Phenotypic and Genomic Assessment of Antimicrobial Resistance and Virulence Factors Determinants in Salmonella Heidelberg Isolated from Broiler Chickens
Abstract
Simple Summary
Abstract
1. Introduction
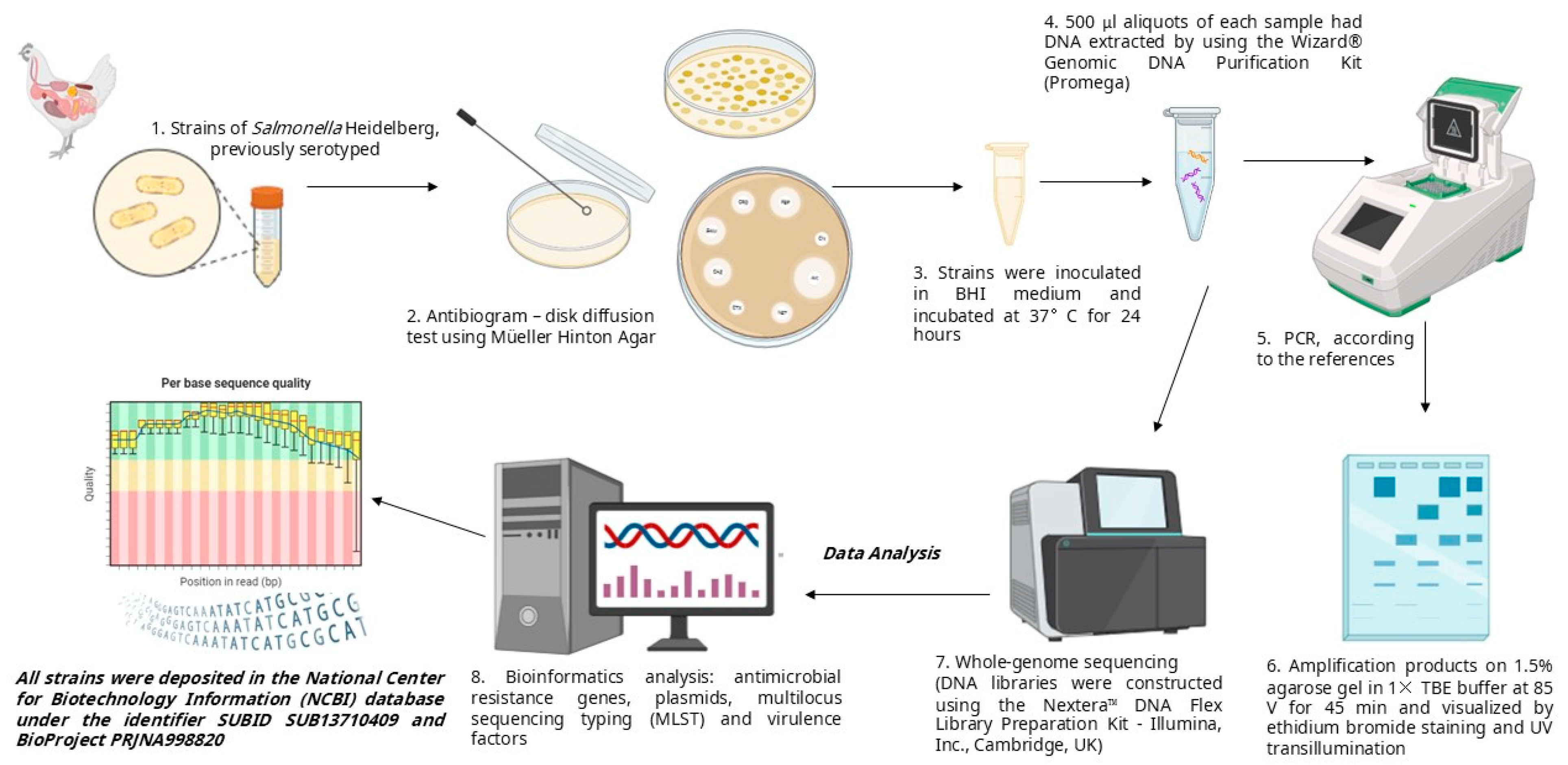
2. Materials and Methods
2.1. Bacterial Strains
2.2. Antibiogram
2.3. DNA Extraction
2.4. Polymerase Chain Reaction
2.5. Whole-Genome Sequencing
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Food Safety Authority (EFSA). The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar] [CrossRef]
- Shah, D.H.; Paul, N.C.; Sischo, W.C.; Crespo, R.; Guard, J. Population Dynamics and Antimicrobial Resistance of the Most Prevalent Poultry-Associated Salmonella Serotypes. Poult. Sci. 2017, 96, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Baptista, D.; Borsoi, A.; Reischak, D.; Nascimento, A.; Montesino, L.; Camillo, S.; Abreu, D.; Pereira, V. Salmonella Serovars Isolated from Poultry Breeding Flocks under the Brazilian Official Control Programme between 2016 and 2018. Braz. J. Poult. Sci. 2023, 25, eRBCA-2022. [Google Scholar] [CrossRef]
- Brasil, Ministério da Agricultura e Pecurária. Instrução Normativa, No 20 de 21 de Outubro de 2016, Brazil. 2016. Available online: https://www.legisweb.com.br/legislacao/?id=330200 (accessed on 21 August 2024).
- Rau, R.B.; Ribeiro, A.R.; dos Santos, A.; Barth, A.L. Antimicrobial Resistance of Salmonella from Poultry Meat in Brazil: Results of a Nationwide Survey. Epidemiol. Infect. 2021, 149, e228. [Google Scholar] [CrossRef]
- Santos, A.; Machado, S.; Dias, T.; Rodrigues, D.; Pereira, V. High Genetic Similarity Among Salmonella Heidelberg Isolated from Poultry Farms, Wild Animals, Beef, Poultry and Pork Meat, and Humans in Brazil. Braz. J. Poult. Sci. 2023, 25, eRBCA-2022. [Google Scholar] [CrossRef]
- Voss-Rech, D.; Kramer, B.; Silva, V.S.; Rebelatto, R.; Abreu, P.G.; Coldebella, A.; Vaz, C.S.L. Longitudinal Study Reveals Persistent Environmental Salmonella Heidelberg in Brazilian Broiler Farms. Vet. Microbiol. 2019, 233, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.M.P.; Ferrari, R.G.; Panzenhagen, P.; Rodrigues, G.L.; Conte-Junior, C.A. Virulence Genes Identification and Characterization Revealed the Presence of the Yersinia High Pathogenicity Island (HPI) in Salmonella from Brazil. Gene 2021, 787, 145646. [Google Scholar] [CrossRef]
- Santos, A.F.d.M.; Amparo, L.F.V.; Machado, S.C.A.; Dias, T.S.; Berto, L.H.; Abreu, D.L.d.C.; Aquino, M.H.C.d.; Rodrigues, D.d.P.; Pereira, V.L.d.A. Salmonella Serovars Associated with Human Salmonellosis in Brazil (2011–2020). Res. Soc. Dev. 2022, 11, e28011830533. [Google Scholar] [CrossRef]
- Gieraltowski, L.; Higa, J.; Peralta, V.; Green, A.; Schwensohn, C.; Rosen, H.; Libby, T.; Kissler, B.; Marsden-Haug, N.; Booth, H.; et al. National Outbreak of Multidrug Resistant Salmonella Heidelberg Infections Linked to a Single Poultry Company. PLoS ONE 2016, 11, e0162369. [Google Scholar] [CrossRef]
- Folster, J.P.; Pecic, G.; Rickert, R.; Taylor, J.; Zhao, S.; Fedorka-Cray, P.J.; Whichard, J.; McDermott, P. Characterization of Multidrug-Resistant Salmonella Enterica Serovar Heidelberg from a Ground Turkey-Associated Outbreak in the United States in 2011. Antimicrob. Agents Chemother. 2012, 56, 3465–3466. [Google Scholar] [CrossRef]
- Crump, J.A.; Medalla, F.M.; Joyce, K.W.; Krueger, A.L.; Hoekstra, R.M.; Whichard, J.M.; Barzilay, E.J. Antimicrobial Resistance among Invasive Nontyphoidal Salmonella Enterica Isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob. Agents Chemother. 2011, 55, 1148–1154. [Google Scholar] [CrossRef] [PubMed]
- FDA. Research to Advance Antimicrobial Resistance Monitoring. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/research-advance-antimicrobial-resistance-monitoring (accessed on 16 July 2024).
- CDC. Antibiotic Resistance Threats in the United States, 2019; CDC: Atlanta, Georgia, 2019. [Google Scholar]
- World Health Organization. 2019 Antibacterial Agents in Clinical Development an Analysis of the Antibacterial Development Pipeline; WHO: Geneva, Switzerland, 2019; ISBN 9789240000193. [Google Scholar]
- Dias, T.S.; Figueira, A.A.; Costa, G.A.; Machado, S.C.A.; da Cunha, N.C.; Abreu, D.L.C.; Pereira, V.L.A.; de Aquino, M.H.C. High Frequency of Non-Susceptibility to Ciprofloxacin in Nontyphoid Salmonella Recovered from Brazilian Broiler Chicken. Br. Poult. Sci. 2022, 64, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, I.; Silva, R.; Menezes, J.; Machado, S.; Rodrigues, D.; Pomba, C.; Abreu, D.; Nascimento, E.; Aquino, M.; Pereira, V. High Prevalence of Multidrug-Resistant Nontyphoidal Salmonella Recovered from Broiler Chickens and Chicken Carcasses in Brazil. Braz. J. Poult. Sci. 2020, 22, eRBCA-2019. [Google Scholar] [CrossRef]
- World Health Organization. Global Genomic Surveillance Strategy 2022–2032; WHO: Geneva, Switzerland, 2022; ISBN 9789240046979. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). M100 Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI: Wayne, PA, USA, 2023. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Aabo, S.; Rasmussen, O.F.; Roseen, L.; Sørensen, P.D.; Olsen, J.E. Salmonella Identification by the Polymerase Chain Reaction. Mol. Cell. Probes 1993, 7, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.; Mourão, J.; Silveira, L.; Saraiva, M.; Correia, C.B.; Maçãs, A.P.; Peixe, L.; Antunes, P. Imported Poultry Meat as a Source of Extended-Spectrum Cephalosporin-Resistant CMY-2-Producing Salmonella Heidelberg and Salmonella Minnesota in the European Union, 2014–2015. Int. J. Antimicrob. Agents 2018, 51, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Collineau, L.; Chapman, B.; Bao, X.; Sivapathasundaram, B.; Carson, C.A.; Fazil, A.; Reid-Smith, R.J.; Smith, B.A. A Farm-to-Fork Quantitative Risk Assessment Model for Salmonella Heidelberg Resistant to Third-Generation Cephalosporins in Broiler Chickens in Canada. Int. J. Food Microbiol. 2020, 330, 108559. [Google Scholar] [CrossRef] [PubMed]
- Melo, R.T.; Galvão, N.N.; Guidotti-Takeuchi, M.; Peres, P.A.B.M.; Fonseca, B.B.; Profeta, R.; Azevedo, V.A.C.; Monteiro, G.P.; Brenig, B.; Rossi, D.A. Molecular Characterization and Survive Abilities of Salmonella Heidelberg Strains of Poultry Origin in Brazil. Front. Microbiol. 2021, 12, 674147. [Google Scholar] [CrossRef] [PubMed]
- Etter, A.J.; West, A.M.; Burnett, J.L.; Wu, S.T.; Veenhuizen, D.R.; Ogas, R.A.; Oliver, H.F. Salmonella Enterica Subsp. Enterica Serovar Heidelberg Food Isolates Associated with a Salmonellosis Outbreak Have Enhanced Stress Tolerance Capabilities. Appl. Environ. Microbiol. 2019, 85, e01065-19. [Google Scholar] [CrossRef] [PubMed]
- Kipper, D.; Orsi, R.H.; Carroll, L.M.; Mascitti, A.K.; Streck, A.F.; Fonseca, A.S.K.; Ikuta, N.; Tondo, E.C.; Wiedmann, M.; Lunge, V.R. Recent Evolution and Genomic Profile of Salmonella Enterica Serovar Heidelberg Isolates from Poultry Flocks in Brazil. Appl. Environ. Microbiol. 2021, 87, e01036-21. [Google Scholar] [CrossRef]
- Saidenberg, A.B.S.; Franco, L.S.; Reple, J.N.; Hounmanou, Y.M.G.; Casas, M.R.T.; Cardoso, B.; Esposito, F.; Lincopan, N.; Dalsgaard, A.; Stegger, M.; et al. Salmonella Heidelberg and Salmonella Minnesota in Brazilian Broilers: Genomic Characterization of Third-Generation Cephalosporin and Fluoroquinolone-Resistant Strains. Environ. Microbiol. Rep. 2023, 15, 119–128. [Google Scholar] [CrossRef]
- Brasil, Ministério da Agricultura e Pecurária. Portaria Do Ministério Da Agricultura, Pecuária e Abastecimento—193, de 19/9/1994. 1994. Available online: https://www.defesa.agricultura.sp.gov.br/legislacoes/portaria-mapa-193-de-19-09-1994,369.html (accessed on 21 August 2024).
- Voss-Rech, D.; Trevisol, I.M.; Brentano, L.; Silva, V.S.; Rebelatto, R.; Jaenisch, F.R.F.; Okino, C.H.; Mores, M.A.Z.; Coldebella, A.; Botton, S.d.A.; et al. Impact of Treatments for Recycled Broiler Litter on the Viability and Infectivity of Microorganisms. Vet. Microbiol. 2017, 203, 308–314. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, R.R.; Dissel, S.; Rapallini, M.L.B.A.; van der Weijden, C.C.; Wit, B.; Heymans, R. Characterization and Whole-genome sequencing of Closely Related Multidrug-Resistant Salmonella Enterica Serovar Heidelberg Isolates from Imported Poultry Meat in the Netherlands. PLoS ONE 2019, 14, e0219795. [Google Scholar] [CrossRef]
- Lyu, N.; Feng, Y.; Pan, Y.; Huang, H.; Liu, Y.; Xue, C.; Zhu, B.; Hu, Y. Genomic Characterization of Salmonella Enterica Isolates From Retail Meat in Beijing, China. Front. Microbiol. 2021, 12, 636332. [Google Scholar] [CrossRef]
- Orlek, A.; Anjum, M.F.; Mather, A.E.; Stoesser, N.; Walker, A.S. Factors Associated with Plasmid Antibiotic Resistance Gene Carriage Revealed Using Large-Scale Multivariable Analysis. Sci. Rep. 2023, 13, 2500. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lynne, A.M.; David, D.E.; Tang, H.; Xu, J.; Nayak, R.; Kaldhone, P.; Logue, C.M.; Foley, S.L. DNA Sequence Analysis of Plasmids from Multidrug Resistant Salmonella Enterica Serotype Heidelberg Isolates. PLoS ONE 2012, 7, e51160. [Google Scholar] [CrossRef] [PubMed]
- Oladeinde, A.; Cook, K.; Orlek, A.; Zock, G.; Herrington, K.; Cox, N.; Plumblee Lawrence, J.; Hall, C. Hotspot Mutations and ColE1 Plasmids Contribute to the Fitness of Salmonella Heidelberg in Poultry Litter. PLoS ONE 2018, 13, e0202286. [Google Scholar] [CrossRef]
- Souza, A.I.S.; Saraiva, M.M.S.; Casas, M.R.T.; Oliveira, G.M.; Cardozo, M.V.; Benevides, V.P.; Barbosa, F.O.; Freitas Neto, O.C.; Almeida, A.M.; Junior, A.B. High Occurrence of β-Lactamase-Producing Salmonella Heidelberg from Poultry Origin. PLoS ONE 2020, 15, e0230676. [Google Scholar] [CrossRef]
- Lemos, M.P.L.; Saraiva, M.M.S.; Leite, E.L.; Silva, N.M.V.; Vasconcelos, P.C.; Giachetto, P.F.; Freitas Neto, O.C.; Givisiez, P.E.N.; Gebreyes, W.A.; Oliveira, C.J.B. The Posthatch Prophylactic Use of Ceftiofur Affects the Cecal Microbiota Similar to the Dietary Sanguinarine Supplementation in Broilers. Poult. Sci. 2020, 99, 6013–6021. [Google Scholar] [CrossRef]
- Touzain, F.; Le Devendec, L.; de Boisséson, C.; Baron, S.; Jouy, E.; Perrin-Guyomard, A.; Blanchard, Y.; Kempf, I. Characterization of Plasmids Harboring BlaCTX-M and BlaCMY Genes in E. Coli from French Broilers. PLoS ONE 2018, 13, e0188768. [Google Scholar] [CrossRef]
- Mahmud, Z.; Shabnam, S.A.; Mishu, I.D.; Johura, F.-T.; Mannan, S.B.; Sadique, A.; Islam, L.N.; Alam, M. Virotyping, Genotyping, and Molecular Characterization of Multidrug Resistant Escherichia Coli Isolated from Diarrheal Patients of Bangladesh. Gene Rep. 2021, 23, 101182. [Google Scholar] [CrossRef]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial Siderophores in Community and Host Interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Petermann, S.R.; Sherwood, J.S.; Logue, C.M. The Yersinia High Pathogenicity Island Is Present in Salmonella Enterica Subspecies I Isolated from Turkeys. Microb. Pathog. 2008, 45, 110–114. [Google Scholar] [CrossRef] [PubMed]
| Antimicrobial | R | I | S |
|---|---|---|---|
| Amoxicillin with clavulanic acid | 71.43 | 7.14 | 21.43 |
| Ampicillin | 85.71 | 0.00 | 14.29 |
| Cephalexin | 78.57 | 0.00 | 21.43 |
| Cefalotin | 78.57 | 0.00 | 21.43 |
| Cefoxitin | 71.43 | 0.00 | 28.57 |
| Ceftazidime | 64.29 | 7.14 | 28.57 |
| Cefotaxime | 78.57 | 0.00 | 21.43 |
| Ceftiofur * | 57.14 | 21.43 | 21.43 |
| Cefepime | 7.14 | 28.57 | 64.29 |
| Imipenem | 0.00 | 0.00 | 100.00 |
| Meropenem | 0.00 | 0.00 | 100.00 |
| Ertapenem | 0.00 | 7.14 | 92.86 |
| Aztreonam | 57.14 | 7.14 | 35.71 |
| Gentamicin | 7.14 | 0.00 | 92.86 |
| Ciprofloxacin | 7.14 | 42.86 | 50.00 |
| Enrofloxacin * | 7.14 | 7.14 | 85.71 |
| Chloramphenicol | 21.43 | 0.00 | 78.57 |
| Tetracycline | 92.86 | 0.00 | 7.14 |
| Sulfonamide | 85.71 | 0.00 | 14.29 |
| Strain | Region | Year | Source | ST | Resistance Genotype | Plasmids | Phenotypic Resistance Profile | Intermediate Resistance | HPI |
|---|---|---|---|---|---|---|---|---|---|
| Heidelberg | South | 2013 | Carcass | 15 | aac(6′)-Iaa, blaCMY-2, tet(A), sul2, gyrA:p.S83F, parC:p.T57S, fosA7 | ColpVC, IncC, IncX1 | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, CTF, ATM, TET, SUL | CPM, CIP | Yes |
| Heidelberg | South | 2013 | Carcass | 15 | aac(6′)-Iaa, blaCMY-2, tet(A), sul2, gyrA:p.S83F, parC:p.T57S, fosA7 | ColpVC, IncC, IncX1 | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, ATM, TET, SUL | CTF, CIP, ENO | No |
| Heidelberg | South | 2013 | Carcass | 15 | aac(6′)-Iaa, blaCMY-2, tet(A), sul2, gyrA:p.S83F, parC:p.T57S, fosA7 | ColpVC, IncC, IncX1, IncI1-I(Alpha) | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, CTF, TET, SUL | CPM, ATM, CIP | No |
| Heidelberg | Midwest | 2016 | Carcass | 15 | aac(6′)-Iaa, blaCMY-2, tet(A), sul2, gyrA:p.S83F, parC:p.T57S, fosA7 | ColpVC, IncC, IncX1 | AMP, CLO, TET | - | No |
| Heidelberg | South | 2017 | Carcass | 15 | aac(6′)-Iaa, gyrA (p.S83F), parC:p.T57S, sul2, tet(A), fosA7, | ColpVC, IncC, IncX1, IncI1-I(Alpha) | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, CTF, ATM, ENO, TET, SUL | CPM, CIP, ENO | Yes |
| Heidelberg | South | 2017 | Carcass | 15 | aac(6′)-Iaa, gyrA (p.S83F), parC:p.T57S, sul2, tet(A), fosA7, | ColpVC, IncC, IncX1 | AMC, AMP, CFE, CFL, CFO, CTX, CTF, ATM, TET, SUL | CAZ | Yes |
| Heidelberg | Southeast | 2017 | Carcass | 15 | aac(6′)-Iaa, gyrA (p.S83F), parC:p.T57S, sul2, tet(A), fosA7, | ColpVC, IncC | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, CTF, ATM, TET, SUL | - | No |
| Heidelberg | Southeast | 2017 | Carcass | 15 | aac(6′)-Iaa, gyrA (p.S83F), parC:p.T57S sul2, tet(A), fosA7 | ColpVC, IncC, IncX1, IncI1-I(Alpha) | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, CTF, ATM, TET, SUL | - | No |
| Heidelberg | Southeast | 2017 | Carcass | 15 | aac(6′)-Iaa, gyrA (p.S83F), parC:p.T57S sul2, tet(A), fosA7 | ColpVC, IncC | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, CTF, ATM, TET, SUL | CPM, ERT, CIP | Yes |
| Heidelberg | Southeast | 2017 | Carcass | 15 | aac(6′)-Iaa, blaCMY-2, tet(A), sul2, gyrA:p.S83F, parC:p.T57S, fosA7 | ColpVC, IncC, IncX1, IncI1-I(Alpha) | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, CTF, ATM, TET, SUL | - | Yes |
| Heidelberg | Southeast | 2017 | Carcass | 15 | aac(6′)-Iaa, gyrA (p.S83F), parC:p.T57S sul2, tet(A), fosA7 | ColpVC, IncC | AMC, AMP, CFE, CFL, CFO, CAZ, CTX, ATM, TET, SUL | CTF, CIP, ENO | Yes |
| Heidelberg | South | 2019 | Carcass | 15 | aac(6′)-Iaa, blaCMY-2, tet(A), sul2, gyrA:p.S83F, parC:p.T57S, fosA7 | ColpVC, IncC, IncX1, IncI1-I(Alpha) | SUL | - | No |
| Heidelberg | Southeast | 2019 | Retail | 15 | aac(6′)-Iaa, blaCMY-2, tet(A), sul2, gyrA:p.S83F, parC:p.T57S, fosA7 | ColpVC, IncC, IncX1, IncI1-I(Alpha) | AMP, CFE, CFL, CTX, CTF, CPM, ATM, GEN, CLO, SUL | AMC, CIP | Yes |
| Heidelberg | South | 2019 | Carcass | 15 | aac(6′)-Iaa, blaCMY-2, tet(A), sul2, gyrA:p.S83F, parC:p.T57S, fosA7 | ColpVC, IncC, IncX1, IncI1-I(Alpha) | CIP, CLO | - | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Almeida Figueira, A.; Salles Dias, T.; Alves Costa, G.; Lima da Costa Abreu, D.; dos Santos Medeiros, L.; Léo de Almeida Pereira, V. Phenotypic and Genomic Assessment of Antimicrobial Resistance and Virulence Factors Determinants in Salmonella Heidelberg Isolated from Broiler Chickens. Animals 2025, 15, 1003. https://doi.org/10.3390/ani15071003
de Almeida Figueira A, Salles Dias T, Alves Costa G, Lima da Costa Abreu D, dos Santos Medeiros L, Léo de Almeida Pereira V. Phenotypic and Genomic Assessment of Antimicrobial Resistance and Virulence Factors Determinants in Salmonella Heidelberg Isolated from Broiler Chickens. Animals. 2025; 15(7):1003. https://doi.org/10.3390/ani15071003
Chicago/Turabian Stylede Almeida Figueira, Arthur, Thomas Salles Dias, Gisllany Alves Costa, Dayse Lima da Costa Abreu, Luciana dos Santos Medeiros, and Virginia Léo de Almeida Pereira. 2025. "Phenotypic and Genomic Assessment of Antimicrobial Resistance and Virulence Factors Determinants in Salmonella Heidelberg Isolated from Broiler Chickens" Animals 15, no. 7: 1003. https://doi.org/10.3390/ani15071003
APA Stylede Almeida Figueira, A., Salles Dias, T., Alves Costa, G., Lima da Costa Abreu, D., dos Santos Medeiros, L., & Léo de Almeida Pereira, V. (2025). Phenotypic and Genomic Assessment of Antimicrobial Resistance and Virulence Factors Determinants in Salmonella Heidelberg Isolated from Broiler Chickens. Animals, 15(7), 1003. https://doi.org/10.3390/ani15071003







