Polyphenol-Containing Feed Additive Polygain™ Reduces Methane Production and Intensity from Grazing Dairy Cows Measured Using an Inverse-Dispersion Technique
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
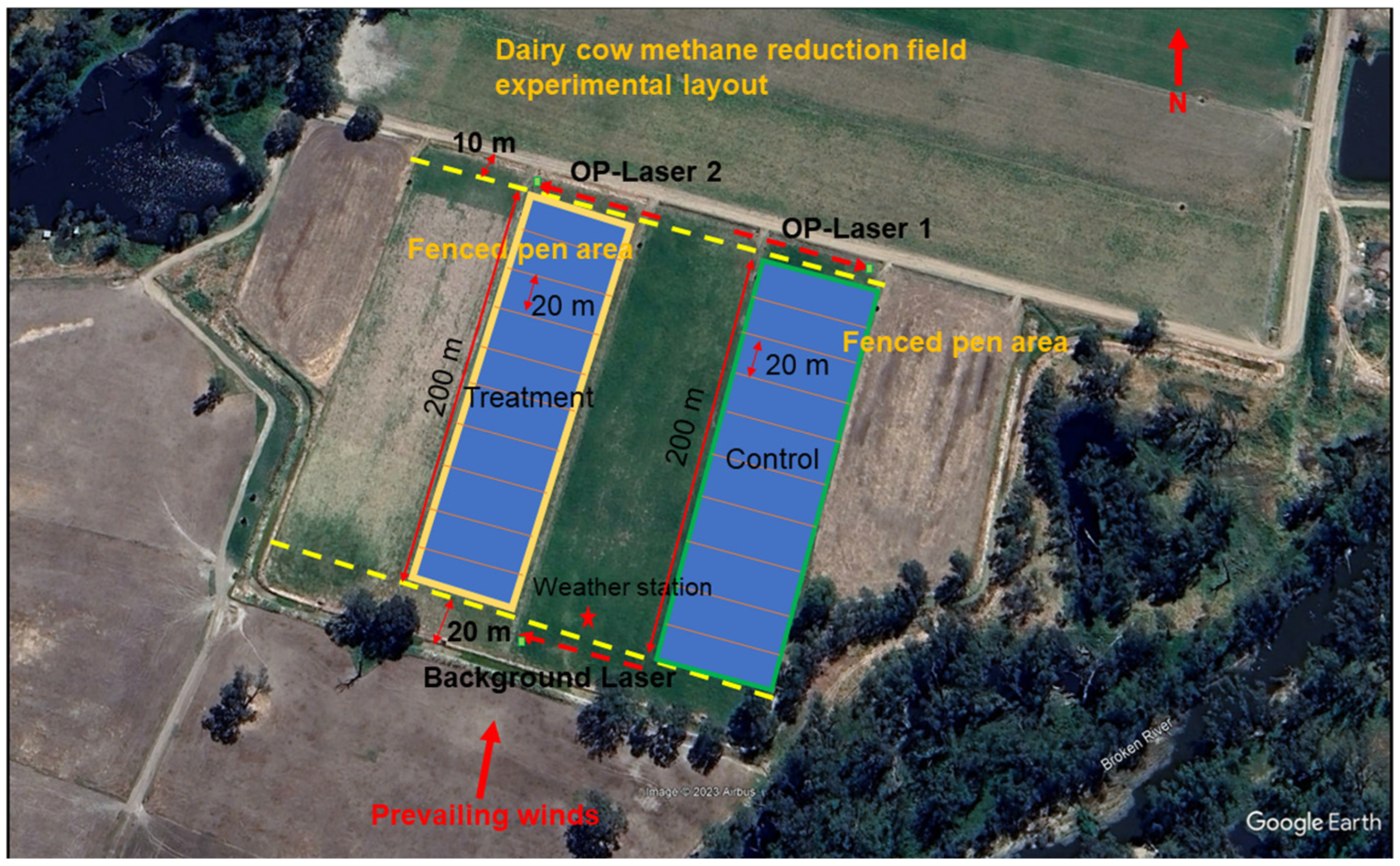
2.1. Animals and Experimental Design
2.2. Methodologies
2.2.1. Inverse-Dispersion Method (IDM)
2.2.2. Instrumentations
2.2.3. Cross-Calibration of Concentration Sensors
2.2.4. Data Filtering
2.2.5. Statistical Analysis
3. Results
3.1. Milk Production, Animal Weight Gain, and Feed Intake
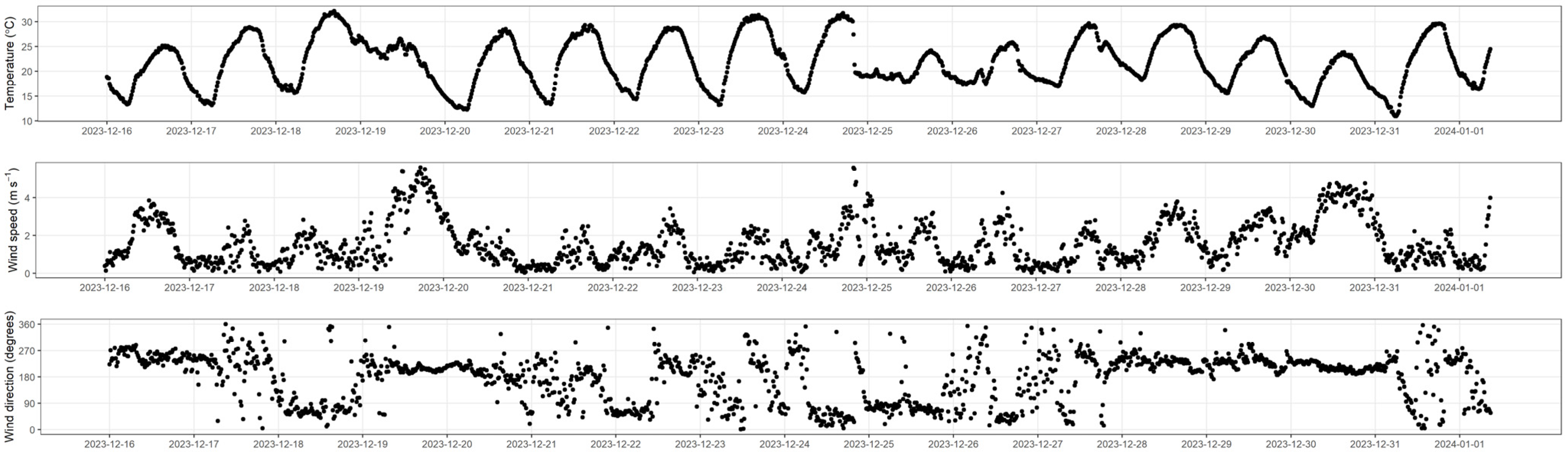
3.2. Climate Data
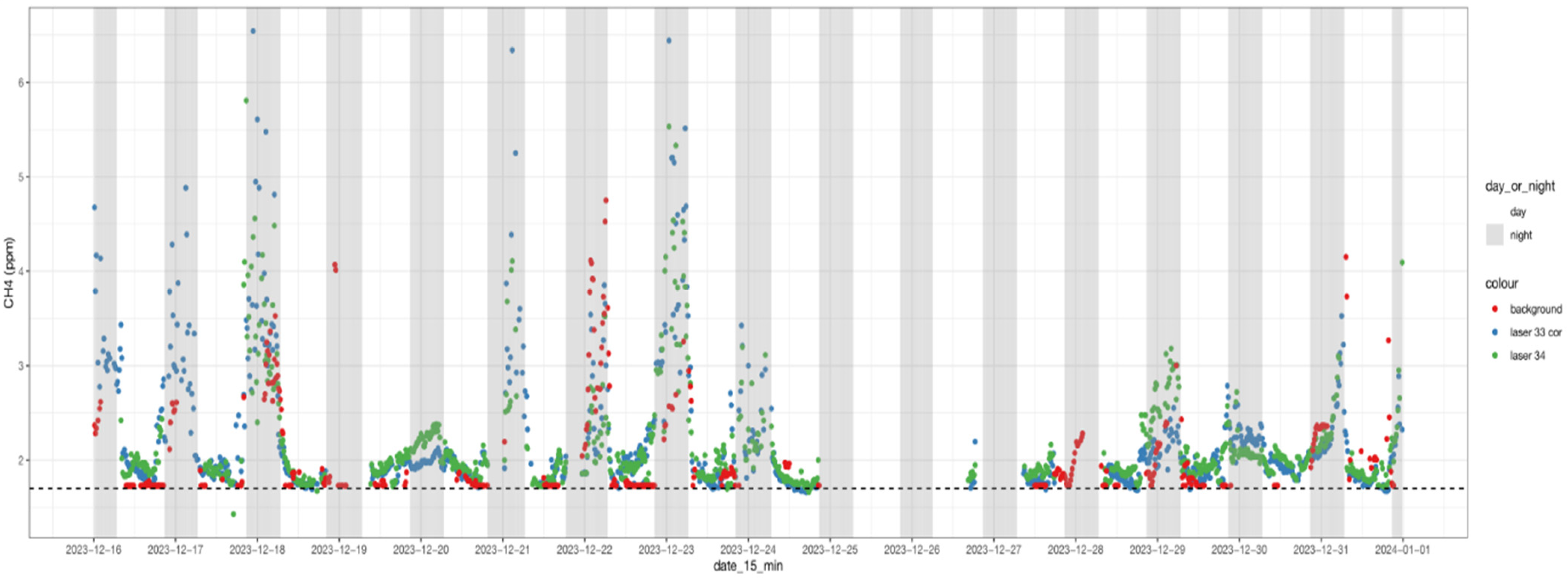
3.3. Fifteen-Minute Average CH4 Concentrations
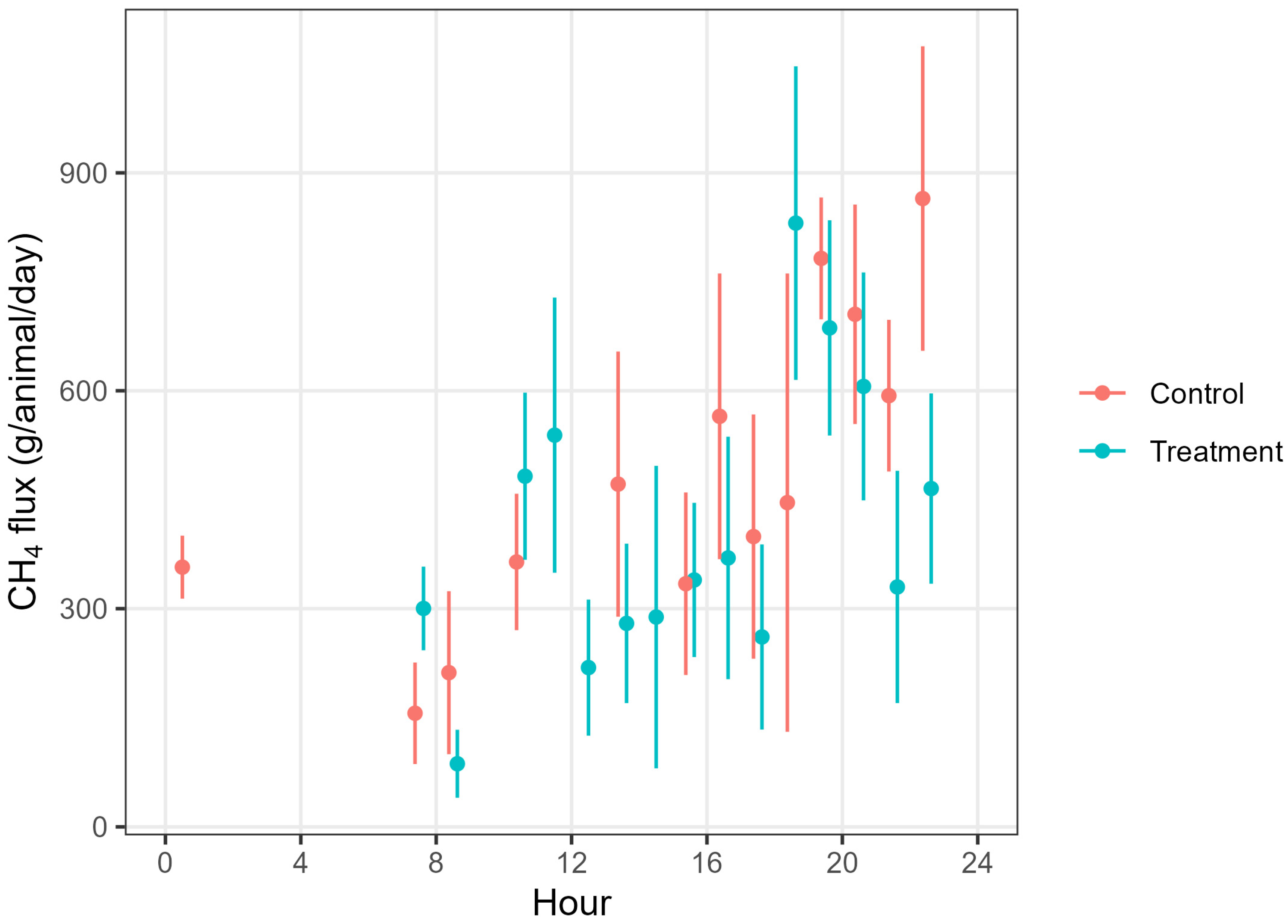
3.4. CH4 Fluxes and Total GHG Emissions (CO2 Equivalents, CO2-e)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GHG | Greenhouse gas emissions |
| CH4 | Methane |
| N2O | Nitrous oxide |
| PRSFM | Polyphenol-rich sugarcane feed material |
| OP-FTIR | Open-path Fourier-transform infrared spectroscopy |
| IDM | Inverse-dispersion model |
| OPL | Open-path laser |
| MP | Methane production |
| MI | Methane intensity |
| DMP | Daily milk production |
| BW | Body weight |
| CO2-e | CO2 equivalents |
References
- FAO. Methane Emissions in Livestock and Rice System Sources, Quantification, Mitigation and Metrics; FAO: Rome, Italy, 2023. [Google Scholar] [CrossRef]
- IPCC. Climate Change: Impacts, Adaptation, and Vulnerability. In Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Portner, H.O., Roberts, D.C., Tignor, M., Poloczanska, E.S., Mintenbeck, K., Alegría, A., Craig, M., Langsdorf, S., Loschke, S., Moller, V., et al., Eds.; Cambridge University Press: Cambridge, UK, 2022. [Google Scholar] [CrossRef]
- EEA. Trends and Projections in Europe. European Environment Agency (EEA) Report 11/2024; EEA: Copenhagen, Denmark, 2024; 66p, Available online: https://www.eea.europa.eu/en/analysis/publications/trends-and-projections-in-europe-2024 (accessed on 5 March 2025).
- Knapp, J.R.; Laur, G.L.; Vadas, P.A.; Weiss, W.P.; Tricarico, J.M. Invited review: Enteric methane in dairy cattle production: Quantifying the opportunities and impact of reducing emissions. J. Dairy Sci. 2014, 97, 3231–3261. [Google Scholar] [PubMed]
- Duan, Y.; Gao, Y.; Zhao, J.; Xue, Y.; Zhang, W.; Wu, W. Agricultural methane emissions in China: Inventories, driving forces and mitigation strategies. Environ. Sci. Tech. 2023, 57, 13292–13303. [Google Scholar] [CrossRef] [PubMed]
- Panchasara, H.; Samrat, N.H.; Islam, N. Greenhouse gas emissions trends and mitigation measures in Australian agriculture sector—A review. Agriculture 2021, 11, 85. [Google Scholar] [CrossRef]
- Dairy Australia. Australian dairy industry. In ‘Focus’; Dairy Australia Ltd.: Melbourne, VIC, Australia, 2025; Available online: https://www.dairyaustralia.com.au/climate-and-environment/greenhouse-gas-emissions/dairy-farm-emissions (accessed on 3 March 2025).
- EPA (US Environ. Prot. Agency). Inventory of U.S. Greenhouse Gas Emissions and Sinks: 1990–2021. 2023. Available online: https://www.epa.gov/ghgemissions/inventory-us-greenhouse-gas-emissions-and-sinks-1990-2021 (accessed on 3 March 2025).
- Rotz, C.A.; Mitloehner, F. United States Dairy Farms and Global Warming. J. Dairy Sci. 2025, 108, 2610–2619. [Google Scholar] [CrossRef]
- Mulatu, K.A.; Nyawira, S.S.; Herold, M.; Carter, S.; Verchot, L. Nationally determined contributions to the 2015 Paris Agreement goals: Transparency in communications from developing country Parties. Clim. Policy 2024, 24, 211–227. [Google Scholar] [CrossRef]
- Li, L.; Hu, X.; Liu, L.; Zhang, Q. Assessing methane emissions and multiple benefits of mitigation through coupled crop-livestock system. J. Clean. Prod. 2024, 477, 143815. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Richardson, C.M.; Post, M.; Amer, P.R.; Nieuwhof, G.J.; Thurn, P.; Shaffer, M. The sustainability index: A new tool to breed for reduced greenhouse-gas emissions intensity in Australian dairy cattle. Anim. Prod. Sci. 2023, 63, 1126–1135. [Google Scholar]
- Garnett, L.M.; Eckard, R.J. Greenhouse-gas abatement on Australian dairy farms: What are the options? Anim. Prod. Sci. 2024, 64, AN24139. [Google Scholar]
- Beauchemin, K.A.; Kebreab, E.; Cain, M.; VandeHaar, M.J. The path to net-zero in dairy production: Are pronounced decreases in enteric methane achievable? Annu. Rev. Anim. Biosci. 2025, 13, 325–341. [Google Scholar]
- Martins, L.F.; Cueva, S.F.; Wasson, D.E.; Almeida, C.V.; Eifert, C.; de Ondarza, M.B.; Tricarico, J.M.; Hristov, A.N. Effects of dose, dietary nutrient composition, and supplementation period on the efficacy of methane mitigation strategies in dairy cows: A meta-analysis. J. Dairy Sci. 2024, 107, 9289–9308. [Google Scholar]
- Beauchemin, K.A.; Eckard, R.J.; Ungerfeld, E.M.; Wang, M. Review: Fifty years of research on rumen methanogenesis: Lessons learned and future challenges for mitigation. Animal 2020, 14, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.L.; Bionaz, M.; MacAdam, J.W.; Beauchemin, K.A.; Naumann, H.D.; Ates, S. Milk production, nitrogen utilization, and methane emissions of dairy cows grazing grass, forb, and legume-based pastures. J. Anim. Sci. 2020, 98, skaa220. [Google Scholar] [CrossRef] [PubMed]
- Munoz, C.; Villalobos, R.; Peralta, A.M.T.; Morales, R.; Urrutia, N.L.; Ungerfeld, E.M. Long-term and carryover effects of supplementation with whole oilseeds on methane emission, milk production and milk fatty acid profile of grazing dairy cows. Animals 2021, 11, 2978. [Google Scholar] [CrossRef]
- Hollmann, M.; Powers, W.J.; Fogiel, A.C.; Liesman, J.S.; Beede, D.K. Response profiles of enteric methane emissions and lactational performance during habituation to dietary coconut oil in dairy cows. J. Dairy Sci. 2013, 96, 1769–1781. [Google Scholar] [CrossRef]
- Ungerfeld, E.M.; Pitta, D. Review: Biological consequences of the inhibition of rumen methanogenesis. Animal 2024, 101170. [Google Scholar] [CrossRef]
- Meo-Filho, P.; Hood, J.; Lee, M.; Fleming, H.; Meethal, M.; Misselbrook, T. Performance and enteric methane emissions from housed beef cattle fed silage produced on pastures with different forage profiles. Animal 2023, 17, 100726. [Google Scholar] [CrossRef]
- Singh, S.; Kaur, I.; Kariyat, R. The multifunctional roles of polyphenols in plant-herbivore interactions. Int. J. Mol. Sci. 2021, 22, 1442. [Google Scholar] [CrossRef]
- Formato, M.; Cimmino, G.; Brahmi-Chendouh, N.; Piccolella, S.; Pacifico, S. Polyphenols for livestock feed: Sustainable perspectives for animal husbandry? Molecules 2022, 27, 7752. [Google Scholar] [CrossRef]
- Yang, C.; Chowdhury, M.K.; Hou, Y.; Gong, J. Phytogenic compounds as alternatives to in-feed antibiotics: Potentials and challenges in application. Pathogens 2015, 4, 137–156. [Google Scholar] [CrossRef]
- Honan, M.; Feng, X.; Tricarico, J.M.; Kebreab, E. Feed additives as a strategic approach to reduce enteric methane production in cattle: Modes of action, effectiveness and safety. Anim. Prod. Sci. 2022, 62, 1303–1317. [Google Scholar] [CrossRef]
- Wang, J.; Deng, L.; Chen, M.; Che, Y.; Li, L.; Zhu, L.; Chen, G.; Feng, T. Phytogenic feed additives as natural antibiotic alternatives in animal health and production: A review of the literature of the last decade. Anim. Nutr. 2024, 17, 244–264. [Google Scholar] [CrossRef] [PubMed]
- Hassan, F.U.; Arshad, M.A.; Ebeid, H.M.; Rehman, M.S.U.; Khan, M.S.; Shahid, S.; Yang, C. Phytogenic additives can modulate rumen microbiome to mediate fermentation kinetics and methanogenesis through exploiting diet–microbe interaction. Front. Vet. Sci. 2020, 7, 575801. [Google Scholar] [CrossRef]
- Sun, X.; Shukla, M.; Wang, W.; Li, S. Unlocking gut-liver-brain axis communication metabolites: Energy metabolism, immunity and barriers. NPJ Biofilms Microbiomes 2024, 10, 136. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K.; Amasheh, S.; Aschenbach, J.R. Modulation of gastrointestinal barrier and nutrient transport function in farm animals by natural plant bioactive compounds–a comprehensive review. Crit. Rev. Food Sci. Nutr. 2019, 59, 3237–3266. [Google Scholar]
- Hassan, F.U.; Arshad, M.A.; Li, M.; Rehman, M.S.U.; Loor, J.J.; Huang, J. Potential of mulberry leaf biomass and its flavonoids to improve production and health in ruminants: Mechanistic insights and prospects. Animals 2020, 10, 2076. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, L.; Riqing, D.; Qu, J.; Chen, J.; Zhandu, D.; Li, B.; Jiang, M. Microbial metagenomes and host transcriptomes reveal the dynamic changes of rumen gene expression, microbial colonization and co-regulation of mineral element metabolism in yaks from birth to adulthood. Animals 2024, 14, 1365. [Google Scholar] [CrossRef]
- McKinlay, J.B. Are bacteria leaky? Mechanisms of metabolite externalization in bacterial cross-feeding. Annu. Rev. Microbiol. 2023, 77, 277–297. [Google Scholar] [CrossRef]
- Makmur, M.; Zain, M.; Sholikin, M.M.; Jayanegara, A. Modulatory effects of dietary tannins on polyunsaturated fatty acid biohydrogenation in the rumen: A meta-analysis. Heliyon 2022, 8, e09828. [Google Scholar]
- Ahmed, M.G.; Elwakeel, E.A.; El-Zarkouny, S.Z.; Al-Sagheer, A.A. Environmental impact of phytobiotic additives on greenhouse gas emission reduction, rumen fermentation manipulation, and performance in ruminants: An updated review. Environ. Sci. Pollut. Res. 2024, 31, 37943–37962. [Google Scholar] [CrossRef]
- Tian, X.; Qin, J.; Luo, Q.; Xu, Y.; Xie, S.; Chen, R.; Wang, X.; Lu, Q. Differences in chemical composition, polyphenol compounds, antioxidant activity, and in vitro rumen fermentation among sorghum stalks. Animals 2024, 14, 415. [Google Scholar] [CrossRef]
- Jo, S.U.; Lee, S.J.; Kim, H.S.; Eom, J.S.; Choi, Y.; Oh, D.S.; Bae, D.; Lee, S.S. Effects of oriental medicinal plants on the reduction of methane production mediated by microbial population. Ital. J. Anim. Sci. 2022, 21, 522–531. [Google Scholar]
- Ahmed, A.; Flavel, M.; Mitchell, S.; Macnab, G.; Dunuarachchige, M.D.; Desai, A. Increased milk yield and reduced enteric methane concentration on a commercial dairy farm associated with dietary inclusion of sugarcane extract (Saccharum officinarum). Animals 2023, 13, 3300. [Google Scholar] [CrossRef] [PubMed]
- Prathap, P.; Chauhan, S.S.; Flavel, M.; Mitchell, S.; Cottrell, J.J.; Leury, B.J. Effects of sugarcane-derived polyphenol supplementation on methane production and rumen microbial diversity of second-cross lambs. Animals 2024, 14, 905. [Google Scholar] [CrossRef]
- Wijesiriwardana, U.A.; Pluske, J.R.; Craig, J.R.; Cottrell, J.J.; Dunshea, F.R. Evaluation of sugarcane-derived polyphenols on the pre-weaning and post-weaning growth of gilt progeny. Animals 2020, 10, 984. [Google Scholar] [CrossRef]
- Prathap, P.; Chauhan, S.S.; Leury, B.J.; Cottrell, J.J.; Joy, A.; Zhang, M. Effects of feeding a commercial starch binding agent during heat stress on enteric methane emission, rumen volatile fatty acid contents, and diet digestibility of merino lambs. Atmosphere 2023, 14, 605. [Google Scholar] [CrossRef]
- Penglase, S.; Ackery, T.; Kitchen, B.; Flavel, M.; Condon, K. The effects of a natural polyphenol extract from sugarcane (Saccharum officinarum) on growth, survival, and feed conversion efficiency of juvenile black tiger shrimp (Penaeus monodon). Appl. Sci. 2022, 12, 8090. [Google Scholar] [CrossRef]
- Tedeschi, L.O.; Abdalla, A.L.; Alvarez, C.; Anuga, S.W.; Arango, J.; Beauchemin, K.A.; Becquet, P.; Berndt, A.; Burns, R.; De Camillis, C.; et al. Quantification of methane emitted by ruminants: A review of methods. J. Anim. Sci. 2022, 100, skac197. [Google Scholar]
- Griffith, D.W.T.; Bryant, G.R.; Hsu, D.; Reisinger, A.R. Methane emissions from free-ranging cattle: Comparison of tracer and integrated horizontal flux techniques. J. Environ. Qual. 2008, 37, 582–591. [Google Scholar]
- Judd, M.J.; Kellier, F.M.; Ulyatt, M.J.; Lassey, K.R.; Tate, K.R.; Shelton, D. Net methane emissions from grazing sheep. Glob. Change Biol. 1999, 5, 647–657. [Google Scholar] [CrossRef]
- Coates, T.W.; Chen, D.; Bai, M. Reducing Greenhouse Gas Emissions from Livestock Production; Burleigh Dodds Science Publishing: Cambridge, UK, 2021. [Google Scholar]
- Laubach, J.; Bai, M.; Pinares-Patino, C.S.; Phillips, F.A.; Naylor, T.A.; Molano, G.; Rocha, E.A.C.; Griffith, D.W. Accuracy of micrometeorological techniques for detecting a change in methane emissions from a herd of cattle. Agric. For. Meteorol. 2013, 176, 50–63. [Google Scholar] [CrossRef]
- Waghorn, G.C.; Jonker, A.; Macdonald, K.A. Measuring methane from grazing dairy cows using GreenFeed. Anim. Prod. Sci. 2016, 56, 252–257. [Google Scholar] [CrossRef]
- Laubach, J.; Kelliher, F.M. Measuring methane emission rates of a dairy cow herd by two micrometeorological techniques. Agric. Forest Meteorol. 2004, 125, 279–303. [Google Scholar] [CrossRef]
- Bai, M.; Velazco, J.I.; Coates, T.W.; Phillips, F.A.; Flesch, T.K.; Hill, J.; Mayer, D.G.; Tomkins, N.W.; Hegarty, R.S.; Chen, D. On-farm beef cattle methane emissions measured with tracer-ratio and inverse-dispersion modelling techniques. Atmos. Meas. Tech. Discuss. 2020, 14, 3469–3479. [Google Scholar] [CrossRef]
- Laubach, J.; Flesch, T.K.; Ammann, C.; Bai, M.; Gao, Z.; Merbold, L.; Campbell, D.I.; Goodrich, J.P.; Graham, S.L.; Hunt, J.E.; et al. Methane emissions from animal agriculture: Micrometeorological solutions for challenging measurement situations. Agric. For. Meteorol. 2024, 350, 109971. [Google Scholar] [CrossRef]
- Flesch, T.K.; Basarab, J.A.; Baron, V.S.; Wilson, J.D.; Tomkins, N.W.; Ohama, A.J. Methane emissions from cattle grazing under diverse conditions: An examination of field configurations appropriate for line-averaging sensors. Agric. For. Meteorol. 2018, 258, 8–17. [Google Scholar] [CrossRef]
- Shakeri, M.; Cottrell, J.J.; Wilkinson, S.; Le, H.H.; Suleria, H.A.; Warner, R.D.; Dunshea, F.R. A dietary sugarcane-derived polyphenol mix reduces the negative effects of cyclic heat exposure on growth performance, blood gas status, and meat quality in broiler chickens. Animals 2020, 10, 1158. [Google Scholar] [CrossRef]
- Iqbal, Y.; Cottrell, J.J.; Suleria, H.A.; Dunshea, F.R. Gut microbiota-polyphenol interactions in chicken: A review. Animals 2020, 10, 1391. [Google Scholar] [CrossRef]
- Weiss, W.P. Brief Introduction to the NASEM (Formerly Known as NRC) of the Nutrient Requirements of Dairy Cattle. 2021. Available online: https://www.txanc.org/Proceedings/2022/2%20-%20Weiss1%20-%20Where%20are%20we%20today%20with%20requirements%20of%20the%20modern%20dairy%20cow.pdf (accessed on 5 March 2025).
- BOM. Australian Government Bureau of Meteorology. 2025. Available online: http://www.bom.gov.au/vic/forecasts/map.shtml?ref=hdr (accessed on 5 March 2025).
- Flesch, T.K.; Wilson, J.D.; Harper, L.A.; Crenna, B.P.; Sharpe, R.R. Deducing ground-to-air emissions from observed trace gas concentrations: A field trial. J. Appl. Meteorol. 2004, 43, 487–502. [Google Scholar] [CrossRef]
- Flesch, T.K.; Wilson, J.D.; Yee, E. Backward-time Lagrangian stochastic dispersion models and their application to estimate gaseous emissions. J. Appl. Meteorol. 1995, 34, 1320–1332. [Google Scholar] [CrossRef]
- Harper, L.A.; Denmead, O.T.; Flesch, T.K. Micrometeorological techniques for measurement of enteric greenhouse gas emissions. Anim. Feed Sci. Tech. 2011, 166, 227–239. [Google Scholar]
- Loh, Z.; Chen, D.; Bai, M.; Naylor, T.; Griffith, D.; Hill, J. Measurement of greenhouse gas emissions from Australian feedlot beef production using open-path spectroscopy and atmospheric dispersion modelling. Aust. J. Exp. Agric. 2008, 48, 244–247. [Google Scholar]
- Samad, H.A.; Kumar Eshwaran, V.; Muquit, S.P.; Sharma, L.; Arumugam, H.; Kant, L.; Fatima, Z.; Sharun, K.; Aradotlu Parameshwarappa, M.; Latheef, S.K.; et al. Sustainable livestock solutions: Addressing carbon footprint challenges from Indian and global perspectives. Sustainability 2025, 17, 2105. [Google Scholar] [CrossRef]
- Necki, J.; Bartyzel, J.; Jagoda, P.; Swolkien, J.; Field, R. Application of TDLAS analysers, challenges and implications for future monitoring of methane emission from the coal mine ventilation shafts. In Proceedings of the EGU General Assembly Conference Abstracts 2024, Vienna, Austria, 14–19 April 2024; p. 17703. [Google Scholar]
- McGinn, S.M.; Beauchemin, K.A.; Coates, T.; Colombatto, D. Methane emissions from beef cattle: Effects of monensin, sunflower oil, enzymes, yeast, and fumaric acid. J. Anim. Sci. 2004, 82, 3346–3356. [Google Scholar] [CrossRef] [PubMed]
- Flesch, T.K.; Harper, L.A.; Powell, J.M.; Wilson, J.D. Inverse-dispersion calculation of ammonia emissions from Wisconsin dairy farms. J. ASABE Trans. ASABE Appl. Eng. 2009, 52, 253–265. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 5 March 2024).
- IPCC. IPCC Guidelines for National Greenhouse Gas Inventories, Prepared by the National Greenhouse Gas Inventories Program; Eggleston, H.S., Buendia, L., Miwa, K., Ngara, T., Tanabe, K., Eds.; IGES: Hayama, Japan, 2006. [Google Scholar]
- Alvarado-Bolovich, V.; Medrano, J.; Haro, J.; Castro-Montoya, J.; Dickhoefer, U.; Gomez, C. Enteric methane emissions from lactating dairy cows grazing cultivated and native pastures in the high Andes of Peru. Live Sci. 2021, 243, 104385. [Google Scholar]
- Lahart, B.; Buckley, F.; Herron, J.; Fitzgerald, R.; Fitzpatrick, E.; Galvin, N. Evaluating enteric methane emissions within a herd of genetically divergent grazing dairy cows. J. Dairy Sci. 2024, 7, 383–397. [Google Scholar]
- Lassey, K.R.; Ulyatt, M.J.; Martin, R.J.; Walker, C.F.; Shelton, I.D. Methane Emissions Measured Directly from Grazing Livestock in New Zealand; Pergamon Press Inc.: Oxford, UK, 1997; p. 2905. [Google Scholar]
- McParland, S.; Frizzarin, M.; Lahart, B.; Kennedy, M.; Shalloo, L.; Egan, M. Predicting methane emissions of individual grazing dairy cows from spectral analyses of their milk samples. J. Dairy Sci. 2024, 7, 978–991. [Google Scholar]
- Lassen, J.; Lovendahl, P. Heritability estimates for enteric methane emissions from Holstein cattle measured using noninvasive methods. J. Dairy Sci. 2016, 99, 1959–1967. [Google Scholar]
- Villanueva, C.; Ibrahim, M.; Castillo, C. Enteric methane emissions in dairy cows with different genetic groups in the humid tropics of Costa Rica. Animals 2023, 13, 730. [Google Scholar] [CrossRef]
- Cavanagh, A.; McNaughton, L.; Clark, H.; Greaves, C.; Gowan, J.M.; Pinares-Patino, C.; Dalley, D.; Vlaming, B.; Molano, G. Methane emissions from grazing Jersey-Friesian dairy cows in mid lactation. Aust. J. Exp. Agric. 2008, 48, 230–233. [Google Scholar] [CrossRef]
- Pedreira, M.D.S.; Primavesi, O.; Lima, M.A.; Frighetto, R.; de Oliveira, S.G.; Berchielli, T.T. Ruminal methane emission by dairy cattle in Southeast Brazil. Sci. Agric. 2009, 66, 743–750. [Google Scholar] [CrossRef]
- Bai, M. Methane Emissions from Livestock Measured by Novel Spectroscopic Techniques. Ph.D. Thesis, University of Wollongong, Wollongong, NSW, Australia, 2010; p. 303. [Google Scholar]
- VanderZaag, A.C.; Flesch, T.K.; Desjardins, R.L.; Balde, H.; Wright, T. Measuring methane emissions from two dairy farms: Seasonal and manure-management effects. Agric. For. Meteorol. 2014, 194, 259–267. [Google Scholar]
- Weerasekara, C.; Morris, L.C.; Malarich, N.A.; Giorgetta, F.R.; Herman, D.I.; Cossel, K.C.; Newbury, N.R.; Owensby, C.E.; Welch, S.M.; Blaga, C.; et al. Using open-path dual-comb spectroscopy to monitor methane emissions from simulated grazing cattle. Atmos. Meas. Tech. 2024, 17, 6107–6117. [Google Scholar]
- Hani, C.; Neftel, A.; Flechard, C.; Ammann, C.; Valach, A.; Kupper, T. Validation of a short-range dispersion and deposition model using field-scale ammonia and methane release experiments. Agric. For. Meteorol. 2024, 353, 110041. [Google Scholar]
- Tomkins, N.W.; McGinn, S.M.; Turner, D.A.; Charmley, E. Comparison of open-circuit respiration chambers with a micrometeorological method for determining methane emissions from beef cattle grazing a tropical pasture. Anim. Feed. Sci. Tech. 2011, 166, 240–247. [Google Scholar]
- McGinn, S.M.; Turner, D.; Tomkins, N.; Charmley, E.; Bishop-Hurley, G.; Chen, D. Methane Emissions from Grazing Cattle Using Point-Source Dispersion. J. Environ. Qual. 2011, 40, 22–27. [Google Scholar] [CrossRef]
- Denmead, O.T.; Harper, L.A.; Freney, J.R.; Griffith, D.W.T.; Leuning, R.; Sharpe, R.R. A mass balance method for non-intrusive measurements of surface-air trace gas exchange. Atmos. Environ. 1998, 32, 3679–3688. [Google Scholar]
- Bai, M.; Suter, H.; Lam, S.K.; Sun, J.; Chen, D. Use of open-path FTIR and inverse dispersion technique to quantify gaseous nitrogen loss from an intensive vegetable production site. Atmos. Environ. 2014, 94, 687–691. [Google Scholar]
- Laubach, J. Testing of a Lagrangian model of dispersion in the surface layer with cattle methane emissions. Agric. For. Meteorol. 2010, 150, 1428–1442. [Google Scholar]
- Manoni, M.; Gschwend, F.; Amelchanka, S.; Terranova, M.; Pinotti, L.; Widmer, F.; Silacci, P.; Tretola, M. Gallic and ellagic acids differentially affect microbial community structures and methane emission when using a rumen simulation technique. J. Agric. Food Chem. 2024, 72, 27163–27176. [Google Scholar]
- Lambo, M.T.; Ma, H.; Liu, R.; Dai, B.; Zhang, Y.; Li, Y. Mechanism, effectiveness, and the prospects of medicinal plants and their bioactive compounds in lowering ruminants’ enteric methane emission. Animal 2024, 18, 101134. [Google Scholar] [CrossRef] [PubMed]
- Roque, B.M.; Salwen, J.K.; Kinley, R.; Kebreab, E. Inclusion of Asparagopsis armata in lactating dairy cows’ diet reduces enteric methane emission by over 50 percent. J. Clean. Prod. 2019, 234, 132–138. [Google Scholar] [CrossRef]
- Eugene, M.; Masse, D.; Chiquette, J.; Benchaar, C. Meta-analysis on the effects of lipid supplementation on methane production in lactating dairy cows. Can. J. Anim. Sci. 2008, 88, 331–337. [Google Scholar] [CrossRef]
- Patra, A.K.; Kamra, D.N.; Bhar, R.; Kumar, R.; Agarwal, N. Effect of Terminalia chebula and Allium sativum on in vivo methane emission by sheep. J. Anim. Physiol. Anim. Nutr. 2011, 95, 187–191. [Google Scholar] [CrossRef]
- Kolling, G.J.; Stivanin, S.C.B.; Gabbi, A.M.; Machado, F.S.; Ferreira, A.L.; Campos, M.M. Performance and methane emissions in dairy cows fed oregano and green tea extracts as feed additives. J. Dairy Sci. 2018, 101, 4221–4234. [Google Scholar] [CrossRef]
- Rezaei Ahvanooei, M.R.; Norouzian, M.A.; Hedayati, M.; Ghaffari, M.H. Comprehensive review of resveratrol as a feed additive in dairy cows: Exploring its potential diverse effects and implications. Vet. Res. Commun. 2023, 47, 1115–1124. [Google Scholar] [CrossRef]
- Kumari, N.; Kala, A.; Agarwal, N.; Chaturvedi, V.B.; Vijayalakshmy, K.; Chaudhary, L.C. Effect of plant-derived feed additives on enteric methane emission, nutrient digestibility and immune status of adult male goats. J. Anim. Physiol. Anim. Nutr. 2025, 1–8. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Diao, Q.; Ma, T.; Tu, Y. In-silico and in vitro studies revealed that osmarinic acid inhibited methanogenesis via regulating composition and function of rumen microbiota. J. Dairy Sci. 2024, 107, 7904–7917. [Google Scholar] [CrossRef]
- Tondini, S.M.; Bayat, A.R.; Khelil-Arfa, H.; Blanchard, A.; Yanez-Ruiz, D.R. Effect of a blend of cinnamaldehyde, eugenol and capsicum oleoresin on methane emission and lactation performance of Nordic Red dairy cows fed grass silage-based diets. Liv. Sci. 2024, 284, 105494. [Google Scholar] [CrossRef]
- Adeniji, Y.A.; Bomberger, R.; Goodall, S.R.; Hristov, A.N.; Stefenoni, H.A.; Harvatine, K.J. Effect of increasing dietary fat by feeding 15% whole cottonseed on milk production, total-tract digestibility, and methane emission in dairy cows. J. Dairy Sci. 2025, 108, 2393–2406. [Google Scholar] [CrossRef]
- Kelly, L.; Pressman, E.M.; Ramirez-Agudelo, J.F.; Chernavsky, H.; Alvarez-Hess, P.; Jacques, S.; Hess, M.; Kebreab, E. The effect of Rumin 8 investigational veterinary product—A bromoform based feed additive–on enteric methane emissions, animal production parameters, and the rumen environment in feedlot cattle. Transl. Anim. Sci. 2025, txaf028. [Google Scholar] [CrossRef]
- Dairy Industry Code, Department of Agriculture, Fisheries and Forestry (DAFF). Available online: https://www.agriculture.gov.au/agriculture-land/farm-food-drought/meat-wool-dairy/dairy-dairycode (accessed on 5 March 2025).
- Prospering in a Low-Emissions World: An Updated Climate Policy Toolkit for Australia. Available online: https://www.climatechangeauthority.gov.au/prospering-low-emissions-world-updated-climate-policy-toolkit-australia (accessed on 5 March 2025).
- Driving Emissions Reduction in the Agriculture and Land Sectors. DAFF. Available online: https://www.agriculture.gov.au/sites/default/files/documents/2024-25-budget-driving-emissions-reduction-in-the-agriculture-and-land-sectors.pdf (accessed on 5 March 2025).
- SBTi Companies Taking Action. 2025. Available online: https://sciencebasedtargets.org/companies-taking-action#dashboard (accessed on 5 March 2025).
- Christie, K.M.; Gourley, C.J.P.; Rawnsley, R.P.; Eckard, R.J.; Awty, I.M. Whole-farm systems analysis of Australian dairy farm greenhouse gas emissions. Anim. Prod. Sci. 2012, 52, 998–1011. [Google Scholar]
- Laubach, J.; Kellier, F.M. Measuring methane emission rates of a dairy cow herd (II): Results from a backward-Lagrangian stochastic model. Agric. For. Meteorol. 2005, 129, 137–150. [Google Scholar]
- Kroliczewska, B.; Pecka-Kielb, E.; Bujok, J. Strategies used to reduce methane emissions from ruminants: Controversies and issues. Agriculture 2023, 13, 602. [Google Scholar] [CrossRef]
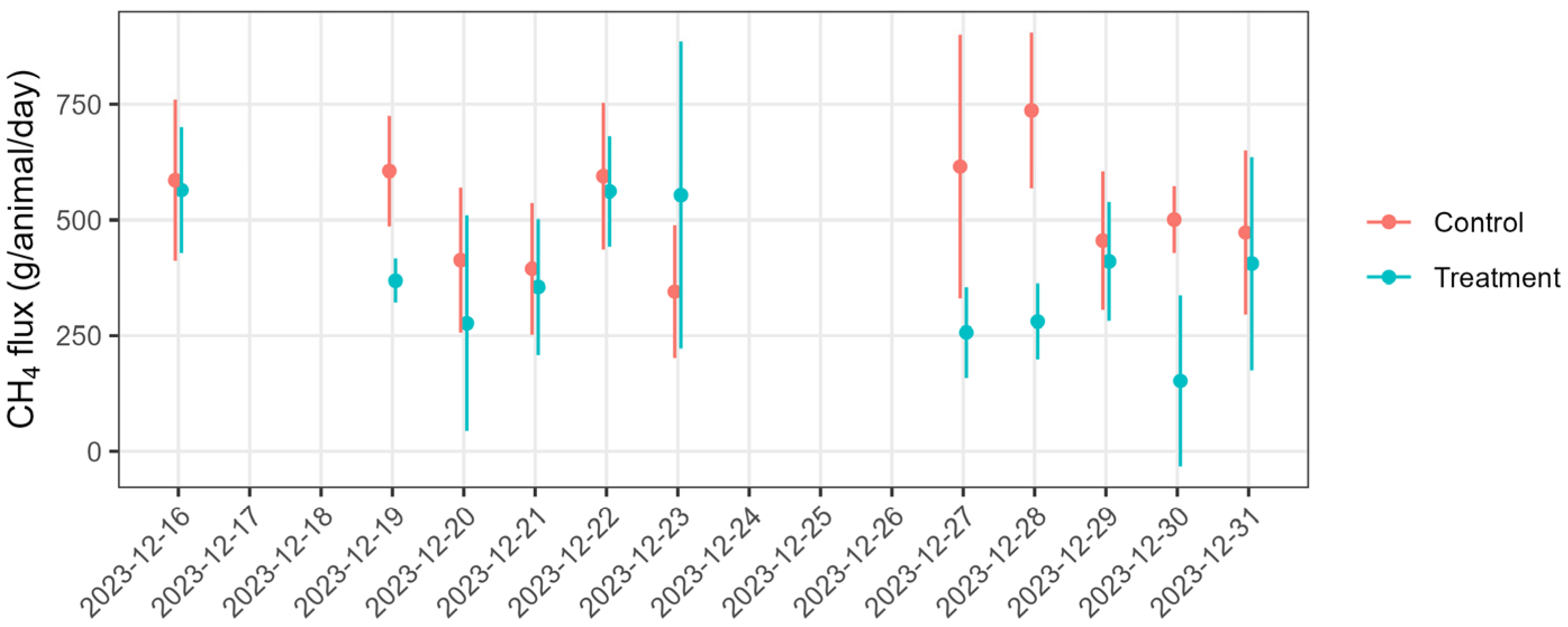
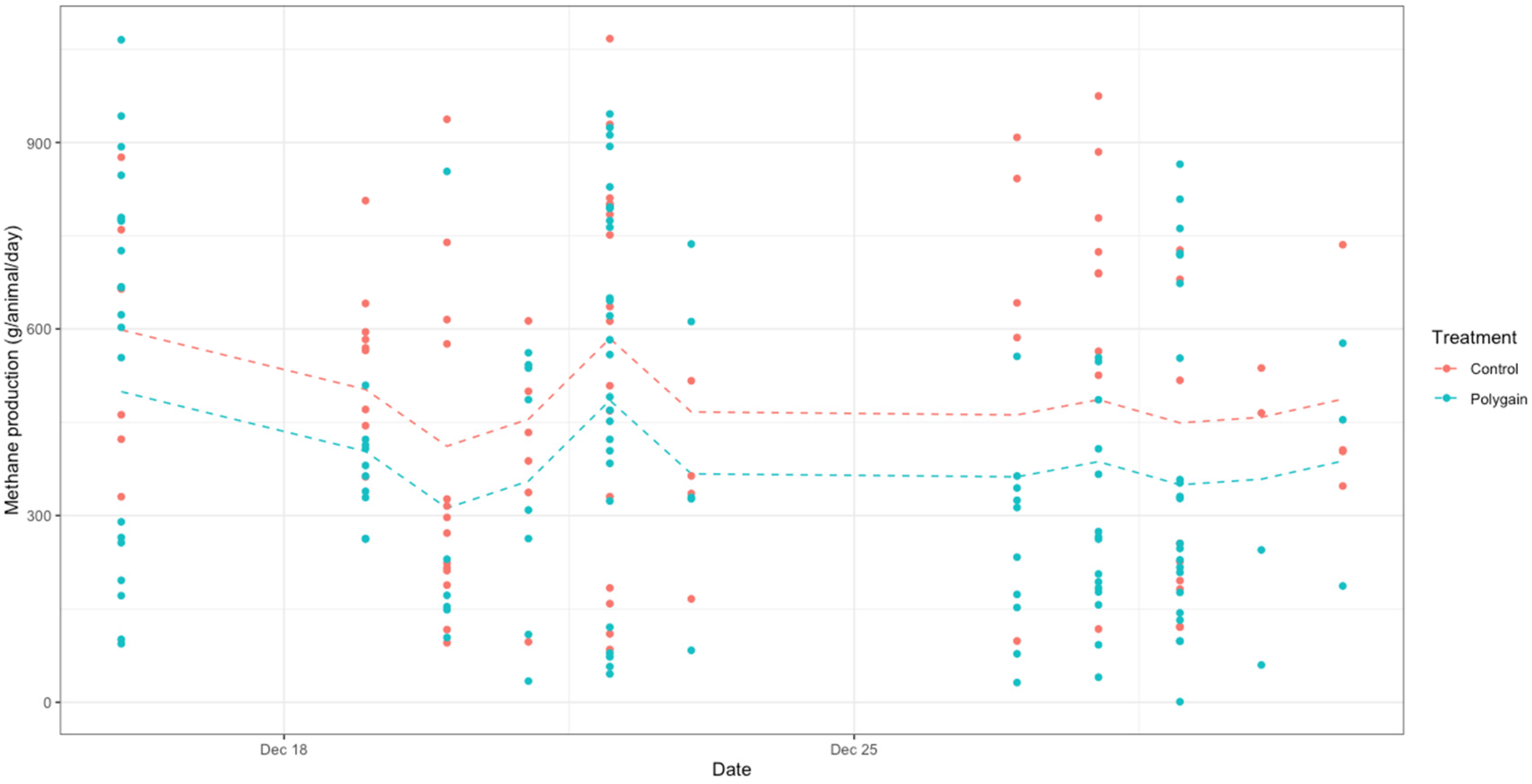
| Parameter | Control (Mean ± SD) | Treatment (Mean ± SD) |
|---|---|---|
| Lactation days | 130.2 ± 81.8 | 121.3 ± 84.6 |
| Lactation no | 3.9 ± 1.1 | 4.1 ± 2.1 |
| Body weight (kg) | 656.61 ± 7.7 | 656.90 ± 5.5 |
| Day production (kg) | 29.45 ± 0.3 | 29.48 ± 0.4 |
| Total concentrate given (kg DM) | 10.8 ± 0.6 | 10.8 ± 0.8 |
| Daily concentrate remaining avg (kg) | 1.4 ± 0.7 | 1.5 ± 0.7 |
| Daily concentrate consumed avg (kg) | 9.4 ± 0.7 | 9.4 ± 0.8 |
| Elements/Nutrients/Vitamins | As Fed |
|---|---|
| Crude protein | 14.25% |
| Crude fibre | 12.0% |
| Crude fat | 1.5% |
| Calcium | 1.0% |
| Phosphorus | 0.5% |
| Magnesium | 0.3% |
| Salt | 0.5% |
| Copper | 45 mg/kg |
| Zinc | 150 mg/kg |
| Selenium | 0.5 mg/kg |
| Manganese | 70 mg/kg |
| Iodine | 1.0 mg/kg |
| Cobalt | 1.5 mg/kg |
| Vitamin A | 6500 iu/kg |
| Vitamin D3 | 800 iu/kg |
| Vitamin E | 15 mg/kg |
| Elements/Nutrients/Vitamins | Composition |
|---|---|
| Dry matter (DM) | 93.5% |
| Moisture | 6.5% |
| Crude protein | 18.3% |
| Acid detergent fibre | 25.7% |
| Neutral detergent fibre | 48.1% |
| Digestibility (DMD) # | 71.3% |
| Digestibility (DOMD-calculated) | 67.2% |
| Metabolizable energy (calculated) | 10.6 MJ/kg DM |
| Water soluble carbohydrates | 6.8% of DM |
| Fat | 5.2% of DM |
| Ash | 13.2% of DM |
| Period | DMP (kg) | BW (kg) | ||
|---|---|---|---|---|
| Treatment | Control | Treatment | Control | |
| week 4 | 29.45 | 29.48 | 656.90 | 656.61 |
| Week 5 | 28.70 | 28.78 | 662.66 | 664.52 |
| Week 6 | 28.68 | 28.85 | 667.95 | 671.97 |
| Average | 28.94 (±0.3) & | 29.04 (±0.2) | 662.50 (±3.2) | 664.37 (±4.4) |
| MP g CH4/animal/day | MI (g CH4/animal/kg milk/day) | |||
| Treatment | Control | Treatment | Control | |
| 376.6 (±12) & | 494.9 (±12) & | 13.01 (±0.4) | 17.04 (±0.4) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, M.; Prathap, P.; Elayadeth-Meethal, M.; Flavel, M.; Eckard, R.; Dunshea, F.R.; Osei-Amponsah, R.; Ashar, M.J.; Chen, D.; Chauhan, S. Polyphenol-Containing Feed Additive Polygain™ Reduces Methane Production and Intensity from Grazing Dairy Cows Measured Using an Inverse-Dispersion Technique. Animals 2025, 15, 926. https://doi.org/10.3390/ani15070926
Bai M, Prathap P, Elayadeth-Meethal M, Flavel M, Eckard R, Dunshea FR, Osei-Amponsah R, Ashar MJ, Chen D, Chauhan S. Polyphenol-Containing Feed Additive Polygain™ Reduces Methane Production and Intensity from Grazing Dairy Cows Measured Using an Inverse-Dispersion Technique. Animals. 2025; 15(7):926. https://doi.org/10.3390/ani15070926
Chicago/Turabian StyleBai, Mei, Pragna Prathap, Muhammed Elayadeth-Meethal, Matthew Flavel, Richard Eckard, Frank R. Dunshea, Richard Osei-Amponsah, Mohammad Javed Ashar, Deli Chen, and Surinder Chauhan. 2025. "Polyphenol-Containing Feed Additive Polygain™ Reduces Methane Production and Intensity from Grazing Dairy Cows Measured Using an Inverse-Dispersion Technique" Animals 15, no. 7: 926. https://doi.org/10.3390/ani15070926
APA StyleBai, M., Prathap, P., Elayadeth-Meethal, M., Flavel, M., Eckard, R., Dunshea, F. R., Osei-Amponsah, R., Ashar, M. J., Chen, D., & Chauhan, S. (2025). Polyphenol-Containing Feed Additive Polygain™ Reduces Methane Production and Intensity from Grazing Dairy Cows Measured Using an Inverse-Dispersion Technique. Animals, 15(7), 926. https://doi.org/10.3390/ani15070926










