholar]
- Xi, L.X.; Chen, Z.R.; Song, H.Y.; Ren, L.; Wen, J.F.; Huang, J.; Yang, R.; Yang, G.Y.; Wang, H.Y.; Yan, Q.G. Identification and drug-resistance, pathogeny characteristics Pseudomonas aeruginosa from captive forest musk deer. J. Microbiol. 2018, 45, 386–394. [Google Scholar]
- Wu, S.Z.; Yang, L.F.; Qian, L.M.; Yang, M.H.; Li, Q.H.; Chen, P.F. Isolation and Identification of Pathogenic Bacteria for Abscess in Captive Forest Musk Deer (Moschus berezovskii). Chin. J. Wildl. 2021, 42, 872–878. [Google Scholar]
- Yeoh, Y.K.; Chan, M.H.; Chen, Z.; Lam, E.W.H.; Wong, P.Y.; Ngai, C.M.; Chan, P.K.S.; Hui, M. The human oral cavity microbiota composition during acute tonsillitis: A cross-sectional survey. BMC Oral Health 2019, 19, 275. [Google Scholar] [CrossRef]
- Kilian, M.; Chapple, I.L.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.L.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef]
- Pan, S.H.; Wan, Y.Y.; Zhang, K.B. Preliminary report on etiological diagnosis of subcutaneous and lymphatic abscess in goats. Guizhou Agric. Sci. 2002, 3, 63. [Google Scholar]
- Pillai, D.K.; Amachawadi, R.G.; Baca, G.; Narayanan, S.K.; Nagaraja, T.G. Leukotoxin production by Fusobacterium necrophorum strains in relation to severity of liver abscesses in cattle. Anaerobe 2021, 69, 102344. [Google Scholar] [CrossRef]
- Jensen, J.L.; Bergem, H.O.; Gilboe, I.M.; Husby, G.; Axéll, T. Oral and ocular sicca symptoms and findings are prevalent in systemic lupus erythematosus. J. Oral Pathol. Med. 1999, 28, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Wade, W.G. The oral microbiome in health and disease. Pharmacol. Res. 2013, 69, 137–143. [Google Scholar] [CrossRef]
- Kobayashi, R.; Ogawa, Y.; Hashizume-Takizawa, T.; Kurita-Ochiai, T. Oral bacteria affect the gut microbiome and intestinal immunity. Pathog. Dis. 2020, 78, ftaa024. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.T.; Li, J.L.; Yang, S.H.; Xia, D.S.; Zhang, C.M.; Wang, S.L. Comparative study on oral floras of minipigs, rats and human being. Beijing J. Stomatol. 2006, 3, 153–156. [Google Scholar]
- Jiang, F.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Cai, Z.; Gao, H.; Chi, X.; Zhang, T. Comparative analysis of gut microbial composition and potential functions in captive forest and alpine musk deer. Appl. Microbiol. Biotechnol. 2022, 106, 1325–1339. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Hu, X.L.; Yang, S.; Zhou, J.T.; Zhang, T.X.; Qi, L.; Sun, X.; Fan, M.; Xu, S.; Cha, M.; et al. Comparative Analysis of the Gut Microbiota Composition between Captive and Wild Forest Musk Deer. Front. Microbiol. 2017, 8, 1705. [Google Scholar] [CrossRef]
- Sims, T.T.; Colbert, L.E.; Zheng, J.; Delgado Medrano, A.Y.; Hoffman, K.L.; Ramondetta, L.; Jazaeri, A.; Jhingran, A.; Schmeler, K.M.; Daniel, C.R.; et al. Gut microbial diversity and genus-level differences identified in cervical cancer patients versus healthy controls. Gynecol. Oncol. 2019, 155, 237–244. [Google Scholar] [CrossRef]
- Rullo, J.; Far, P.M.; Quinn, M.; Sharma, N.; Bae, S.; Irrcher, I.; Sharma, S. Local oral and nasal microbiome diversity in age-related macular degeneration. Sci. Rep. 2020, 10, 3862. [Google Scholar] [CrossRef]
- Tadepalli, S.; Narayanan, S.K.; Stewart, G.C.; Chengappa, M.M.; Nagaraja, T.G. Fusobacterium necrophorum: A ruminal bacterium that invades liver to cause abscesses in cattle. Anaerobe 2009, 15, 36–43. [Google Scholar] [CrossRef]
- Shanthalingam, S.; Narayanan, S.; Batra, S.A.; Jegarubee, B.; Srikumaran, S. Fusobacterium necrophorum in North American Bighorn Sheep (Ovis canadensis) Pneumonia. J. Wildl. Dis. 2016, 52, 616–620. [Google Scholar] [CrossRef]
- Farooq, S.; Wani, S.A.; Hassan, M.N.; Aalamgeer, S.; Kashoo, Z.A.; Magray, S.N.; Bhat, M. The detection and prevalence of leukotoxin gene variant strains of Fusobacterium necrophorum in footrot lesions of sheep in Kashmir, India. Anaerobe 2018, 51, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.W.; Kumar, A.; Narayanan, S.; Myers, S.; Brown, K.; Nagaraja, T.G.; Jayarao, B.M. Characterization of Fusobacterium isolates from the respiratory tract of white-tailed deer (Odocoileus virginianus). J. Vet. Diagn. Investig. 2014, 26, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.J.; Zhang, H.Y.; Chen, L.Z.; Liu, X.Y.; Cheng, S.P.; Xu, M.; Miao, L.G.; Zhang, H.T.; Qian, G.C.; Yang, F.H.; et al. Isolation and Identification of the Causative agent of Necrobacillosis in Deer. Chin. J. Prev. Vet. Med. 2000, 84–87. [Google Scholar]
- Kjærulff, A.M.; Thomsen, M.K.; Ovesen, T.; Klug, T.E. Clinical and biochemical characteristics of patients with Fusobacterium necrophorum-positive acute tonsillitis. Eur. Arch. Oto-Rhino-Laryngol. 2015, 272, 1457–1463. [Google Scholar] [CrossRef]
- Narayanan, S.K.; Nagaraja, T.G.; Chengappa, M.M.; Stewart, G.C. Leukotoxins of gram-negative bacteria. Vet. Microbiol. 2002, 84, 337–356. [Google Scholar] [CrossRef] [PubMed]
- Batty, A.; Wren, M.W.; Gal, M. Fusobacterium necrophorum as the cause of recurrent sore throat: Comparison of isolates from persistent sore throat syndrome and Lemierre’s disease. J. Infect. 2005, 51, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Alfreijat, M. A case of I emierre’s Syndrome with a brief literature review. J. Infect. Public Health 2016, 9, 681–683. [Google Scholar] [CrossRef]
- Riordan, T. Human infection with Fusobacterium necrophorum (Necrobacillosis), with a focus on Lemierre’s syndrome. Clin. Microbiol. Rev. 2007, 20, 622–659. [Google Scholar] [CrossRef]
- Wang, Q.J.; Li, W.X.; Jin, H.; Zhang, X.Y.; Cai, S.; Jian, Y.N.; Xu, L.; Li, X.P.; Chao, S.Y.; Ma, L.Q. Diagnose of pathogens on liver abscess from lamb in Haixi area of QingHai Province. Chin. Qinghai J. Anim. Vet. Sci. 2017, 47, 21–23. [Google Scholar]
- Rzewuska, M.; Kwiecień, E.; Chrobak-Chmiel, D.; Kizerwetter-Świda, M.; Stefańska, I.; Gieryńska, M. Pathogenicity and Virulence of Trueperella pyogenes: A Review. Int. J. Mol. Sci. 2019, 20, 2737. [Google Scholar] [CrossRef]
- Zhou, Y.; Shao, Z.; Dai, G.; Li, X.; Xiang, Y.; Jiang, S.; Zhang, Z.; Ren, Y.; Zhu, Z.; Fan, C.; et al. Pathogenic infection characteristics and risk factors for bovine respiratory disease complex based on the detection of lung pathogens in dead cattle in Northeast China. J. Dairy Sci. 2023, 106, 589–606. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.G.; Risseti, R.M.; Bolaños, C.A.; Caffaro, K.A.; de Morais, A.C.; Lara, G.H.; Zamprogna, T.O.; Paes, A.C.; Listoni, F.J.; Franco, M.M.; et al. Trueperella pyogenes multispecies infections in domestic animals: A retrospective study of 144 cases (2002 to 2012). Vet. Q. 2015, 35, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Basis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. Mbio 2015, 6, e00037. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, T.M.; Juncos, V.A.; Zimmermann, K. Interactions between the Gut Microbiome, Lung Conditions, and Coronary Heart Disease and How Probiotics Affect These. Int. J. Mol. Sci. 2021, 22, 9700. [Google Scholar] [CrossRef]
- Sun, J.H.; Ying, J.; Li, X.L.; Hou, B.X.; Zhang, Z.T. Characteristics of oral microbiome in gastric cancer patients. J. Beijing J. Stomatol. Beijing J. Stom. 2018, 26, 5–10. [Google Scholar]
- Liu, B.; Zhang, A.H.; Zhang, Z. Study on the Relationship between UCA1, Nuclear Factor, Oral Flora and the Occurrence and Development of Oral Cancer. J. Pract. J. Cancer 2017, 32, 1423–1426. [Google Scholar]
- Yang, J.J.; Zhang, J.M.; Chen, B.; Meng, Q.S.; Shi, F.; Shi, G.S.; Zhang, L. Analysis of the oral microbiota and its potential association with gastric cancer. J. Pathog. Biol. 2017, 12, 631–634. [Google Scholar]
- Plata, M.R.; Contento, A.M.; Villaseñor, M.J.; Cabezas, M.L.; Ríos, A. Development of a novel biotoxicity screening assay for analytical use. Chemosphere 2009, 76, 959–966. [Google Scholar] [CrossRef]
- Pakbin, B.; Brück, W.M.; Rossen, J.W.A. Virulence Factors of Enteric Pathogenic Escherichia coli: A Review. Int. J. Mol. Sci. 2021, 22, 9922. [Google Scholar] [CrossRef]
- Veetilvalappil, V.V.; Manuel, A.; Aranjani, J.M.; Tawale, R.; Koteshwara, A. Pathogenic arsenal of Pseudomonas aeruginosa: An update on virulence factors. Future Microbiol. 2022, 17, 465–481. [Google Scholar] [CrossRef]
- Holt, S.C.; Kesavalu, L.; Walker, S.; Genco, C.A. Virulence factors of Porphyromonas gingivalis. Periodontology 2000 1999, 20, 168–238. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Watarai, M.; Cheun, H.I.; Shirahata, T.; Uchida, I. Effect of the lower molecular capsule released from the cell surface of Bacillus anthracis on the pathogenesis of anthrax. J. Infect. Dis. 2002, 186, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Vecchiarelli, A.G.; Havey, J.C.; Ing, L.L.; Wong, E.O.; Waples, W.G.; Funnell, B.E. Dissection of the ATPase active site of P1 ParA reveals multiple active forms essential for plasmid partition. J. Biol. Chem. 2013, 288, 17823–17831. [Google Scholar] [CrossRef]
- Strakova, N.; Korena, K.; Karpiskova, R. Klebsiella pneumoniae producing bacterial toxin colibactin as a risk of colorectal cancer development—A systematic review. Toxicon 2021, 197, 126–135. [Google Scholar] [CrossRef] [PubMed]
| Name | Sequence (5′–3′) |
|---|---|
| F | ACTCCTACGGGAGGCAGCA |
| R | GGACTACHVGGGTWTCTAAT |
| Sample ID | Clean Data Base (bp) | Number of Reads | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| AMP1 | 1,037,601,201 | 3,399,457 | 43.24 | 96.84 | 92.55 |
| AMP2 | 1,103,460,085 | 3,593,644 | 45.26 | 96.25 | 91.46 |
| AMP3 | 1,024,944,773 | 3,340,425 | 44.52 | 96.45 | 91.79 |
| AMO1 | 1,216,288,821 | 3,978,145 | 42.94 | 96.94 | 92.53 |
| AMO2 | 1,215,037,665 | 3,974,587 | 42.49 | 97.13 | 92.9 |
| AMO3 | 841,226,856 | 2,708,702 | 46.57 | 96.43 | 91.88 |
| HMO4 | 769,195,329 | 2,466,201 | 43.65 | 96.24 | 91.48 |
| HMO5 | 1,440,773,547 | 4,732,485 | 41.62 | 96.95 | 92.43 |
| HMO6 | 1,444,528,949 | 4,746,220 | 43.95 | 97.27 | 93.23 |
| Sample ID | Raw Reads | Clean Reads | Denoised Reads | Merged Reads | Nonchimeric Reads |
| HMO4 | 79,834 | 79,572 | 78,409 | 77,227 | 71,391 |
| HMO5 | 80,328 | 80,073 | 79,386 | 78,940 | 75,743 |
| HMO6 | 80,156 | 79,857 | 78,035 | 76,232 | 70,728 |
| HMO7 | 79,927 | 79,651 | 77,793 | 75,756 | 70,498 |
| HMO8 | 80,051 | 79,801 | 78,544 | 76,761 | 71,292 |
| HMO9 | 80,131 | 79,858 | 78,184 | 76,142 | 71,194 |
| HMO10 | 79,981 | 79,731 | 79,048 | 78,515 | 72,828 |
| HMO11 | 80,035 | 79,762 | 78,962 | 78,601 | 72,107 |
| HMO12 | 79,940 | 79,664 | 78,201 | 76,634 | 68,675 |
| HMO13 | 79,909 | 79,658 | 79,067 | 78,758 | 75,252 |
| AMO4 | 80,349 | 80,120 | 79,813 | 79,704 | 79,685 |
| AMO5 | 79,953 | 79,704 | 78,430 | 77,336 | 75,599 |
| AMO6 | 80,018 | 79,762 | 78,012 | 76,522 | 73,992 |
| AMO7 | 80,083 | 79,842 | 79,563 | 79,474 | 79,458 |
| AMO8 | 60,624 | 60,448 | 60,057 | 59,898 | 58,629 |
| AMO9 | 79,429 | 79,169 | 78,982 | 78,934 | 77,530 |
| AMO10 | 79,812 | 79,591 | 79,284 | 78,872 | 67,972 |
| AMO11 | 79,944 | 79,698 | 79,491 | 79,429 | 78,005 |
| AMO12 | 79,740 | 79,497 | 77,661 | 75,727 | 75,780 |
| ID | VF_Name | VF_Function |
|---|---|---|
| VF0268 * | HitABC | HitABC(fbpABC) operon |
| VF0573 * | Colibactin | Inducing DNA damage |
| VF0272 | FbpABC | periplasmic Fe3+ binding protein |
| VF0091 * | Alginate | Forming biofilm |
| VF0228 * | Enterobactin | Iron uptake |
| VF0467 | Acinetobactin | High-affinity catechol-hydroxamate siderophore |
| VF0323 * | Capsule | Bacterial survival, persistance, evasion of host immune response |
| VF0136 * | Yersiniabactin | FyuA/Psn-Irp system uses yersiniabactin |
| VF0227 * | Chu | Iron uptake |
| VF0560 | Capsule | Assisting in evading the host immune system |
| VF0106 * | MgtBC | Intracellular survival or for virulence |
| VF0361 * | Capsule | Contributes to host immune evasion |
| VF0436 * | Capsule I | Key virulence determinant |
| VF0392 * | O-antigen | LPS O antigen mutants |
| VF0367 * | LPS | Early survival inside macrophage |
| VF0455 * | MntABC | Quenches ROS |
| VF0072 | ClpC | ATPase promoting early escape |
| VF0256 * | Shu | Shu heme utilization locus |
| VF0025 * | FHA | Adherence to ciliated respiratory epithelial cells |
| VF0274 | Capsule | Inhibits the complement factor C3b |
| Others |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; An, X.; Yang, P.; Tan, R.; Chen, T.; Chen, J.; Tao, Y.; Li, X.; Sun, R.; Zhang, S.; et al. Analysis of the Primary Pathogenic Bacteria in Abscess Disease of Musk Deer Using Metagenomic Approaches. Animals 2025, 15, 1105. https://doi.org/10.3390/ani15081105
Hu J, An X, Yang P, Tan R, Chen T, Chen J, Tao Y, Li X, Sun R, Zhang S, et al. Analysis of the Primary Pathogenic Bacteria in Abscess Disease of Musk Deer Using Metagenomic Approaches. Animals. 2025; 15(8):1105. https://doi.org/10.3390/ani15081105
Chicago/Turabian StyleHu, Jingyao, Xian An, Pengcheng Yang, Rongzeng Tan, Taoyue Chen, Jiatong Chen, Yifan Tao, Xuxin Li, Runbin Sun, Shouyun Zhang, and et al. 2025. "Analysis of the Primary Pathogenic Bacteria in Abscess Disease of Musk Deer Using Metagenomic Approaches" Animals 15, no. 8: 1105. https://doi.org/10.3390/ani15081105
APA StyleHu, J., An, X., Yang, P., Tan, R., Chen, T., Chen, J., Tao, Y., Li, X., Sun, R., Zhang, S., Liu, S., & Yang, L. (2025). Analysis of the Primary Pathogenic Bacteria in Abscess Disease of Musk Deer Using Metagenomic Approaches. Animals, 15(8), 1105. https://doi.org/10.3390/ani15081105




