Application of Omics in Donkey Meat Research: A Review
Simple Summary
Abstract
1. Introduction
2. Literature Search Methodology
3. Characteristics of Donkey Meat
3.1. Donkey Meat Compositions
3.2. Physical and Chemical Properties of Donkey Meat
3.3. Health Benefits
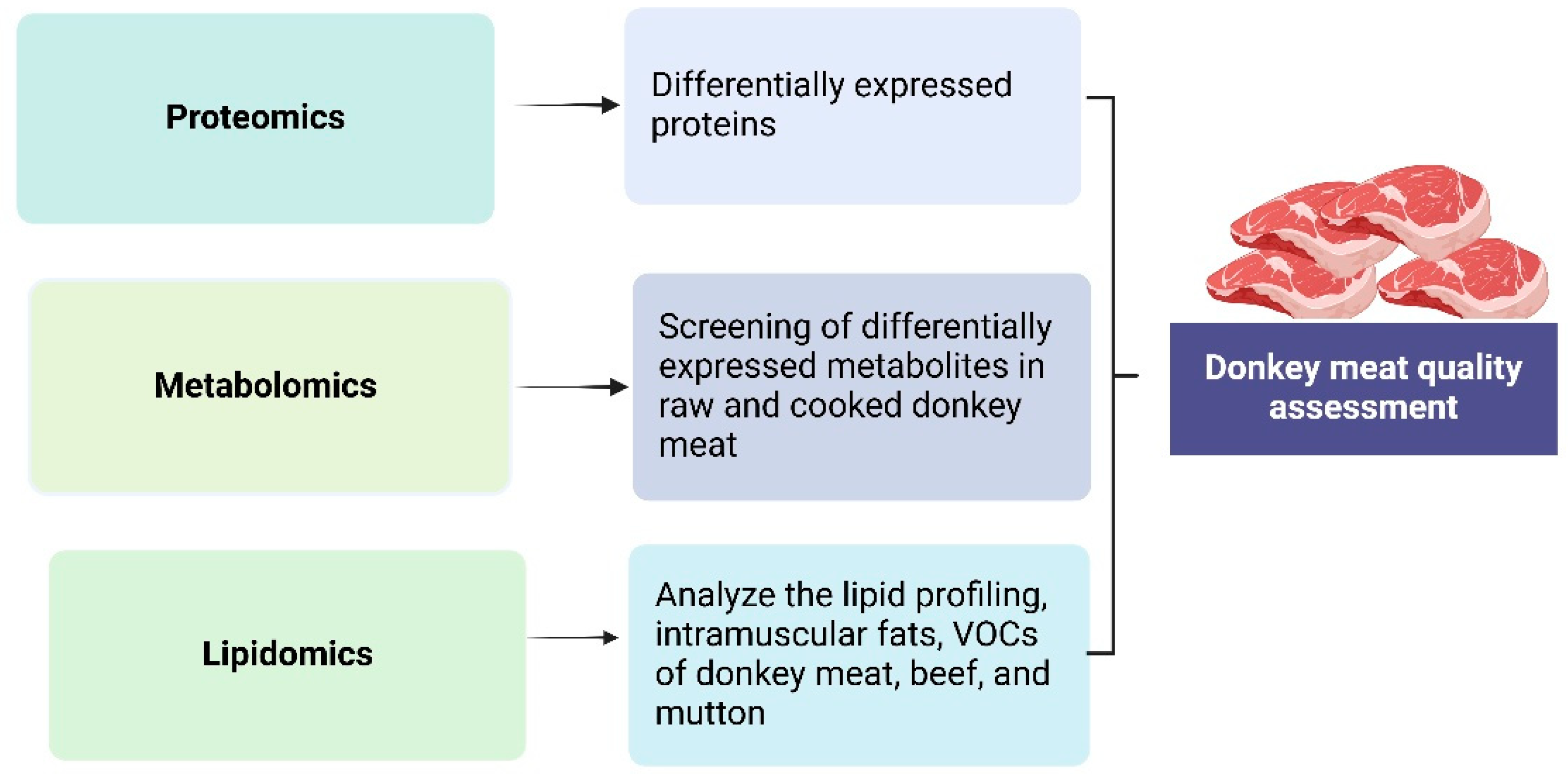
4. Omic Applications in Donkey Meat Research
4.1. Proteomic Applications in Donkey Meat Research
4.2. Lipidomic Applications in Donkey Meat Research
4.3. Metabolomic Applications in Donkey Meat Research
4.4. Genomic and Transcriptomic Approaches for Screening Potential Candidate Genes Associated with Meat Phenotypic Traits
5. Authentication Methods for Donkey Meat
6. Influencing Factors of Donkey Meat Quality and Nutritional Value
6.1. Breed Variation
6.2. Age Effects
6.3. Feeding Management Impact
6.4. Storage and Processing Considerations
7. Research Gaps
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Todd, E.T.; Tonasso-Calvière, L.; Chauvey, L.; Schiavinato, S.; Fages, A.; Seguin-Orlando, A.; Clavel, P.; Khan, N.; Pérez Pardal, L.; Patterson Rosa, L.; et al. The genomic history and global expansion of domestic donkeys. Science 2022, 377, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Stine, R.; Fiona, M.; Joris, P.; Tom, P.; DAM, O.; David, C. Domestication of the donkey: Timing, processes, and indicators. Proc. Natl. Acad. Sci. USA 2008, 105, 3715–3720. [Google Scholar]
- Seyiti, S.; Kelimu, A. Donkey industry in China: Current aspects, suggestions and future challenges. J. Equine Vet. Sci. 2021, 102, 103642. [Google Scholar] [CrossRef]
- Aroua, M.; Haj Koubaier, H.; Rekik, C.; Fatica, A.; Ben Said, S.; Malek, A.; Mahouachi, M.; Salimei, E. Comparative study of carcass characteristics and meat quality of local mediterranean donkey breeds. Foods 2024, 13, 942. [Google Scholar] [CrossRef] [PubMed]
- Camillo, F.; Rota, A.; Biagini, L.; Tesi, M.; Fanelli, D.; Panzani, D. The Current Situation and Trend of Donkey Industry in Europe. J. Equine Vet. Sci. 2018, 65, 44–49. [Google Scholar] [CrossRef]
- Khan, M.Z.; Chen, W.; Li, M.; Ren, W.; Huang, B.; Kou, X.; Ullah, Q.; Wei, L.; Wang, T.; Khan, A. Is there sufficient evidence to support the health benefits of including donkey milk in the diet? Front. Nutr. 2024, 11, 1404998. [Google Scholar] [CrossRef]
- Huang, B.; Khan, M.Z.; Chai, W.; Ullah, Q.; Wang, C. Exploring genetic markers: Mitochondrial dna and genomic screening for biodiversity and production traits in donkeys. Animals 2023, 13, 2725. [Google Scholar] [CrossRef]
- Guo, H.Y.; Pang, K.; Zhang, X.Y.; Zhao, L.; Chen, S.W.; Dong, M.L.; Ren, F.Z. Composition, Physiochemical Properties, Nitrogen Fraction Distribution, and Amino Acid Profile of Donkey Milk. J. Dairy Sci. 2007, 90, 1635–1643. [Google Scholar] [CrossRef]
- Claeys, W.L.; Verraes, C.; Cardoen, S.; De Block, J.; Huyghebaert, A.; Raes, K.; Dewettinck, K.; Herman, L. Consumption of raw or heated milk from different species: An evaluation of the nutritional and potential health benefits. Food Control 2014, 42, 188–201. [Google Scholar] [CrossRef]
- Changfa, W.; Haijing, L.; Yu, G.; Jinming, H.; Yan, S.; Jiumeng, M.; Jinpeng, W.; Xiaodong, F.; Zicheng, Z.; Shuai, W. Donkey genomes provide new insights into domestication and selection for coat color. Nat. Commun. 2020, 11, 6014. [Google Scholar]
- Min, W.; Haijing, L.; Xinhao, Z.; Li, Y.; Yu, L.; Shuqin, L.; Yujiang, S.; Chunjiang, Z. An analysis of skin thickness in the Dezhou donkey population and identification of candidate genes by RNA-seq. Anim. Genet. 2022, 53, 368–379. [Google Scholar]
- Wang, X.; Peng, Y.; Liang, H.; Khan, M.Z.; Ren, W.; Huang, B.; Chen, Y.; Xing, S.; Zhan, Y.; Wang, C. Comprehensive transcriptomic analysis unveils the interplay of mRNA and LncRNA expression in shaping collagen organization and skin development in Dezhou donkeys. Front. Genet. 2024, 15, 1335591. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ren, W.; Peng, Y.; Khan, M.Z.; Liang, H.; Zhang, Y.; Liu, X.; Chen, Y.; Kou, X.; Wang, L.; et al. Elucidating the Role of Transcriptomic Networks and DNA Methylation in Collagen Deposition of Dezhou Donkey Skin. Animals 2024, 14, 1222. [Google Scholar] [CrossRef]
- Yan, L.; Qingshan, M.; Xiaoyuan, S.; Wenmin, Y.; Guiqin, L.; Changfa, W. Comparative Transcriptome Analysis of Slow-Twitch and Fast-Twitch Muscles in Dezhou Donkeys. Genes 2022, 13, 1610. [Google Scholar] [CrossRef]
- Jiafei, S.; Jie, Y.; Xuelei, D.; Mei, L.; Gang, W.; Ningbo, C.; Hong, C.; Chuzhao, L.; Ruihua, D. Genomic analyses reveal distinct genetic architectures and selective pressures in Chinese donkeys. J. Genet. Genom. 2021, 48, 737–745. [Google Scholar]
- Bingjian, H.; Zahoor, K.M.; Yinghui, C.; Huili, L.; Xiyan, K.; Xinrui, W.; Wei, R.; Changfa, W.; Zhenwei, Z. Yeast polysaccharide supplementation: Impact on lactation, growth, immunity, and gut microbiota in Dezhou donkeys. Front. Microbiol. 2023, 14, 1289371. [Google Scholar]
- Liang, R.; Le Xu Fan, C.; Cao, L.; Guo, X. Structural Characteristics and Antioxidant Mechanism of Donkey-Hide Gelatin Peptides by Molecular Dynamics Simulation. Molecules 2023, 28, 7975. [Google Scholar] [CrossRef]
- Qingshan, M.; Xiyan, K.; Youyou, Y.; Yunshuang, Y.; Weihai, X.; Xiaohui, F.; Guiqin, L.; Changfa, W.; Yan, L. Comparison of Lipids and Volatile Compounds in Dezhou Donkey Meat with High and Low Intramuscular Fat Content. Foods 2023, 12, 3269. [Google Scholar] [CrossRef]
- Marino, R.; Albenzio, M.; Malva, A.D.; Muscio, A.; Sevi, A. Nutritional properties and consumer evaluation of donkey bresaola and salami: Comparison with conventional products. Meat Sci. 2015, 101, 19–24. [Google Scholar]
- Ahmed, A.M.; Sanka, J.S.; Sani, A. Observations on the Phenotyphic Characteristics and Management of Donkey in Sokoto, Northwestern Nigeria. Sch. J. Agric. Vet. Sci. 2018, 5, 1–5. [Google Scholar]
- Wang, Y.; Hua, X.; Shi, X.; Wang, C. Origin, Evolution, and Research Development of Donkeys. Genes 2022, 13, 1945. [Google Scholar] [CrossRef] [PubMed]
- Matlhola, D.M.; Chen, R. Telecoupling of the Trade of Donkey-Hides between Botswana and China: Challenges and Opportunities. Sustainability 2020, 12, 1730. [Google Scholar] [CrossRef]
- Ivanković, A.; Bittante, G.; Šubara, G.; Šuran, E.; Ivkić, Z.; Pećina, M.; Konjačić, M.; Kos, I.; Kelava Ugarković, N.; Ramljak, J. Genetic and Population Structure of Croatian Local Donkey Breeds. Diversity 2022, 14, 322. [Google Scholar] [CrossRef]
- Davis, E. Donkey and Mule Welfare. Vet. Clin. N. Am. Equine Pract. 2019, 35, 481–491. [Google Scholar]
- Legarra, A.; Christensen, O.F. Genomic evaluation methods to include intermediate correlated features such as high-throughput or omics phenotypes* *Presented as part of the Breeding and Genetics Symposium: Beyond Genetic Markers—Additional Data to Improve Long-Term Selection held at the ADSA Annual Meeting, June 2022. JDS Commun. 2023, 4, 55–60. [Google Scholar]
- Planell, N.; Lagani, V.; Sebastian-Leon, P.; van der Kloet, F.; Ewing, E.; Karathanasis, N.; Urdangarin, A.; Arozarena, I.; Jagodic, M.; Tsamardinos, I. STATegra: Multi-Omics Data Integration—A conceptual scheme with a bioinformatics pipeline. Front. Genet. 2021, 12, 620453. [Google Scholar]
- Krassowski, M.; Das, V.; Sahu, S.K.; Misra, B.B. State of the field in multi-omics research: From computational needs to data mining and sharing. Front. Genet. 2020, 11, 610798. [Google Scholar]
- Yugi, K.; Kubota, H.; Hatano, A.; Kuroda, S. trans-omics: How to reconstruct biochemical networks across multiple ‘Omic’ layers. Trends. Biotechnol. 2016, 34, 276–290. [Google Scholar]
- Gong, Y.; Li, Y.; Liu, X.; Ma, Y.; Jiang, L. A review of the pangenome: How it affects our understanding of genomic variation, selection and breeding in domestic animals? J. Anim. Sci. Biotechnol. 2023, 14, 73. [Google Scholar]
- Tan, X.; He, Z.; Fahey, A.G.; Zhao, G.; Liu, R.; Wen, J. Research progress and applications of genome-wide association study in farm animals. Anim. Res. One Health 2023, 1, 56–77. [Google Scholar]
- Wang, Y.; Miao, X.; Zhao, Z.; Wang, Y.; Li, S.; Wang, C. Transcriptome atlas of 16 donkey tissues. Front. Genet. 2021, 12, 682734. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Jiang, H.; Ma, F.; Li, S.; Guo, Y.; Zhu, L.; Shi, C.; Na, R.; Wang, Y.; Zhang, W. Multi-Omics Analysis Revealed the Molecular Mechanisms Affecting Average Daily Gain in Cattle. Int. J. Mol. Sci. 2025, 26, 2343. [Google Scholar] [CrossRef] [PubMed]
- Parsad, R.; Bagiyal, M.; Ahlawat, S.; Arora, R.; Gera, R.; Chhabra, P.; Sharma, U. Unraveling the genetic and physiological potential of donkeys: Insights from genomics, proteomics, and metabolomics approaches. Mamm. Genome 2024, 36, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Behren, L.E.; König, S.; May, K. Genomic Selection for Dairy Cattle Behaviour Considering Novel Traits in a Changing Technical Production Environment. Genes 2023, 14, 1933. [Google Scholar] [CrossRef]
- Kertz, N.C.; Banerjee, P.; Dyce, P.W.; Diniz, W.J.S. Harnessing Genomics and Transcriptomics Approaches to Improve Female Fertility in Beef Cattle—A Review. Animals 2023, 13, 3284. [Google Scholar] [CrossRef]
- Ashokan, M.; Rana, E.; Sneha, K.; Namith, C.; Naveen Kumar, G.S.; Azharuddin, N.; Elango, K.; Jeyakumar, S.; Ramesha, K.P. Metabolomics—A powerful tool in livestock research. Anim. Biotechnol. 2023, 34, 3237–3249. [Google Scholar] [CrossRef]
- Fabrile, M.P.; Ghidini, S.; Conter, M.; Varrà, M.O.; Ianieri, A.; Zanardi, E. Filling gaps in animal welfare assessment through metabolomics. Front. Vet. Sci. 2023, 10, 1129741. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, S.; Wang, C.; Xu, R.; Wang, W.; Jiang, Y.; Wang, M.; Jiang, L.; Dai, L.; Wang, J. Pangenome obtained by long-read sequencing of 11 genomes reveal hidden functional structural variants in pigs. iScience 2023, 26, 106119. [Google Scholar] [CrossRef]
- Lai, X.; Zhang, Z.; Zhang, Z.; Liu, S.; Bai, C.; Chen, Z.; Qadri, Q.R.; Fang, Y.; Wang, Z.; Pan, Y. Integrated microbiome-metabolome-genome axis data of Laiwu and Lulai pigs. Sci. Data 2023, 10, 280. [Google Scholar] [CrossRef]
- Zeng, H.; Zhang, W.; Lin, Q.; Gao, Y.; Teng, J.; Xu, Z.; Cai, X.; Zhong, Z.; Wu, J.; Liu, Y. PigBiobank: A valuable resource for understanding genetic and biological mechanisms of diverse complex traits in pigs. Nucleic Acids Res. Acids Res. 2023, 52, D980–D989. [Google Scholar] [CrossRef]
- Qiao, C.; He, M.; Wang, S.; Jiang, X.; Wang, F.; Li, X.; Tan, S.; Chao, Z.; Xin, W.; Gao, S. Multi-omics analysis reveals substantial linkages between the oral-gut microbiomes and inflamm-aging molecules in elderly pigs. Front. Microbiol. 2023, 14, 1250891. [Google Scholar] [CrossRef] [PubMed]
- Kasper, C.; Ribeiro, D.; Almeida, A.M.D.; Larzul, C.; Liaubet, L.; Murani, E. Omics Application in Animal Science—A Special Emphasis on Stress Response and Damaging Behaviour in Pigs. Genes 2020, 11, 920. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fu, Y.; Su, T.; Wang, Y.; Soladoye, O.P.; Huang, Y.; Zhao, Z.; Zhao, Y.; Wu, W. A Role of Multi-Omics Technologies in Sheep and Goat Meats: Progress and Way Ahead. Foods 2023, 12, 4069. [Google Scholar] [CrossRef] [PubMed]
- Rice, E.S.; Alberdi, A.; Alfieri, J.; Athrey, G.; Balacco, J.R.; Bardou, P.; Blackmon, H.; Charles, M.; Cheng, H.H.; Fedrigo, O. A pangenome graph reference of 30 chicken genomes allows genotyping of large and complex structural variants. BMC Biol. 2023, 21, 267. [Google Scholar]
- Jia, W.; Di, C.; Shi, L. Applications of lipidomics in goat meat products: Biomarkers, structure, nutrition interface and future perspectives. J. Proteom. 2023, 270, 104753. [Google Scholar] [CrossRef]
- Mao, X.; Bassey, A.P.; Sun, D.; Yang, K.; Shan, K.; Li, C. Overview of omics applications in elucidating the underlying mechanisms of biochemical and biological factors associated with meat safety and nutrition. J. Proteom. 2023, 276, 104840. [Google Scholar]
- Ramanathan, R.; Kiyimba, F.; Suman, S.P.; Mafi, G.G. The potential of metabolomics in meat science: Current applications, trends, and challenges. J. Proteomics 2023, 283–284, 104926. [Google Scholar] [CrossRef]
- Jiang, N.; Wu, R.; Wu, C.; Wang, R.; Wu, J.; Shi, H. Multi-omics approaches to elucidate the role of interactions between microbial communities in cheese flavor and quality. Food Rev. Int. 2023, 39, 5446–5458. [Google Scholar]
- Yuan, L.; Dai, H.; He, G.; Yang, Z.; Jiao, X. Invited review: Current perspectives for analyzing the dairy biofilms by integrated multiomics. J. Dairy. Sci. 2023, 106, 8181–8192. [Google Scholar]
- Polidori, P.; Cavallucci, C.; Beghelli, D.; Vincenzetti, S. Physical and chemical characteristics of donkey meat from Martina Franca breed. Meat Sci. 2009, 82, 469–471. [Google Scholar] [CrossRef]
- Khan, M.Z.; Chen, W.; Wang, X.; Liang, H.; Wei, L.; Huang, B.; Kou, X.; Liu, X.; Zhang, Z.; Chai, W.; et al. A review of genetic resources and trends of omics applications in donkey research: Focus on China. Front. Vet. Sci. 2024, 11, 1366128. [Google Scholar] [CrossRef] [PubMed]
- Polidori, P.; Pucciarelli, S.; Ariani, A.; Polzonetti, V.; Vincenzetti, S. A comparison of the carcass and meat quality of Martina Franca donkey foals aged 8 or 12months. Meat Sci. 2015, 106, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Chiofalo, B.; Dugo, P.; Bonaccorsi, I.L.; Mondello, L. Comparison of major lipid components in human and donkey milk: New perspectives for a hypoallergenic diet in humans. Immunopharmacol. Immunotoxicol. 2011, 33, 633–644. [Google Scholar]
- Garhwal, R.; Bhardwaj, A.; Sangwan, K.; Mehra, R.; Pal, Y.; Nayan, V.; Iquebal, M.A.; Jaiswal, S.; Kumar, H. Milk from Halari Donkey Breed: Nutritional Analysis, Vitamins, Minerals, and Amino Acids Profiling. Foods 2023, 12, 853. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, D.; Chai, W.; Zhu, M.; Wang, Y.; Liu, Y.; Wei, Q.; Fan, D.; Lv, M.; Jiang, X. Chemical and physical properties of meat from Dezhou black donkey:Original papers. Food Sci. Technol. Res. 2022, 28, 87–94. [Google Scholar]
- Frank, D.; Kaczmarska, K.; Paterson, J.; Piyasiri, U.; Warner, R. Effect of marbling on volatile generation, oral breakdown and in mouth flavor release of grilled beef. Meat Sci. 2017, 133, 61–68. [Google Scholar]
- Li, M.; Sun, L.; Du, X.; Ren, W.; Man, L.; Chai, W.; Zhu, M.; Liu, G.; Wang, C. Characterization of lipids and volatile compounds in boiled donkey meat by lipidomics and volatilomics. J. Food Sci. 2024, 89, 3445–3454. [Google Scholar]
- Polidori, P.; Cammertoni, N.; Santini, G.; Klimanova, Y.; Zhang, J.; Vincenzetti, S. Effects of Donkeys Rearing System on Performance Indices, Carcass, and Meat Quality. Foods 2021, 10, 3119. [Google Scholar] [CrossRef]
- Pereira, P.M.D.C.; Vicente, A.F.D.R. Meat nutritional composition and nutritive role in the human diet. Meat Sci. 2013, 93, 586–592. [Google Scholar]
- Paolo, P.; Paola, D.G.; Silvia, V. Vitamins and Minerals in Raw and Cooked Donkey Meat; Chhabi, L.R., Ed.; IntechOpen: Rijeka, Croatia, 2021. [Google Scholar]
- Ivanković, A.; Šubara, G.; Bittante, G.; Šuran, E.; Amalfitano, N.; Aladrović, J.; Kelava Ugarković, N.; Pađen, L.; Pećina, M.; Konjačić, M. Potential of Endangered Local Donkey Breeds in Meat and Milk Production. Animals 2023, 13, 2146. [Google Scholar] [CrossRef]
- Salazar-Pressler, F.; Melo-Ruíz, V.; Sánchez-Herrera, K.; Gazga-Urioste, F.L.A.C. Mineral Composition of the Donkey (Equus asinus) Muscle Meat. J. Life Sci. 2018, 12, 100–104. [Google Scholar] [CrossRef]
- Campo, M.M.; Romero, J.V.; Guerrero, A.; Bouzaida, M.D.; Resconi, V.C.; Tesniere, G.; Santolaria, P.; Olleta, J.L. Nutrient composition of beef from the pyrenees. J. Food Compos. Anal. 2024, 133, 106452. [Google Scholar] [CrossRef]
- Mortensen, E.G.; Fuerniss, H.F.; Legako, J.F.; Thompson, L.D.; Woerner, D.R. Nutrient Analysis of Raw and Cooked USDA Prime Beef Cuts. Nutrients 2024, 16, 2912. [Google Scholar] [CrossRef]
- Gamage, N.H.; Giotto, F.; Fonseca, M.A.; de Mello, A. PSX-14 Effects of Forage and Grain-Based Finishing Diets on Fatty Acid Profile of Angus Steers Backgrounded Either in a Moderate Or in a High Plane of Nutrition. J. Anim. Sci. 2023, 101, 513–514. [Google Scholar] [CrossRef]
- Wang, Z.; You, W.; Hu, X.; Cheng, H.; Song, E.; Hu, Z.; Jiang, F. Effects of Capsicum oleoresin on the Growth Performance, Nutrient Digestibility and Meat Quality of Fattening Beef Cattle. Ruminants 2025, 5, 5. [Google Scholar] [CrossRef]
- Vicente, F.; Pereira, P.C. Pork Meat Composition and Health: A Review of the Evidence. Foods 2024, 13, 1905. [Google Scholar] [CrossRef] [PubMed]
- Dragica, N.; Sasa, J.; Nenad, P.; Vladimir, K.; Nikola, S.; Lato, P.; Mila, L. Nutrient Composition of Three Mangulica Pork Cuts from Serbia. Biol. Trace Elem. Res. 2017, 184, 369–377. [Google Scholar]
- Sun, Z.; Liu, D.; An, S.; Wu, X.; Zhang, J.; Miao, Z. Effects of Acorns on Fatty Acid Composition and Lipid Metabolism in Adipose Tissue of Yuxi Black Pigs. Animals 2024, 14, 3271. [Google Scholar] [CrossRef]
- Vulić, A.; Cvetnić, Ž.; Kos, I.; Vnučec, I.; Vahčić, N.; Lešić, T.; Simonović, D.; Kudumija, N.; Pleadin, J. Comparison of the Nutritional Composition of Meat Products Derived from Croatian Indigenous Pig Breeds. Foods 2024, 13, 4175. [Google Scholar] [CrossRef]
- Zduńczyk, W.; Tkacz, K.; Fiećko, R.P.; Bottari, B.; Kapituła, M.M. Pork as a source of nutrients in a human diet—Comparison of meat obtained from conventional and organic systems offered in the Polish market. Nfs J. 2024, 37, 100199. [Google Scholar]
- Qiangqiang, C.; Wei, Z.; Lixia, X.; Qian, S.; Fen, W.; Guoliang, L.; Yuan, W.; Yuchun, P.; Qishan, W.; Jinzhi, Z. Multi-Omics Reveals the Effect of Crossbreeding on Some Precursors of Flavor and Nutritional Quality of Pork. Foods 2023, 12, 3237. [Google Scholar] [CrossRef]
- Dos Santos, I.J.; Dias Junior, P.C.G.; Alvarenga, T.I.R.C.; Pereira, I.G.; Gallo, S.B.; Alvarenga, F.A.P.; Furusho-Garcia, I.F. Impact of Feeding Systems on Performance, Blood Parameters, Carcass Traits, Meat Quality, and Gene Expressions of Lambs. Agriculture 2024, 14, 957. [Google Scholar] [CrossRef]
- Sun, L.; Jiang, H. Research on meat quality of Qianhua sheep Merino sheep and Small-tail Han sheep. Open Life Sci. 2022, 17, 1315–1323. [Google Scholar] [CrossRef]
- Kong, D.; Han, R.; Yuan, M.; Xi, Q.; Du, Q.; Li, P.; Yang, Y.; Rahman, S.M.E.; Wang, J. Slightly acidic electrolyzed water as a novel thawing media combined with ultrasound for improving thawed sheep quality, nutrients and microstructure. Food Chem. X 2023, 18, 100630. [Google Scholar] [PubMed]
- Xu, X.; Guo, T.; Zhang, Q.; Liu, H.; Wang, X.; Li, N.; Wang, Y.; Wei, L.; Hu, L.; Xu, S. Comparative Evaluation of the Nutrient Composition and Lipidomic Profile of Different Parts of Muscle in the Chaka Sheep. Food Sci. Anim. Resour. 2024, 44, 1305–1326. [Google Scholar] [PubMed]
- Kamal, B.; Farid, M.; Abdessamad, B.M.; Marianne, S.; Laure, F.M.; Mohamed, B.; Hana, S.C.; Ahmed, E. Proximate Composition, Amino Acid Profile, and Mineral Content of Four Sheep Meats Reared Extensively in Morocco: A Comparative Study. Sci. World J. 2021, 2021, 6633774. [Google Scholar]
- Li, M.; Zhu, M.; Chai, W.; Wang, Y.; Fan, D.; Lv, M.; Jiang, X.; Liu, Y.; Wei, Q.; Wang, C. Determination of lipid profiles of Dezhou donkey meat using an LC-MS-based lipidomics method. J. Food Sci. 2021, 86, 4511–4521. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, Z.; Zhang, H.; Xie, L.; Vincenzetti, S.; Polidori, P.; Li, L.; Liu, G. Changes in the Physical–Chemical Properties and Volatile Flavor Components of Dry-Cured Donkey Leg during Processing. Foods 2022, 11, 3542. [Google Scholar] [CrossRef]
- Marino, R.; Malva, A.D.; Gliatta, G.; Muscio, A.; Sevi, A. Quality of donkey bresaola. Ital. J. Anim. Sci. 2016, 8, 9–717. [Google Scholar]
- Zhang, W.; Zhang, M.; Sun, Y.; Liu, S. Factors affecting the quality and nutritional value of donkey meat: A comprehensive review. Front. Vet. Sci. 2024, 11, 1460859. [Google Scholar]
- Karina, R.; Isabell, C.; Matthias, H.; Gerald, G.; Sucharit, B. Unsaturated fatty acids drive disintegrin and metalloproteinase (ADAM)-dependent cell adhesion, proliferation, and migration by modulating membrane fluidity. J. Biol. Chem. 2011, 286, 26931–26942. [Google Scholar]
- Cheung, S.N.; Lieberman, H.R.; Pasiakos, S.M.; Fulgoni, V.L.; Berryman, C.E. Associations between Essential Amino Acid Intake and Functional Health Outcomes in Older Adults: Analysis of the National Health and Nutrition Examination Survey, 2001–2018. Curr. Dev. Nutr. 2024, 8, 104411. [Google Scholar] [CrossRef] [PubMed]
- Jihyun, I.; Hyoungsu, P.; Kyong, P. Higher Intake of Total Dietary Essential Amino Acids Is Associated with a Lower Prevalence of Metabolic Syndrome among Korean Adults. Nutrients 2022, 14, 4771. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Rinalducci, S.; Marrocco, C.; Zolla, V.; Napolitano, F.; Zolla, L. Love me tender: An Omics window on the bovine meat tenderness network. J. Proteom. 2012, 75, 4360–4380. [Google Scholar]
- D’Alessandro, A.; Zolla, L. Meat science: From proteomics to integrated omics towards system biology. J. Proteom. 2013, 78, 558–577. [Google Scholar]
- Gagaoua, M. Recent Advances in OMICs Technologies and Application for Ensuring Meat Quality, Safety and Authenticity. Foods 2022, 11, 2532. [Google Scholar] [CrossRef]
- YoungHwa, H.; EunYeong, L.; HyenTae, L.; SeonTea, J. Multi-Omics Approaches to Improve Meat Quality and Taste Characteristics. Food Sci. Anim. Resour. 2023, 43, 1067–1086. [Google Scholar]
- Purslow, P.P.; Gagaoua, M.; Warner, R.D. Insights on meat quality from combining traditional studies and proteomics. Meat Sci. 2021, 174, 108423. [Google Scholar]
- Gobert, M.; Sayd, T.; Gatellier, P.; Santé-Lhoutellier, V. Application to proteomics to understand and modify meat quality. Meat Sci. 2014, 98, 539–543. [Google Scholar]
- Nxumalo, N.; Rhode, C.; Kunene, N.; Molotsi, A. A review on omics approaches, towards understanding environmental resilience of indigenous Nguni sheep: Implications for their conservation and breeding programs in South Africa. Ecol. Genet. Genom. 2024, 33, 100305. [Google Scholar]
- Chaudhary, P.; Choudhary, K.; Rajput, R. A Review of Omics Technologies in Seafood Safety and Quality Control: Integrating Modern Approaches with Traditional Practices. Eur. J. Nutr. Food Saf. 2024, 16, 221–239. [Google Scholar]
- Sun, Y.; Wang, Y.; Li, Y.; Li, H.; Wang, C.; Zhang, Q. Comparative transcriptome and proteome analyses of the longissimus dorsi muscle for explaining the difference between donkey meat and other meats. Anim. Biotechnol. 2023, 34, 3085–3098. [Google Scholar] [PubMed]
- Chai, W.; Xu, J.; Qu, H.; Ma, Q.; Zhu, M.; Li, M.; Zhan, Y.; Wang, T.; Gao, J.; Yao, H. Differential proteomic analysis to identify potential biomarkers associated with quality traits of Dezhou donkey meat using a data-independent acquisition (DIA) strategy. LWT 2022, 166, 113792. [Google Scholar]
- Wang, L.; Qu, H.; Wang, X.; Wang, T.; Ma, Q.; Khan, M.Z.; Zhu, M.; Wang, C.; Liu, W.; Chai, W. Data-Independent Acquisition Method for In-Depth Proteomic Screening of Donkey Meat. Agriculture 2024, 14, 2102. [Google Scholar] [CrossRef]
- Li, M.; Zhu, M.; Chai, W.; Wang, Y.; Song, Y.; Liu, B.; Cai, C.; Song, Y.; Sun, X.; Xue, P. Determination of the heterogeneity of intramuscular fat and visceral adipose tissue from Dezhou donkey by lipidomics and transcriptomics profiling. Front. Nutr. 2021, 8, 746684. [Google Scholar]
- Zhao, J.; Li, K.; Yang, Q.; Du, M.; Liu, X.; Cao, G. Enhanced adipogenesis in Mashen pigs compared with Large White pigs. Ital. J. Anim. Sci. 2017, 16, 217–225. [Google Scholar]
- Romani, A.; Ieri, F.; Urciuoli, S.; Noce, A.; Marrone, G.; Nediani, C.; Bernini, R. Health Effects of Phenolic Compounds Found in Extra-Virgin Olive Oil, By-Products, and Leaf of Olea europaea L. Nutrients 2019, 11, 1776. [Google Scholar] [CrossRef] [PubMed]
- Man, L.; Ren, W.; Qin, H.; Sun, M.; Yuan, S.; Zhu, M.; Liu, G.; Wang, C.; Li, M. Characterization of the relationship between lipids and volatile compounds in donkey, bovine, and sheep meat by UHPLC–ESI–MS and SPME–GC–MS. LWT 2023, 175, 114426. [Google Scholar]
- Li, M.; Ren, W.; Chai, W.; Zhu, M.; Man, L.; Zhan, Y.; Qin, H.; Sun, M.; Liu, J.; Zhang, D. Comparing the Profiles of Raw and Cooked Donkey Meat by Metabonomics and Lipidomics Assessment. Front. Nutr. 2022, 9, 851761. [Google Scholar]
- Khan, M.I.; Jo, C.; Tariq, M.R. Meat flavor precursors and factors influencing flavor precursors—A systematic review. Meat Sci. 2015, 110, 278–284. [Google Scholar]
- Maggiolino, A.; Lorenzo, J.M.; Centoducati, G.; Domínguez, R.; Dinardo, F.R.; Marino, R.; Malva, A.D.; Bragaglio, A.; De Palo, P. How Volatile Compounds, Oxidative Profile and Sensory Evaluation Can Change with Vacuum Aging in Donkey Meat. Animals 2020, 10, 2126. [Google Scholar] [CrossRef]
- Xiu, L.I.; Amadou, I.; Zhou, G.Y.; Qian, L.Y.; Cheng, X.R. Flavor Components Comparison between the Neck Meat of Donkey, Swine, Bovine, and Sheep. Food Sci. Anim. Resour. 2020, 40, 527–540. [Google Scholar]
- Man, L.; Ren, W.; Sun, M.; Du, Y.; Chen, H.; Qin, H.; Chai, W.; Zhu, M.; Liu, G.; Wang, C. Characterization of donkey-meat flavor profiles by GC–IMS and multivariate analysis. Front. Nutr. 2023, 10, 1079799. [Google Scholar]
- Mario, E.; David, M.; Sonia, V.; Ramón, C. Analysis of volatiles in meat from Iberian pigs and lean pigs after refrigeration and cooking by using SPME-GC-MS. J. Agric. Food. Chem. 2003, 51, 3429–3435. [Google Scholar]
- Polidori, P.; Santini, G.; Klimanova, Y.; Zhang, J.; Vincenzetti, S. Effects of Ageing on Donkey Meat Chemical Composition, Fatty Acid Profile and Volatile Compounds. Foods 2022, 11, 821. [Google Scholar] [CrossRef]
- Yu, X.; Feng, Y.; Ma, W.; Xiao, X.; Liu, J.; Dong, W.; Hu, Y.; Liu, H. Ultrasound combined with Adenosine 5'-Monophosphate Treatment: A Strategic Approach for enhancing the tenderness of chicken wooden breast meat. Ultrason Sonochem. 2025, 114, 107284. [Google Scholar] [PubMed]
- Ribeiro, F.A.; Lau, S.K.; Furbeck, R.A.; Herrera, N.J.; Calkins, C.R. Ultimate pH effects on dry-aged beef quality. Meat Sci. 2021, 172, 108365. [Google Scholar]
- de Alvarenga, E.S.; Isac, M.F.; Rosa, A.F.; Silva, S.L.; Nassu, R.T.; Barretto, A.C.D.S. Effects of medium voltage electrical stimulation on initial pH decline and quality parameters during ageing and frozen storage of Nellore beef. Meat Sci. 2024, 212, 109464. [Google Scholar] [PubMed]
- Chai, W.; Wang, L.; Li, T.; Wang, T.; Wang, X.; Yan, M.; Zhu, M.; Gao, J.; Wang, C.; Ma, Q. Liquid Chromatography–Mass Spectrometry-Based Metabolomics Reveals Dynamic Metabolite Changes during Early Postmortem Aging of Donkey Meat. Foods 2024, 13, 1466. [Google Scholar] [CrossRef]
- Liu, X.; Chen, W.; Huang, B.; Wang, X.; Peng, Y.; Zhang, X.; Chai, W.; Khan, M.Z.; Wang, C. Advancements in copy number variation screening in herbivorous livestock genomes and their association with phenotypic traits. Front. Vet. Sci. 2024, 10, 1334434. [Google Scholar]
- Khan, M.Z.; Chen, W.; Huang, B.; Liu, X.; Wang, X.; Liu, Y.; Chai, W.; Wang, C. Advancements in Genetic Marker Exploration for Livestock Vertebral Traits with a Focus on China. Animals 2024, 14, 594. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.; Qu, H.; Ma, Q.; Zhu, M.; Li, M.; Zhan, Y.; Liu, Z.; Xu, J.; Yao, H.; Li, Z. RNA-seq analysis identifies differentially expressed gene in different types of donkey skeletal muscles. Anim. Biotechnol. 2023, 34, 1786–1795. [Google Scholar]
- Yang, G.; Sun, M.; Wang, Z.; Hu, Q.; Guo, J.; Yu, J.; Lei, C.; Dang, R. Comparative Genomics Identifies the Evolutionarily Conserved Gene TPM3 as a Target of eca-miR-1 Involved in the Skeletal Muscle Development of Donkeys. Int. J. Mol. Sci. 2023, 24, 15440. [Google Scholar] [CrossRef]
- Yu, J.; Yang, G.; Li, S.; Li, M.; Ji, C.; Liu, G.; Wang, Y.; Chen, N.; Lei, C.; Dang, R. Identification of Dezhou donkey muscle development-related genes and long non-coding RNA based on differential expression analysis. Anim. Biotechnol. 2023, 34, 2313–2323. [Google Scholar] [PubMed]
- Pj, B.; Guo, X.; Pei, J.; Yan, P. Characterization of the complete mitochondrial genome of the Liangzhou donkey (Equus asinus). Mitochondrial DNA Part B 2019, 4, 1846–1847. [Google Scholar]
- Liu, L.; Chen, B.; Chen, S.; Liu, W. A Genome-Wide Association Study of the Chest Circumference Trait in Xinjiang Donkeys Based on Whole-Genome Sequencing Technology. Genes 2023, 14, 1081. [Google Scholar] [CrossRef]
- Yu, J.; Wang, Z.; Wang, F.; Yang, G.; Cheng, J.; Ji, C.; Li, M.; Liu, B.; Wang, Y.; Dang, R. Analysis of lncRNA and mRNA Expression Profiling in Immature and Mature DeZhou donkey (equine Taurus) Testes. Reprod. Domest. Anim. = Zuchthyg. 2023, 58, 646–656. [Google Scholar]
- Li, W.; Wang, X. Transcriptomic analysis of different intramuscular fat contents on the flavor of the longissimus dorsi tissues from Guangling donkey. Genomics 2024, 116, 110905. [Google Scholar]
- Peng, Y.; Zhu, M.; Gong, Y.; Wang, C. Identification and functional prediction of lncRNAs associated with intramuscular lipid deposition in Guangling donkeys. Front. Vet. Sci. 2024, 11, 1410109. [Google Scholar] [CrossRef]
- Wufeng, L.; Lixia, Q.; Jiawei, G.; Yutong, S.; Jingwei, Z.; Du, M. Comparative transcriptome analysis of longissimus dorsi tissues with different intramuscular fat contents from Guangling donkeys. Bmc Genom. 2022, 23, 644. [Google Scholar]
- Li, W.; Guan, J.; Qiu, L.; Sun, Y.; Du, M. Study on the Molecular Mechanism of Regulating Tenderness of Longissimus Dorsi Muscle of Donkey Based on Transcriptomics and Metabolomics. Acta Vet. et Zootech. Sin. 2022, 53, 743–754. [Google Scholar]
- Li, B.; Feng, C.; Zhu, S.; Zhang, J.; Irwin, D.M.; Zhang, X.; Zhe, W.; Zhang, S. Identification of Candidate Circular RNAs Underlying Intramuscular Fat Content in the Donkey. Front. Genet. 2020, 11, 587559. [Google Scholar]
- Wang, T.; Wang, X.; Liu, Z.; Shi, X.; Ren, W.; Huang, B.; Liang, H.; Wang, C.; Chai, W. Genotypes and haplotype combination of DCAF7 gene sequence variants are associated with number of thoracolumbar vertebrae and carcass traits in Dezhou donkey. J. Appl. Anim. Res. 2023, 51, 31–39. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Z.; Wang, X.; Li, Y.; AKHTAR, F.; Li, M.; Zhang, Z.; Zhan, Y.; Shi, X.; Ren, W. Polymorphism detection of PRKG2 gene and its association with the number of thoracolumbar vertebrae and carcass traits in Dezhou donkey. Bmc Genom. Data 2023, 24, 2. [Google Scholar]
- Sun, Y.; Li, Y.; Zhao, C.; Teng, J.; Wang, Y.; Wang, T.; Shi, X.; Liu, Z.; Li, H.; Wang, J. Genome-wide association study for numbers of vertebrae in Dezhou donkey population reveals new candidate genes. J. Integr. Agric. 2023, 22, 3159–3169. [Google Scholar]
- Liu, Z.; Gao, Q.; Wang, T.; Chai, W.; Zhan, Y.; Akhtar, F.; Zhang, Z.; Li, Y.; Shi, X.; Wang, C. Multi-Thoracolumbar Variations and NR6A1 Gene Polymorphisms Potentially Associated with Body Size and Carcass Traits of Dezhou Donkey. Animals 2022, 12, 1349. [Google Scholar] [CrossRef]
- Shuang, S.; Shiwei, W.; Nan, L.; Siyu, C.; Shizhen, D.; Yajun, G.; Xuan, W.; Yuanweilu, C.; Shenming, Z. Genome-wide association study to identify SNPs and candidate genes associated with body size traits in donkeys. Front. Genet. 2023, 14, 1112377. [Google Scholar]
- Wang, X.; Wang, T.; Liang, H.; Wang, L.; Akhtar, F.; Shi, X.; Ren, W.; Huang, B.; Kou, X.; Chen, Y. A novel SNP in NKX1-2 gene is associated with carcass traits in Dezhou donkey. Bmc Genom. Data 2023, 24, 41. [Google Scholar]
- Liu, Z.; Wang, T.; Shi, X.; Wang, X.; Ren, W.; Huang, B.; Wang, C. Identification of LTBP2 gene polymorphisms and their association with thoracolumbar vertebrae number, body size, and carcass traits in Dezhou donkeys. Front. Genet. 2022, 13, 969959. [Google Scholar]
- Shi, X.; Li, Y.; Wang, T.; Ren, W.; Huang, B.; Wang, X.; Liu, Z.; Liang, H.; Kou, X.; Chen, Y. Association of HOXC8 Genetic Polymorphisms with Multi-Vertebral Number and Carcass Weight in Dezhou Donkey. Genes 2022, 13, 2175. [Google Scholar] [CrossRef]
- Wang, T.; Shi, X.; Liu, Z.; Ren, W.; Wang, X.; Huang, B.; Kou, X.; Liang, H.; Wang, C.; Chai, W. A Novel A > G Polymorphism in the Intron 1 of LCORL Gene Is Significantly Associated with Hide Weight and Body Size in Dezhou Donkey. Animals 2022, 12, 2581. [Google Scholar] [CrossRef]
- Lai, Z.; Wu, F.; Li, M.; Bai, F.; Gao, Y.; Yu, J.; Li, H.; Lei, C.; Dang, R. Tissue expression profile, polymorphism of IGF1 gene and its effect on body size traits of Dezhou donkey. Gene 2021, 766, 145118. [Google Scholar] [PubMed]
- Tingjin, C.; Mei, L.; Xiaoya, A.; Fuxia, B.; Fuwen, W.; Jie, Y.; Chuzhao, L.; Ruihua, D. Association analysis of IGF2 gene polymorphisms with growth traits of Dezhou donkey. Anim. Biotechnol. 2021, 34, 11. [Google Scholar]
- Wang, G.; Li, M.; Zhou, J.; An, X.; Bai, F.; Gao, Y.; Yu, J.; Li, H.; Lei, C.; Dang, R. A novel A > G polymorphism in the intron 2 of TBX3 gene is significantly associated with body size in donkeys. Gene 2021, 785, 145602. [Google Scholar] [PubMed]
- Fuwen, W.; Gang, W.; Baligen, D.; Zhaofei, W.; Tingjin, C.; Ge, Y.; Chuzhao, L.; Ruihua, D. A novel 31bp deletion within the CDKL5 gene is significantly associated with growth traits in Dezhou donkey. Anim. Biotechnol. 2021, 34, 503–507. [Google Scholar]
- Au-aff, O.A. Expression profiles and polymorphic identification of the ACSL1 gene and their association with body size traits in Dezhou donkeys. Arch. Anim. Breed. 2020, 63, 377–386. [Google Scholar]
- Lai, Z.; Li, S.; Wu, F.; Zhou, Z.; Gao, Y.; Yu, J.; Lei, C.; Dang, R. Genotypes and haplotype combination of ACSL3 gene sequence variants is associated with growth traits in Dezhou donkey. Gene 2020, 743, 144600. [Google Scholar]
- Shi, T.; Hu, W.; Hou, H.; Zhao, Z.; Shang, M.; Zhang, L. Identification and Comparative Analysis of Long Non-Coding RNA in the Skeletal Muscle of Two Dezhou Donkey Strains. Genes 2020, 11, 508. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Wang, M.; Zhang, X.; Yang, L.; Zhao, C.; Wu, C. Genetic architectures and selection signatures of body height in Chinese indigenous donkeys revealed by next-generation sequencing. Anim. Genet. 2022, 53, 487–497. [Google Scholar]
- Wang, G.; Wang, F.; Pei, H.; Li, M.; Bai, F.; Lei, C.; Dang, R. Genome-wide analysis reveals selection signatures for body size and drought adaptation in Liangzhou donkey. Genomics 2022, 114, 110476. [Google Scholar]
- Tan, X.; He, Y.; Qin, Y.; Yan, Z.; Chen, J.; Zhao, R.; Zhou, S.; Irwin, D.M.; Li, B.; Zhang, S. Comparative analysis of differentially abundant proteins between high and low intramuscular fat content groups in donkeys. Front. Vet. Sci. 2022, 9, 951168. [Google Scholar]
- Siddiqui, M.A.; Khir, M.H.M.; Witjaksono, G.; Ghumman, A.S.M.; Junaid, M.; Magsi, S.A.; Saboor, A. Multivariate Analysis Coupled with M-SVM Classification for Lard Adulteration Detection in Meat Mixtures of Beef, Lamb, and Chicken Using FTIR Spectroscopy. Foods 2021, 10, 2405. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, L.; Xiang, J.; Fu, Q.; Wang, J.; Wang, K.; Sun, X.; Ai, L.; Xu, X.; Wang, J. Identification of horse, donkey and pig ingredients by species-specific ERA-based methods to assess the authenticity of meat products. Food Biosci. 2023, 53, 102827. [Google Scholar]
- Atef, O.; Yassin, N.; Hamed, R.; Hariri, M.E.; Elyazeed, H.A.; Ella, H.A.; Soliman, R. Development and evaluation of a lateral flow immunochromatographic assay for the rapid detection of donkey meat in beef as a tool for meat adulteration identification. J. Consum. Prot. Food Saf. 2024, 20, 29–39. [Google Scholar]
- Liu, H.; Cao, T.; Wang, J.; Yuan, Y.; Li, H.; He, K.; Chen, H.; Wang, L. Accurate and simultaneous detection of pork and horse meat adulteration by double tailed recombinase polymerase amplification integrated with SERS based two-color lateral flow nucleic acid hybridization strip. J. Food Compos. Anal. 2024, 134, 106562. [Google Scholar]
- Zhu, T.; Zhou, X.; Zhang, W.; Wu, Y.; Yang, J.; Xu, L.; Chen, M.; Dong, W.; Xu, H. Multiplex and real-time PCR for qualitative and quantitative donkey meat adulteration. J. Food Meas. Charact. 2020, 15, 1161–1168. [Google Scholar]
- Edris, S.; Mutwakil, M.H.Z.; Abuzinadah, O.A.; Mohammed, H.E.; Ramadan, A.; Gadalla, N.O.; Shokry, A.M.; Hassan, S.M.; Shoaib, R.M.; El-Domyati, F.M.; et al. Conventional multiplex polymerase chain reaction (PCR) versus real-time PCR for species-specific meat authentication. Life Sci. J. 2012, 9, 26552–26558. [Google Scholar]
- Zhao, L.; Hua, M.Z.; Li, S.; Liu, J.; Zheng, W.; Lu, X. Identification of donkey meat in foods using species-specific PCR combined with lateral flow immunoassay. Rsc Adv. 2019, 9, 26552–26558. [Google Scholar]
- Vadera, N.; Dhanekar, S. Classification and Prediction of VOCs Using an IoT-Enabled Electronic Nose System-Based Lab Prototype for Breath Sensing Applications. ACS Sensors 2025, 10, 439–447. [Google Scholar]
- Zaukuu, J.Z.; Gillay, Z.; Kovacs, Z. Standardized Extraction Techniques for Meat Analysis with the Electronic Tongue: A Case Study of Poultry and Red Meat Adulteration. Sensors 2021, 21, 481. [Google Scholar] [CrossRef]
- Qiu, W.; Zhang, X.; Zhang, H.; Liang, C.; Xu, J.; Gao, H.; Ai, L.; Zhao, S.; Wang, Y.; Yang, Y. Discrimination of meat from fur-producing and food-providing animals using mass spectrometry-based proteomics. Food Res. Int. 2020, 137, 109446. [Google Scholar] [PubMed]
- Fengou, L.; Tsakanikas, P.; Nychas, G.E. Rapid detection of minced pork and chicken adulteration in fresh, stored and cooked ground meat. Food Control 2021, 125, 108002. [Google Scholar]
- Akhtar, M.T.; Samar, M.; Shami, A.A.; Mumtaz, M.W.; Mukhtar, H.; Tahir, A.; Shahzad-ul-Hussan, S.; Chaudhary, S.U.; Kaka, U. 1H-NMR-Based Metabolomics: An Integrated Approach for the Detection of the Adulteration in Chicken, Chevon, Beef and Donkey Meat. Molecules 2021, 26, 4643. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, L.; Wei, H.; Luo, Q.; Yang, P.; Liu, M.; Wang, Z.; Zou, X.; Zhu, H.; Zha, G. Design and development of a rapid meat detection system based on RPA-CRISPR/Cas12a-LFD. Curr. Res. Food Sci. 2023, 7, 100609. [Google Scholar] [PubMed]
- De Palo, P.; Tateo, A.; Maggiolino, A.; Marino, R.; Ceci, E.; Nisi, A.; Lorenzo, J.M. Martina Franca donkey meat quality: Influence of slaughter age and suckling technique. Meat Sci. 2017, 134, 128–134. [Google Scholar] [PubMed]
- Domínguez, R.; Crecente, S.; Borrajo, P.; Agregán, R.; Lorenzo, J.M. Effect of slaughter age on foal carcass traits and meat quality. Animal 2015, 9, 1713–1720. [Google Scholar]
- Nogalski, Z.; Pogorzelska-Przybyłek, P.; Sobczuk-Szul, M.; Modzelewska-Kapituła, M. Effects of Rearing System and Fattening Intensity on the Chemical Composition, Physicochemical Properties and Sensory Attributes of Meat from Young Crossbred (Holstein-Friesian × Hereford) Bulls. Animal 2022, 12, 933. [Google Scholar]
- Lorenzo, J.M.; Fuciños, C.; Purriños, L.; Franco, D. Intramuscular fatty acid composition of “Galician Mountain” foals breed: Effect of sex, slaughtered age and livestock production system. Meat Sci. 2010, 86, 825–831. [Google Scholar]
- Scollan, N.; Hocquette, J.; Nuernberg, K.; Dannenberger, D.; Richardson, I.; Moloney, A. Innovations in beef production systems that enhance the nutritional and health value of beef lipids and their relationship with meat quality. Meat Sci. 2006, 74, 17–33. [Google Scholar]
- Smet, S.D.; Raes, K.; Demeyer, D. Meat fatty acid composition as affected by fatness and genetic factors: A review. Anim. Res. 2004, 53, 81–98. [Google Scholar]
- De Palo, P.; Maggiolino, A.; Milella, P.; Centoducati, N.; Papaleo, A.; Tateo, A. Artificial suckling in Martina Franca donkey foals: Effect on in vivo performances and carcass composition. Trop. Anim. Health Prod. 2016, 48, 167–173. [Google Scholar] [CrossRef] [PubMed]
- De Palo, P.; Maggiolino, A.; Albenzio, M.; Casalino, E.; Neglia, G.; Centoducati, G.; Tateo, A. Survey of biochemical and oxidative profile in donkey foals suckled with one natural and one semi-artificial technique. PLoS ONE 2018, 13, e198774. [Google Scholar] [CrossRef] [PubMed]
- Piccione, G.; Casella, S.; Giannetto, C.; Bazzano, M.; Giudice, E.; Fazio, F. Oxidative stress associated with road transportation in ewes. Small Rumin. Res. 2013, 112, 235–238. [Google Scholar] [CrossRef]
- Falowo, A.B.; Fayemi, P.O.; Muchenje, V. Natural antioxidants against lipid–protein oxidative deterioration in meat and meat products: A review. Food Res. Int. 2014, 64, 171–181. [Google Scholar] [CrossRef]
- Webb, E.C.; Erasmus, L.J. The effect of production system and management practices on the quality of meat products from ruminant livestock. S. Afr. J. Anim. Sci. 2013, 43, 413–423. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, H.; Liu, M.; Zhao, X.; Luo, H. Effect of Breed on the Volatile Compound Precursors and Odor Profile Attributes of Lamb Meat. Foods 2020, 9, 1178. [Google Scholar] [CrossRef]
- Maggiolino, A.; Lorenzo, J.M.; Marino, R.; Malva, A.D.; Centoducati, P.; De Palo, P. Foal meat volatile compounds: Effect of vacuum ageing on semimembranosus muscle. J. Sci. Food Agric. 2018, 99, 1660–1667. [Google Scholar] [CrossRef]
- Polidori, P.; Ariani, A.; Micozzi, D.; Vincenzetti, S. The effects of low voltage electrical stimulation on donkey meat. Meat Sci. 2016, 119, 160–164. [Google Scholar] [CrossRef]
| Nutrients | Donkey Meat [58,59,60,61,62] | Beef [63,64,65,66] | Pork [67,68,69,70,71,72] | Sheep [58,73,74,75,76,77] |
|---|---|---|---|---|
| Protein (g/100g) | 23.56 | 23.50 | 18.60 | 20.70 |
| Fat (g/100g) | 1.77 | 4.53 | 23.80 | 8.85 |
| Ash% | 1.13 | 1.13 | 0.90–1.00 | 1.62 |
| Vitamin B12 (μg/100 g) | 1.90 | 6.53 | 1.00 | 2.08 |
| Sodium (mg/100g) | 36.80–83.60 | 64.80 | 53.00 | 85.70 |
| Phosphorus (mg/100g) | 185.00–335.00 | 182.00 | 190.00 | 611.36 |
| Iron (mg/100g) | 2.86–4.77 | 1.76 | 1.05 | 2.93 |
| Zinc (mg/100g) | 2.99–4.71 | 3.27 | 1.90 | 3.23 |
| Calcium (mg/100g) | 7.95 | 3.72 | 7.00 | 21.34 |
| Potassium (mg/100g) | 353.00 | 391.00 | 330.00 | 280.00 |
| Cholesterol (mg/100 g) | 66.70 | 63.00 | 77.00 | 133.28 |
| Polyunsaturated fatty acids (PUFA)/saturated fatty acids | 0.73 | 0.15 | 0.29 | 0.09 |
| Genes | Association with Meat Quality and Growth Traits | Breed | Omics Techniques /Instruments | Reference |
|---|---|---|---|---|
| ACTN3, TPM2, TPM3 |
| Dezhou Donkeys | Transcriptomics | [8] |
| NCAPG, LCORL |
| Guanzhong, Taihang, Dezhou, Huaibeihui, Biyang, and Qingyang Qinghai, Guoluo, Xinjiang, XizangGuanzhong, Taihang, Dezhou, Huaibeihui, Biyang, Qingyang | Genomics | [9] |
| KRT10, KRT1, CLDN9 |
| Dezhou Donkeys | Transcriptomics | [11] |
| ARF6, IQGAP, AGPAT1 |
| Dezhou Donkeys | Proteomics | [93] |
| MYH1, MYH7, TNNC1 |
| Dezhou Donkeys | Transcriptomics | [113] |
| NFATC2, PROP1 |
| Xinjiang donkeys | Genomics | [117] |
| SCD, LEPR, CIDEA |
| Guangling donkeys | Transcriptomics | [121] |
| miR-429, miR-224, miR-125a-5p, miR-223 |
| Liaoxi donkey | Transcriptomics | [123] |
| DCAF7 |
| Dezhou donkey | Targeted sequencing Sanger sequencing | [124] |
| PRKG2 |
| Dezhou donkey | Targeted sequencing | [125] |
| NLGN1, DCC, SLC26A7, LCORL, BMP7, Wnt7a |
| Dezhou donkeys | Genomics | [126] |
| NR6A1 |
| Dezhou donkeys | Genomics | [127] |
| SMPD4, RPS6KA6 |
| Yangyuan donkeys | Genomics | [128] |
| NKX1-2 |
| Dezhou donkeys | Genomics | [129] |
| LTBP2 |
| Dezhou donkeys | Genomics | [130] |
| HOXC8 |
| Dezhou donkeys | Genomics | [131] |
| LCORL |
| Dezhou donkeys | Targeted sequencing | [132] |
| IGF-1, IGF-2 |
| Dezhou donkeys | TranscriptomicsGenomics | [133,134] |
| TBX3 |
| Dezhou donkeys | Genomics | [135] |
| CDKL5 |
| Dezhou donkeys | Genomics | [136] |
| ACSL1 |
| Dezhou donkeys | Transcriptomics, Genomics | [137] |
| ACSL3 |
| Dezhou donkeys | Genomics | [138] |
| ACTN1, CDON, FMOD, BMPR1B |
| Dezhou donkeys | Transcriptomics | [139] |
| LCORL/NCAPG, FAM184B, TBX3, IHH |
| Biyang, Dezhou, Guangling, Hetian, Jiami, Kulun, Qingyang, Turfan, Tibetan, Xinjiang, Yunnan, Zamorano~Leonés and Andalusian | Whole genome resequencing technology | [140] |
| NCAPG, LCORL, CYP4A11 |
| Liangzhou donkeys | Whole genome resequencing technology | [141] |
| SPAG8, RPL27A, TPM1, |
| Liaoxi donkeys | Proteomics | [142] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Q.; Peng, Y.; Liu, X.; Chen, W.; Geng, M.; Na, J.; Khan, M.Z.; Wang, C. Application of Omics in Donkey Meat Research: A Review. Animals 2025, 15, 991. https://doi.org/10.3390/ani15070991
Zhu Q, Peng Y, Liu X, Chen W, Geng M, Na J, Khan MZ, Wang C. Application of Omics in Donkey Meat Research: A Review. Animals. 2025; 15(7):991. https://doi.org/10.3390/ani15070991
Chicago/Turabian StyleZhu, Qifei, Yongdong Peng, Xiaotong Liu, Wenting Chen, Mingyang Geng, Jincheng Na, Muhammad Zahoor Khan, and Changfa Wang. 2025. "Application of Omics in Donkey Meat Research: A Review" Animals 15, no. 7: 991. https://doi.org/10.3390/ani15070991
APA StyleZhu, Q., Peng, Y., Liu, X., Chen, W., Geng, M., Na, J., Khan, M. Z., & Wang, C. (2025). Application of Omics in Donkey Meat Research: A Review. Animals, 15(7), 991. https://doi.org/10.3390/ani15070991










