Genetic Associations of ACOX2 Gene with Milk Yield and Composition Traits in Chinese Holstein Cows
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Phenotypic Data Collection
2.2. Genomic DNA Extraction
2.3. SNP Identification and Genotyping
2.4. Linkage Disequilibrium (LD) Estimation
2.5. Association Analyses
2.6. Prediction of the Changes in Transcription Factor Binding Sites
3. Results
3.1. SNP Identification in ACOX2 Gene
3.2. Genetic Associations Between SNPs and Milk Yield and Composition Traits
3.3. Genetic Associations Between Haplotypes and Milk Yield and Composition Traits
3.4. Transcription Factor Binding Site Changes Caused by SNPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pereira, P.C. Milk Nutritional Composition and Its Role in Human Health. Nutrition 2014, 30, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Jauhiainen, T.; Korpela, R. Milk Peptides and Blood Pressure. J. Nutr. 2007, 137, 825S–829S. [Google Scholar] [CrossRef]
- Rumbold, P.; McCullogh, N.; Boldon, R.; Haskell-Ramsay, C.; James, L.; Stevenson, E.; Green, B. The Potential Nutrition-, Physical- and Health-Related Benefits of Cow’s Milk for Primary-School-Aged Children. Nutr. Res. Rev. 2022, 35, 50–69. [Google Scholar] [CrossRef]
- Thomasen, J.R.; Liu, H.; Sørensen, A.C. Genotyping More Cows Increases Genetic Gain and Reduces Rate of True Inbreeding in a Dairy Cattle Breeding Scheme Using Female Reproductive Technologies. J. Dairy Sci. 2020, 103, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, L.R. Strategy for Applying Genome-Wide Selection in Dairy Cattle. J. Anim. Breed. Genet. 2006, 123, 218–223. [Google Scholar] [CrossRef]
- Liu, A.; Lund, M.S.; Boichard, D.; Karaman, E.; Fritz, S.; Aamand, G.P.; Nielsen, U.S.; Wang, Y.; Su, G. Improvement of Genomic Prediction by Integrating Additional Single Nucleotide Polymorphisms Selected from Imputed Whole Genome Sequencing Data. Heredity 2020, 124, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ober, U.; Erbe, M.; Zhang, H.; Gao, N.; He, J.; Li, J.; Simianer, H. Improving the Accuracy of Whole Genome Prediction for Complex Traits Using the Results of Genome Wide Association Studies. PLoS ONE 2014, 9, e93017. [Google Scholar] [CrossRef]
- de Las Heras-Saldana, S.; Lopez, B.I.; Moghaddar, N.; Park, W.; Park, J.-E.; Chung, K.Y.; Lim, D.; Lee, S.H.; Shin, D.; van der Werf, J.H.J. Use of Gene Expression and Whole-Genome Sequence Information to Improve the Accuracy of Genomic Prediction for Carcass Traits in Hanwoo Cattle. Genet. Sel. Evol. 2020, 52, 54. [Google Scholar] [CrossRef]
- Cole, J.B.; VanRaden, P.M.; O’Connell, J.R.; Van Tassell, C.P.; Sonstegard, T.S.; Schnabel, R.D.; Taylor, J.F.; Wiggans, G.R. Distribution and Location of Genetic Effects for Dairy Traits. J. Dairy Sci. 2009, 92, 2931–2946. [Google Scholar] [CrossRef]
- Grisart, B.; Farnir, F.; Karim, L.; Cambisano, N.; Kim, J.-J.; Kvasz, A.; Mni, M.; Simon, P.; Frère, J.-M.; Coppieters, W.; et al. Genetic and Functional Confirmation of the Causality of the DGAT1 K232A Quantitative Trait Nucleotide in Affecting Milk Yield and Composition. Proc. Natl. Acad. Sci. USA 2004, 101, 2398–2403. [Google Scholar] [CrossRef]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Cambisano, N.; Mni, M.; Reid, S.; Simon, P.; et al. Positional Candidate Cloning of a QTL in Dairy Cattle: Identification of a Missense Mutation in the Bovine DGAT1 Gene with Major Effect on Milk Yield and Composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Blott, S.; Kim, J.-J.; Moisio, S.; Schmidt-Küntzel, A.; Cornet, A.; Berzi, P.; Cambisano, N.; Ford, C.; Grisart, B.; Johnson, D.; et al. Molecular Dissection of a Quantitative Trait Locus: A Phenylalanine-to-Tyrosine Substitution in the Transmembrane Domain of the Bovine Growth Hormone Receptor Is Associated with a Major Effect on Milk Yield and Composition. Genetics 2003, 163, 253–266. [Google Scholar] [CrossRef]
- Cohen-Zinder, M.; Seroussi, E.; Larkin, D.M.; Loor, J.J.; Everts-van der Wind, A.; Lee, J.-H.; Drackley, J.K.; Band, M.R.; Hernandez, A.G.; Shani, M.; et al. Identification of a Missense Mutation in the Bovine ABCG2 Gene with a Major Effect on the QTL on Chromosome 6 Affecting Milk Yield and Composition in Holstein Cattle. Genome Res. 2005, 15, 936–944. [Google Scholar] [CrossRef]
- Xu, L.; Shi, L.; Liu, L.; Liang, R.; Li, Q.; Li, J.; Han, B.; Sun, D. Analysis of Liver Proteome and Identification of Critical Proteins Affecting Milk Fat, Protein, and Lactose Metabolism in Dariy Cattle with iTRAQ. Proteomics 2019, 19, e1800387. [Google Scholar] [CrossRef] [PubMed]
- Monte, M.J.; Alonso-Peña, M.; Briz, O.; Herraez, E.; Berasain, C.; Argemi, J.; Prieto, J.; Marin, J.J.G. ACOX2 Deficiency: An Inborn Error of Bile Acid Synthesis Identified in an Adolescent with Persistent Hypertransaminasemia. J. Hepatol. 2017, 66, 581–588. [Google Scholar] [CrossRef]
- Sharma, V.; Hiller, M. Loss of Enzymes in the Bile Acid Synthesis Pathway Explains Differences in Bile Composition among Mammals. Genome Biol. Evol. 2018, 10, 3211–3217. [Google Scholar] [CrossRef]
- Ichimura-Shimizu, M.; Watanabe, S.; Kashirajima, Y.; Nagatomo, A.; Wada, H.; Tsuneyama, K.; Omagari, K. Dietary Cholic Acid Exacerbates Liver Fibrosis in NASH Model of Sprague-Dawley Rats Fed a High-Fat and High-Cholesterol Diet. Int. J. Mol. Sci. 2022, 23, 9268. [Google Scholar] [CrossRef] [PubMed]
- Schettini, G.P.; Peripolli, E.; Alexandre, P.A.; Dos Santos, W.B.; da Silva Neto, J.B.; Pereira, A.S.C.; de Albuquerque, L.G.; Curi, R.A.; Baldi, F. Transcriptomic Profile of Longissimus Thoracis Associated with Fatty Acid Content in Nellore Beef Cattle. Anim. Genet. 2022, 53, 264–280. [Google Scholar] [CrossRef]
- Rodriguez-Zas, S.L.; Southey, B.R.; Heyen, D.W.; Lewin, H.A. Detection of Quantitative Trait Loci Influencing Dairy Traits Using a Model for Longitudinal Data. J. Dairy Sci. 2002, 85, 2681–2691. [Google Scholar] [CrossRef]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Matukumalli, L.K.; et al. Genome-Wide Association Analysis of Thirty One Production, Health, Reproduction and Body Conformation Traits in Contemporary U.S. Holstein Cows. BMC Genom. 2011, 12, 408. [Google Scholar] [CrossRef]
- Garrick, D.J.; Taylor, J.F.; Fernando, R.L. Deregressing Estimated Breeding Values and Weighting Information for Genomic Regression Analyses. Genet. Sel. Evol. 2009, 41, 55. [Google Scholar] [CrossRef]
- Yousuf, U.; Sofi, S.; Makhdoomi, A.; Mir, M.A. Identification and Analysis of Dysregulated Fatty Acid Metabolism Genes in Breast Cancer Subtypes. Med. Oncol. 2022, 39, 256. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Su, F.; Yu, X.; Geng, N.; Li, L.; Wang, R.; Zhang, M.; Liu, J.; Liu, Y.; Han, B. RNA-Seq Whole Transcriptome Analysis of Bovine Mammary Epithelial Cells in Response to Intracellular Staphylococcus Aureus. Front. Vet. Sci. 2020, 7, 642. [Google Scholar] [CrossRef]
- Gharib-Naseri, K.; de Las Heras-Saldana, S.; Kheravii, S.; Qin, L.; Wang, J.; Wu, S.-B. Necrotic Enteritis Challenge Regulates Peroxisome Proliferator-1 Activated Receptors Signaling and β-Oxidation Pathways in Broiler Chickens. Anim. Nutr. 2021, 7, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Ferdinandusse, S.; Denis, S.; van Roermund, C.W.T.; Preece, M.A.; Koster, J.; Ebberink, M.S.; Waterham, H.R.; Wanders, R.J.A. A Novel Case of ACOX2 Deficiency Leads to Recognition of a Third Human Peroxisomal Acyl-CoA Oxidase. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 952–958. [Google Scholar] [CrossRef]
- Stayrook, K.R.; Bramlett, K.S.; Savkur, R.S.; Ficorilli, J.; Cook, T.; Christe, M.E.; Michael, L.F.; Burris, T.P. Regulation of Carbohydrate Metabolism by the Farnesoid X Receptor. Endocrinology 2005, 146, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chiang, J.Y.L. Bile Acid Signaling in Metabolic Disease and Drug Therapy. Pharmacol. Rev. 2014, 66, 948–983. [Google Scholar] [CrossRef]
- Kumar, V.; Westra, H.-J.; Karjalainen, J.; Zhernakova, D.V.; Esko, T.; Hrdlickova, B.; Almeida, R.; Zhernakova, A.; Reinmaa, E.; Võsa, U.; et al. Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression. PLoS Genet. 2013, 9, e1003201. [Google Scholar] [CrossRef]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic Variation in microRNA Networks: The Implications for Cancer Research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef]
- Fernández-Cortés, M.; Andrés-León, E.; Oliver, F.J. The PARP Inhibitor Olaparib Modulates the Transcriptional Regulatory Networks of Long Non-Coding RNAs during Vasculogenic Mimicry. Cells 2020, 9, 2690. [Google Scholar] [CrossRef]
- Imai, K.; Okamoto, T. Transcriptional Repression of Human Immunodeficiency Virus Type 1 by AP-4. J. Biol. Chem. 2006, 281, 12495–12505. [Google Scholar] [CrossRef]
- Ku, W.-C.; Chiu, S.-K.; Chen, Y.-J.; Huang, H.-H.; Wu, W.-G.; Chen, Y.-J. Complementary Quantitative Proteomics Reveals That Transcription Factor AP-4 Mediates E-Box-Dependent Complex Formation for Transcriptional Repression of HDM2. Mol. Cell Proteom. 2009, 8, 2034–2050. [Google Scholar] [CrossRef]
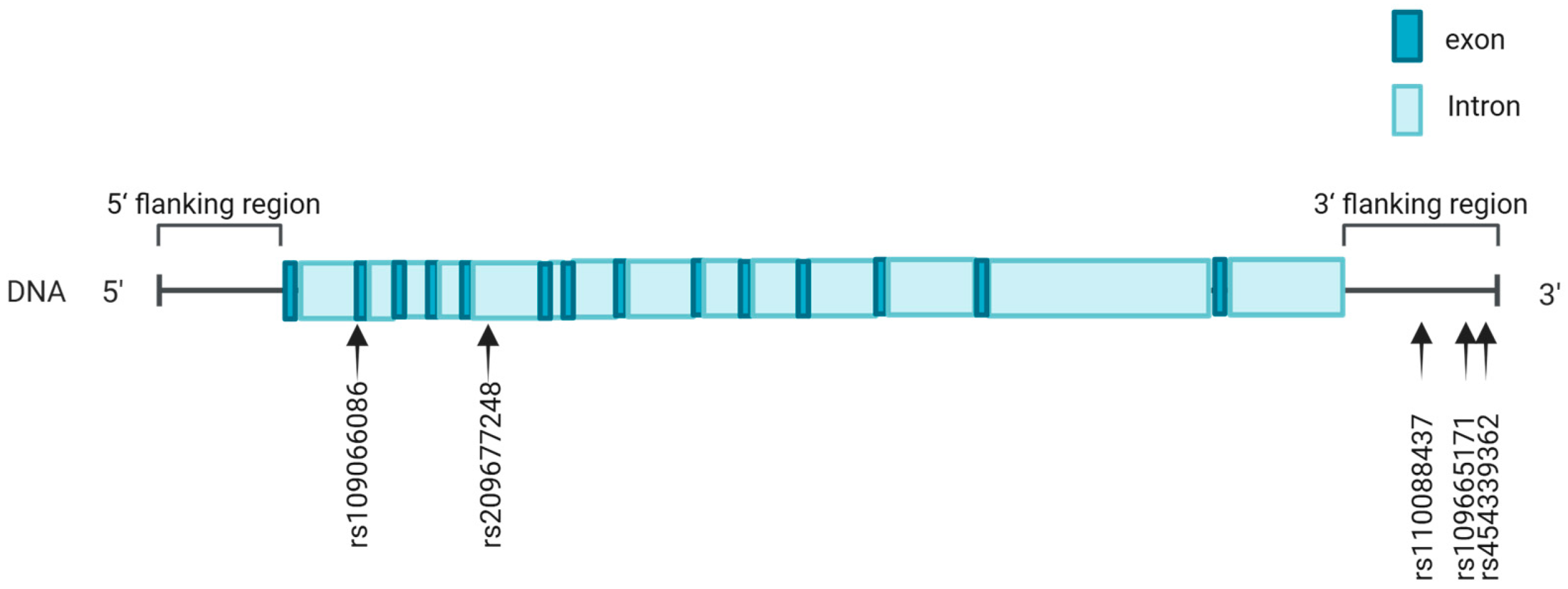
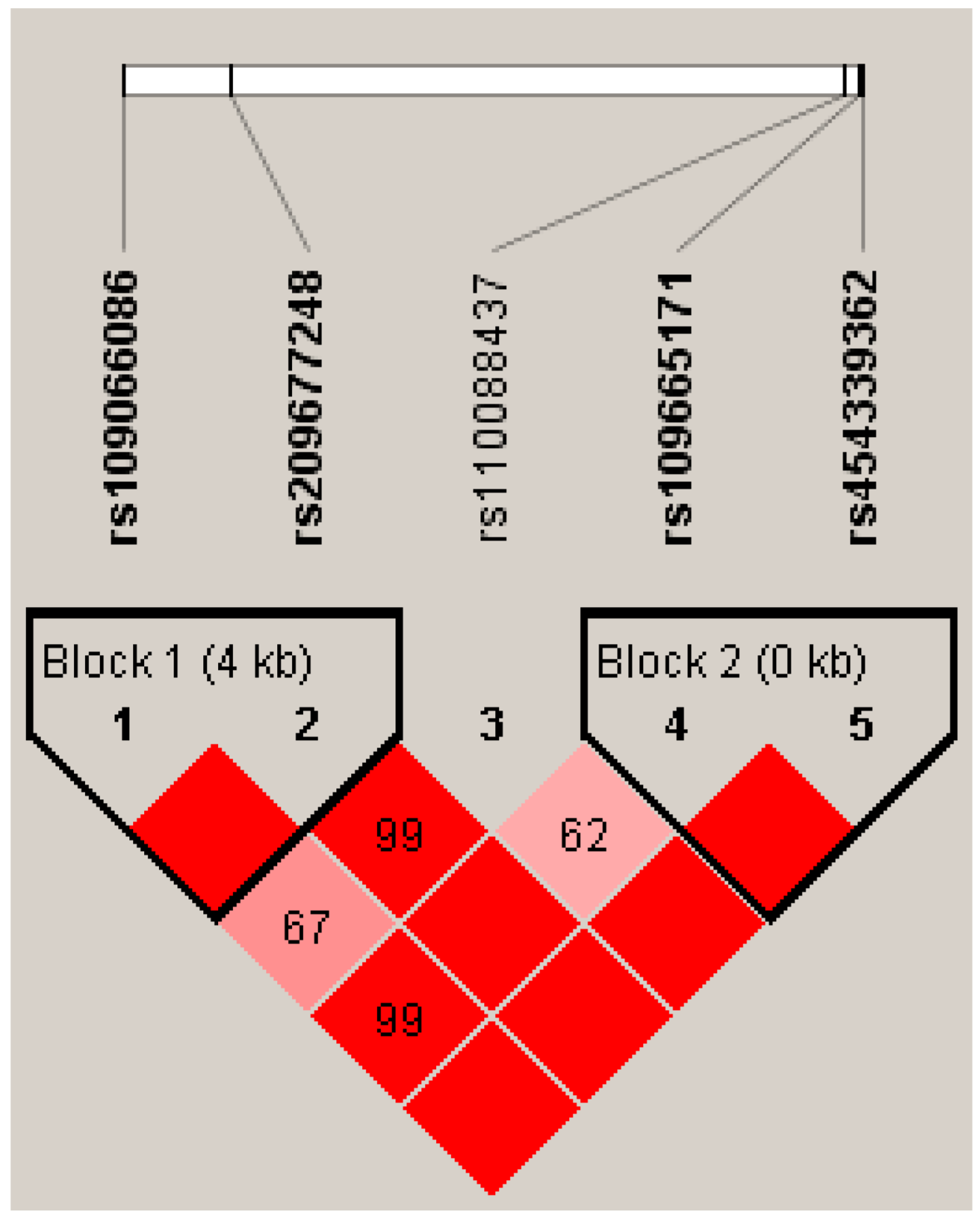
| Gene Region | SNPs | Genotype | Genotypic Frequency | Allele | Allelic Frequency |
|---|---|---|---|---|---|
| 5′ UTR | rs109066086 | CC | 0.51 | C | 0.71 |
| TC | 0.40 | T | 0.29 | ||
| TT | 0.09 | ||||
| Intron | rs209677248 | CC | 0.56 | C | 0.76 |
| CG | 0.40 | G | 0.24 | ||
| GG | 0.04 | ||||
| 3′ flanking region | rs110088437 | TT | 0.26 | T | 0.52 |
| TC | 0.52 | C | 0.48 | ||
| CC | 0.22 | ||||
| rs109665171 | CC | 0.43 | C | 0.67 | |
| TC | 0.47 | T | 0.33 | ||
| TT | 0.10 | ||||
| rs454339362 | TT | 0.82 | T | 0.90 | |
| TC | 0.17 | C | 0.10 | ||
| CC | 0.01 |
| SNP | Lactation | Genotype (No.) | Milk Yield (kg) | Fat Yield (kg) | Fat Percentage (%) | Protein Yield (kg) | Protein Percentage (%) |
|---|---|---|---|---|---|---|---|
| rs109066086 | 1 | CC (470) | 10,322 ± 64.60 Aa | 341.81 ± 2.88 a | 3.34 ± 0.03 | 306.73 ± 2.10 Aa | 2.98 ± 0.02 |
| TC (367) | 10,239 ± 66.11 Aa | 342.02 ± 2.92 a | 3.37 ± 0.03 | 304.42 ± 2.13 Aa | 2.99 ± 0.02 | ||
| TT (85) | 10,004 ± 98.15 B | 333.07 ± 4.13 b | 3.35 ± 0.04 | 296.24 ± 3.00 B | 2.98 ± 0.03 | ||
| p | 2.60 × 10−3 | 4.03 × 10−2 | 0.42 | 6.00 × 10−4 | 0.88 | ||
| 2 | CC (347) | 9830.93 ± 68.73 Aa | 342.61 ± 3.07 Aa | 3.56 ± 0.03 | 284.34 ± 2.23 Aa | 2.97 ± 0.02 | |
| TC (270) | 9722.67 ± 71.23 Aa | 339.07 ± 3.14 Aa | 3.60 ± 0.03 | 280.55 ± 2.29 Aa | 2.99 ± 0.02 | ||
| TT (64) | 9231.82 ± 114.87 B | 322.55 ± 4.82 B | 3.64 ± 0.05 | 263.76 ± 3.52 B | 2.98 ± 0.03 | ||
| p | <1.00 × 10−4 | <1.00 × 10−4 | 0.16 | <1.00 × 10−4 | 0.49 | ||
| rs209677248 | 1 | CC (520) | 10,226 ± 62.51 | 339.48 ± 2.79 | 3.35 ± 0.03 | 304.21 ± 2.03 | 2.99 ± 0.02 |
| CG (366) | 10,292 ± 65.91 | 342.72 ± 2.92 | 3.35 ± 0.03 | 305.17 ± 2.12 | 2.98 ± 0.02 | ||
| GG (36) | 10,423 ± 135.32 | 349.55 ± 5.58 | 3.38 ± 0.05 | 310.03 ± 4.07 | 2.99 ± 0.03 | ||
| p | 0.19 | 7.43 × 10−2 | 0.80 | 0.29 | 0.57 | ||
| 2 | CC (378) | 9615.03 ± 67.60 Bb | 332.53 ± 3.02 C | 3.58 ± 0.03 | 277.13 ± 2.20 C | 2.99 ± 0.02 Aa | |
| CG (276) | 9751.25 ± 70.30 Ba | 342.15 ± 3.11 B | 3.59 ± 0.03 | 281.93 ± 2.27 B | 2.98 ± 0.02 ABa | ||
| GG (27) | 10,930 ± 165.42 A | 389.53 ± 6.79 A | 3.56 ± 0.07 | 315.14 ± 4.95 A | 2.88 ± 0.04 Bb | ||
| p | <1.00 × 10−4 | <1.00 × 10−4 | 0.77 | <1.00 × 10−4 | 3.37 × 10−2 | ||
| rs110088437 | 1 | TT (239) | 10,192 ± 71.72 b | 338.70 ± 3.13 | 3.36 ± 0.03 | 303.04 ± 2.28 | 2.99 ± 0.02 |
| TC (477) | 10,253 ± 62.91 ab | 341.87 ± 2.81 | 3.36 ± 0.03 | 304.98 ± 2.04 | 2.99 ± 0.02 | ||
| CC (206) | 10,363 ± 75.61 a | 342.12 ± 3.28 | 3.32 ± 0.03 | 306.55 ± 2.39 | 2.97 ± 0.02 | ||
| p | 5.82 × 10−2 | 0.37 | 0.39 | 0.26 | 0.48 | ||
| 2 | TT (171) | 9415.33 ± 80.37 C | 328.31 ± 3.49 C | 3.59 ± 0.03 | 270.06 ± 2.54 C | 2.99 ± 0.02 | |
| TC (361) | 9695.53 ± 66.54 B | 336.34 ± 2.97 B | 3.58 ± 0.03 | 279.55 ± 2.16 B | 2.98 ± 0.02 | ||
| CC (149) | 10,184 ± 83.28 A | 359.39 ± 3.60 A | 3.58 ± 0.03 | 296.75 ± 2.62 A | 2.96 ± 0.02 | ||
| p | <1.00 × 10−4 | <1.00 × 10−4 | 0.88 | <1.00 × 10−4 | 0.53 | ||
| rs109665171 | 1 | CC (396) | 10,210 ± 65.14 b | 340.44 ± 340.44 | 3.36 ± 0.03 | 302.83 ± 2.10 Bb | 2.98 ± 0.02 |
| TC (436) | 10,279 ± 64.14 ab | 341.29 ± 341.29 | 3.35 ± 0.03 | 305.88 ± 2.08 ab | 2.99 ± 0.02 | ||
| TT (90) | 10,415 ± 97.54 a | 343.56 ± 343.56 | 3.33 ± 0.04 | 309.78 ± 3.00 Aa | 2.99 ± 0.03 | ||
| p | 6.14 × 10−2 | 0.68 | 0.74 | 1.65 × 10−2 | 0.73 | ||
| 2 | CC (292) | 9677.97 ± 70.60 B | 341.78 ± 3.12 Aa | 3.62 ± 0.03 Aa | 279.88 ± 2.27 Aa | 2.98 ± 0.02 | |
| TC (322) | 9864.84 ± 68.12 A | 340.04 ± 3.04 Aa | 3.55 ± 0.03 Bb | 284.41 ± 2.21 Ab | 2.97 ± 0.02 | ||
| TT (67) | 9160.17 ± 110.68 C | 320.03 ± 4.66 B | 3.58 ± 0.04 ab | 263.55 ± 3.40 B | 3.00 ± 0.03 | ||
| p | <1.00 × 10−4 | <1.00 × 10−4 | 1.17 × 10−2 | <1.00 × 10−4 | 0.42 | ||
| rs454339362 | 1 | TT (754) | 10,304 ± 59.87 Aa | 341.1 ± 2.70 | 3.34 ± 0.02 Bb | 306.04 ± 1.96 Aa | 2.98 ± 0.02 |
| TC (159) | 10,034 ± 81.43 Bb | 340.6 ± 3.50 | 3.42 ± 0.03 Aa | 298.66 ± 2.55 Bb | 3.00 ± 0.02 | ||
| CC (9) | 10,328 ± 245.37 ab | 345.68 ± 9.94 | 3.33 ± 0.10 ab | 304.25 ± 7.25 ab | 2.96 ± 0.06 | ||
| p | 4.00 × 10−4 | 0.87 | 9.30 × 10−3 | 1.30 × 10−3 | 0.62 | ||
| 2 | TT (558) | 9776.45 ± 62.12 Aa | 341.2 ± 2.82 Aa | 3.58 ± 0.03 | 282.46 ± 2.05 Aa | 2.97 ± 0.02 | |
| TC (115) | 9520.22 ± 92.82 Bb | 330.42 ± 3.97 Bb | 3.60 ± 0.038 | 272.99 ± 2.89 Bb | 2.98 ± 0.03 | ||
| CC (8) | 9658.28 ± 270.74 ab | 335.08 ± 10.99 ab | 3.57 ± 0.11 | 284.62 ± 8.02 ab | 3.04 ± 0.07 | ||
| p | 8.00 × 10−3 | 5.50 × 10−3 | 0.79 | 5.00 × 10−4 | 0.60 |
| Block | Lactation | Haplotype Combination (No.) | Milk Yield (kg) | Fat Yield (kg) | Fat Percentage (%) | Protein Yield (kg) | Protein Percentage (%) |
|---|---|---|---|---|---|---|---|
| Block 1 | 1 | H1H1 (195) | 10,262 ± 76.92 AaBb | 337.48 ± 3.33 ABb | 3.32 ± 0.03 | 305.66 ± 2.43 Aab | 2.99 ± 0.02 |
| H1H2 (240) | 10,279 ± 72.28 Aab | 343.53 ± 3.16 Aa | 3.37 ± 0.03 | 306.08 ± 2.30 Aab | 2.99 ± 0.02 | ||
| H1H3 (239) | 10,361 ± 73.07 Aa | 344.49 ± 3.19 Aa | 3.35 ± 0.03 | 307.36 ± 2.32 Aa | 2.98 ± 0.02 | ||
| H2H2 (85) | 10,011 ± 98.21 Bc | 333.51 ± 4.13 Bb | 3.35 ± 0.04 | 296.43 ± 3.01 Bc | 2.98 ± 0.03 | ||
| H2H3 (127) | 10,179 ± 84.35 ABbc | 340.18 ± 3.60 ab | 3.36 ± 0.03 | 301.76 ± 2.62 ABbc | 2.98 ± 0.02 | ||
| H3H3 (36) | 10,429 ± 135.36 Aab | 349.71 ± 5.59 Aa | 3.38 ± 0.05 | 310.28 ± 4.07 Aa | 2.99 ± 0.03 | ||
| p | 6.90 × 10−3 | 8.60 × 10−3 | 0.65 | 1.40 × 10−3 | 0.89 | ||
| 2 | H1H1 (139) | 9568.54 ± 86.65 bC | 328.45 ± 3.73 CcDd | 3.54 ± 0.04 b | 276.60 ± 2.72 bC | 2.99 ± 0.02 ABb | |
| H1H2 (175) | 9784.78 ± 79.70 aBC | 339.57 ± 3.46 aBb | 3.58 ± 0.03 ab | 282.20 ± 2.52 aBCc | 2.98 ± 0.02 ABb | ||
| H1H3 (181) | 9856.28 ± 80.13 aB | 345.62 ± 3.50 aB | 3.58 ± 0.03 ab | 285.28 ± 2.55 Bc | 2.97 ± 0.02 ABb | ||
| H2H2 (64) | 9210.82 ± 115.00 D | 321.36 ± 4.83 Dd | 3.64 ± 0.05 a | 263.15 ± 3.52 D | 2.99 ± 0.03 ABb | ||
| H2H3 (95) | 9587.56 ± 94.51 bC | 336.91 ± 4.00 BbCc | 3.62 ± 0.04 ab | 276.83 ± 2.92 abC | 2.99 ± 0.03 Ab | ||
| H3H3 (27) | 10,925 ± 165.48 A | 389.24 ± 6.80 A | 3.56 ± 0.07 ab | 315.08 ± 4.96 A | 2.88 ± 0.04 Ba | ||
| p | <1.00 × 10−4 | <1.00 × 10−4 | 0.38 | <1.00 × 10−4 | 0.16 | ||
| Block 2 | 1 | H1H1 (90) | 10,553 ± 95.65 Aa | 348.40 ± 4.04 | 3.33 ± 0.04 ab | 315.57 ± 2.94 Aa | 2.99 ± 0.03 |
| H1H3 (384) | 10,416 ± 64.06 Aab | 345.31 ± 2.84 | 3.33 ± 0.03 b | 310.34 ± 2.07 ABb | 2.98 ± 0.02 | ||
| H2H2 (9) | 10,429 ± 246.10 ac | 343.57 ± 9.97 | 3.29 ± 0.10 ab | 306.92 ± 7.27 abc | 2.95 ± 0.06 | ||
| H2H3 (107) | 10,192 ± 89.13 Bc | 345.23 ± 3.79 | 3.40 ± 0.04 a | 304.09 ± 2.76 Cc | 2.99 ± 0.02 | ||
| H3H3 (280) | 10,344 ± 69.79 ABbc | 342.45 ± 3.06 | 3.33 ± 0.03 b | 307.59 ± 2.23 BbCc | 2.98 ± 0.02 | ||
| p | 1.03 × 10−2 | 0.54 | 0.22 | 3.20 × 10−3 | 0.92 | ||
| 2 | H1H1 (67) | 8942.88 ± 108.87 C | 304.18 ± 4.57 Dd | 3.55 ± 0.04 abc | 254.06 ± 3.33 BC | 3.03 ± 0.03 | |
| H1H2 (35) | 9855.98 ± 140.50 A | 327.54 ± 5.80 ABbCc | 3.49 ± 0.06 ABc | 279.74 ± 4.23 A | 2.96 ± 0.04 | ||
| H1H3 (287) | 9709.93 ± 70.33 A | 329.43 ± 3.13 Bb | 3.54 ± 0.03 Bc | 276.85 ± 2.28 A | 2.99 ± 0.02 | ||
| H2H3 (80) | 9100.98 ± 101.99 BC | 316.08 ± 4.31 CcD | 3.63 ± 0.04 AaBb | 258.64 ± 3.14 B | 3.00 ± 0.03 | ||
| H3H3 (204) | 9726.49 ± 76.24 A | 340.41 ± 3.33 Aa | 3.62 ± 0.03 Aa | 279.76 ± 2.43 A | 2.98 ± 0.02 | ||
| p | <1.00 × 10−4 | <1.00 × 10−4 | 9.10 × 10−3 | <1.00 × 10−4 | 0.29 |
| GENE | SNP | Allele | Transcription Factor | Relative Score | Predicted Binding Site Sequence |
|---|---|---|---|---|---|
| ACOX2 | rs109066086 | C | — | — | — |
| T | NR2C2 | 0.91 | CAGGTGAT | ||
| TFAP4 | 0.90 | GCCAGGTGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, H.; Wang, Z.; Xu, L.; Han, B.; Sun, D. Genetic Associations of ACOX2 Gene with Milk Yield and Composition Traits in Chinese Holstein Cows. Animals 2025, 15, 953. https://doi.org/10.3390/ani15070953
Cao H, Wang Z, Xu L, Han B, Sun D. Genetic Associations of ACOX2 Gene with Milk Yield and Composition Traits in Chinese Holstein Cows. Animals. 2025; 15(7):953. https://doi.org/10.3390/ani15070953
Chicago/Turabian StyleCao, Hui, Zhe Wang, Lingna Xu, Bo Han, and Dongxiao Sun. 2025. "Genetic Associations of ACOX2 Gene with Milk Yield and Composition Traits in Chinese Holstein Cows" Animals 15, no. 7: 953. https://doi.org/10.3390/ani15070953
APA StyleCao, H., Wang, Z., Xu, L., Han, B., & Sun, D. (2025). Genetic Associations of ACOX2 Gene with Milk Yield and Composition Traits in Chinese Holstein Cows. Animals, 15(7), 953. https://doi.org/10.3390/ani15070953






