The Stalk and 1B Domains Are Required for Porcine Deltacoronavirus Helicase NSP13 to Separate the Double-Stranded Nucleic Acids, and the Deletion of the ZBD Impairs This Activity
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of the Recombinant Plasmids
2.2. Expression of PDCoV Helicase NSP13WT and Deletion Mutants
2.3. Purification of Recombinant Proteins
2.4. Preparation of Double-Stranded Substrates
2.5. Electrophoretic Unwinding Assay
2.6. ATPase Assay of NSP13WT and Mutants
2.7. Fluorescence Resonance Energy Transfer (FRET)-Based Unwinding Assay
2.8. Sequence Alignment and Structure Prediction
3. Results
3.1. The Sequences of Coronaviruses Helicase NSP13WT Are Highly Conservative
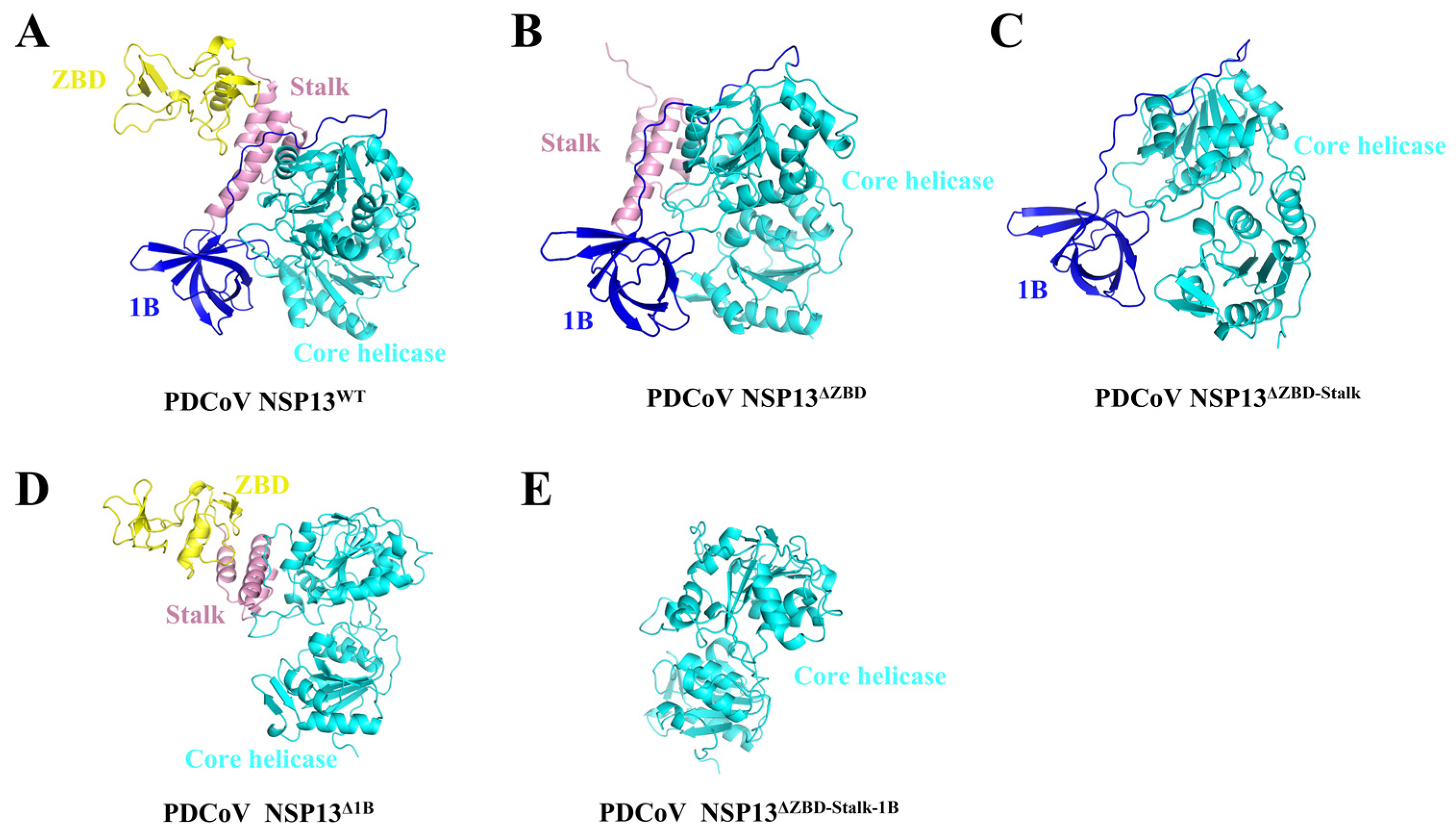
3.2. The Structures of Helicase NSP13WT and Various Mutants
3.3. Determination of the Unwinding and ATPase Activities of PDCoV NSP13WT
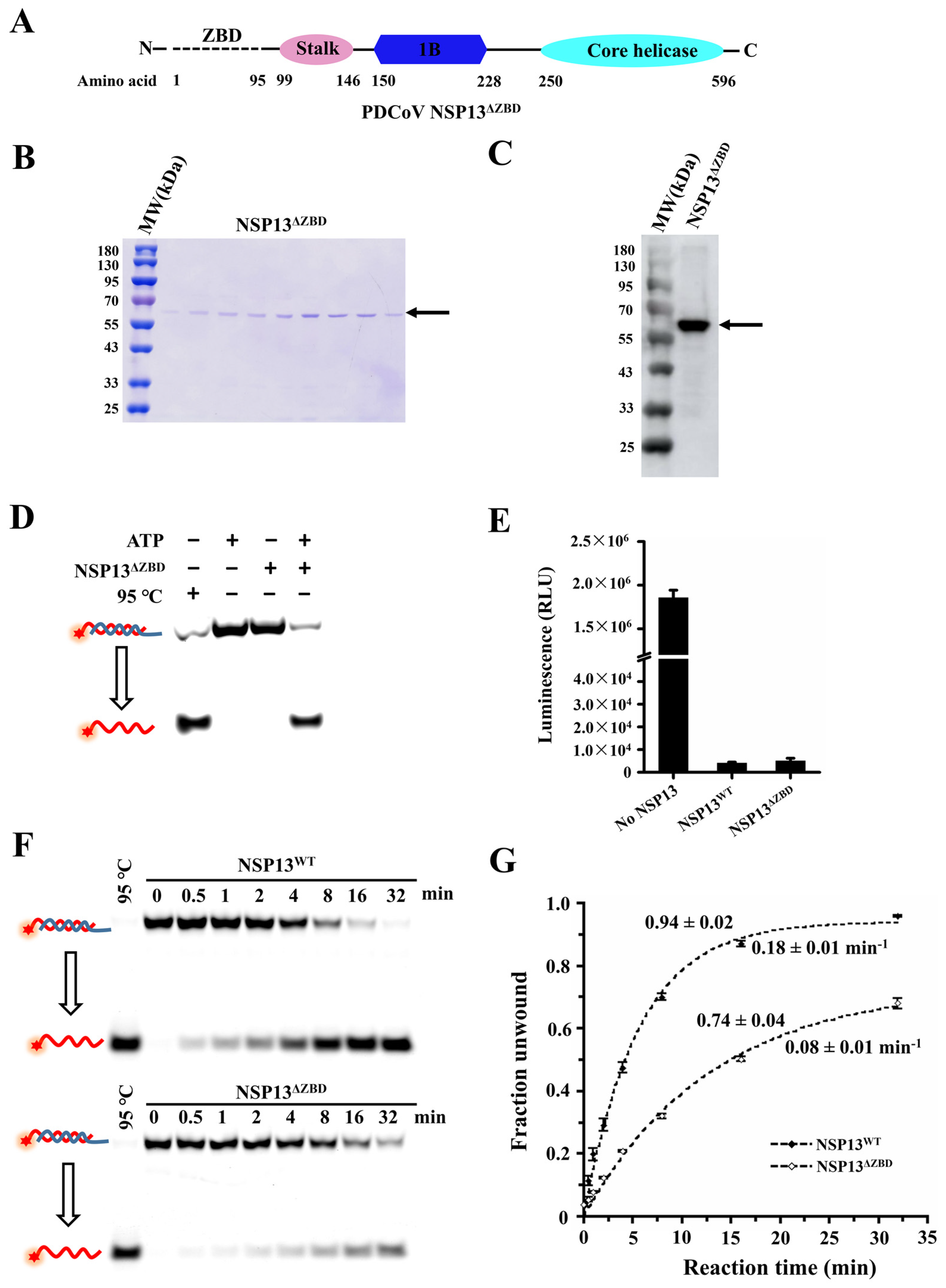
3.4. The Deletion of the ZBD Impaired the Unwinding Activity of PDCoV Helicase NSP13WT
3.5. The Stalk and 1B Domains Are Essential for Unwinding Activity
3.6. The Independent Core Helicase Domain Retains Only the Ability to Hydrolyze ATP
3.7. The Unwinding Activities of NSP13WT and Mutants Were Tested Using FRET-Based Assays
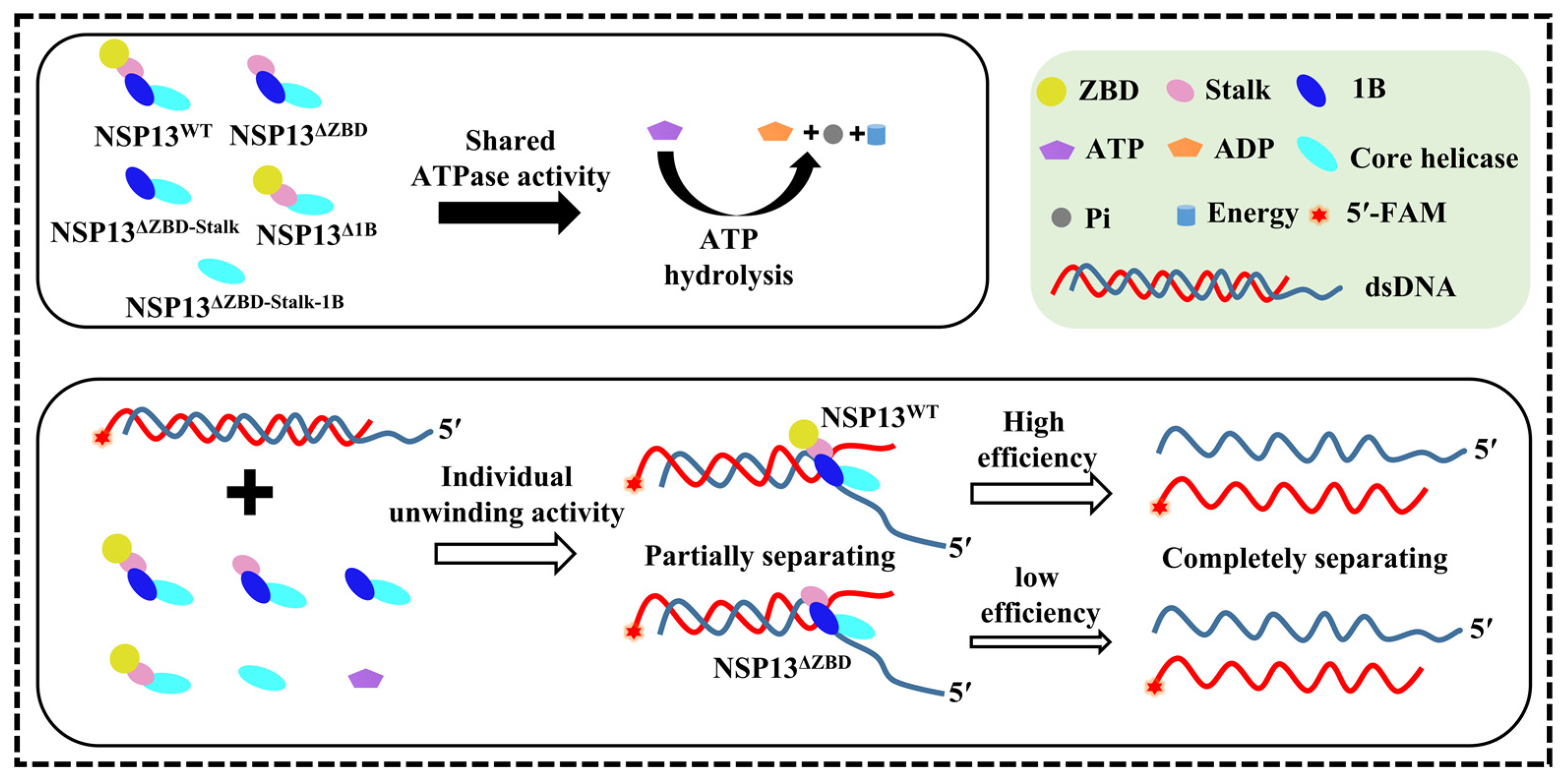
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| BSA | Bovine serum albumin |
| 3CLpro | 3-chymotrypsin-like protease |
| DEPC | Diethylpyrocarbonate |
| dsDNA | Double-stranded DNA |
| DTT | Dithiothreitol |
| EAV | Equine arteritis virus |
| EDTA | Ethylenediaminetetraacetic acid |
| FAM | Carboxyfluorescein |
| FRET | Fluorescence resonance energy transfer |
| HPLC | High-performance liquid chromatography |
| IBV | Infectious bronchitis virus |
| LB | Luria–Bertani |
| MERS-CoV | Middle East respiratory syndrome coronavirus |
| NCBI | National Center for Biotechnology Information |
| Ni-NTA | Nickel-Nitrilotriacetic acid |
| NSP13 | Nonstructural protein 13 |
| NTP | Nucleotide trisphosphate |
| PAGE | Polyacrylamide gel electrophoresis |
| PBS | Phosphate-buffer saline |
| PDCoV | Pocine deltacoronavirus |
| PEDV | Porcine epidemic diarrhea virus |
| PLpro | Papain-like protease |
| PMT | Photomultiplier tube |
| Rec1A | Recombination-like domain 1A |
| SARS-CoV | Severe acute respiratory syndrome coronavirus |
| SDS | Sodium dodecyl sulfate |
| ssDNA | Single-stranded DNA |
| TAMRA | Tetramethylrhodamine |
| TBE | Tris-borate-ethylene diamine tetraacetie acid |
| TGEV | Transmissible gastroenteritis virus |
| Upf1 | Upstream frameshift protein 1 |
| ZBD | Zinc-binding domain |
References
- Duan, C. An Updated Review of Porcine Deltacoronavirus in Terms of Prevalence, Pathogenicity, Pathogenesis and Antiviral Strategy. Front. Vet. Sci. 2021, 8, 811187. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gao, Y.; Ma, Y.; Shi, K.; Shi, Y.; Feng, S.; Yin, Y.; Long, F.; Sun, W. Genetic and Evolutionary Analysis of Porcine Deltacoronavirus in Guangxi Province, Southern China, from 2020 to 2023. Microorganisms 2024, 12, 416. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar]
- Jung, K.; Hu, H.; Saif, L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017, 162, 2357–2362. [Google Scholar] [CrossRef] [PubMed]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef]
- Boley, P.A.; Alhamo, M.A.; Lossie, G.; Yadav, K.K.; Vasquez-Lee, M.; Saif, L.J.; Kenney, S.P. Porcine Deltacoronavirus Infection and Transmission in Poultry, United States. Emerg. Infect. Dis. 2020, 26, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Fang, L.; Liu, X.; Hong, Y.; Wang, Y.; Dong, N.; Ma, P.; Bi, J.; Wang, D.; Xiao, S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology 2016, 499, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Mou, C.; Xie, S.; Zhu, L.; Cheng, Y.; Pan, S.; Zhang, C.; Chen, Z. Porcine deltacoronavirus NS7a antagonizes JAK/STAT pathway by inhibiting the interferon-stimulated gene factor 3 (ISGF3) formation. Int. J. Biol. Macromol. 2024, 264, 130693. [Google Scholar] [CrossRef]
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216. [Google Scholar] [CrossRef]
- Kanaan, J.; Raj, S.; Decourty, L.; Saveanu, C.; Croquette, V.; Le Hir, H. UPF1-like helicase grip on nucleic acids dictates processivity. Nat. Commun. 2018, 9, 3752. [Google Scholar] [CrossRef]
- Meir, A.; Greene, E.C. Srs2 and Pif1 as Model Systems for Understanding Sf1a and Sf1b Helicase Structure and Function. Genes 2021, 12, 1319. [Google Scholar] [CrossRef]
- Lehmann, K.C.; Snijder, E.J.; Posthuma, C.C.; Gorbalenya, A.E. What we know but do not understand about nidovirus helicases. Virus Res. 2015, 202, 12–32. [Google Scholar] [CrossRef]
- Adedeji, A.O.; Marchand, B.; Te Velthuis, A.J.; Snijder, E.J.; Weiss, S.; Eoff, R.L.; Singh, K.; Sarafianos, S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE 2012, 7, e36521. [Google Scholar] [CrossRef]
- Adedeji, A.O.; Lazarus, H. Biochemical Characterization of Middle East Respiratory Syndrome Coronavirus Helicase. mSphere 2016, 1, 10–1128. [Google Scholar] [CrossRef]
- Yazdi, A.K.; Pakarian, P.; Perveen, S.; Hajian, T.; Santhakumar, V.; Bolotokova, A.; Li, F.; Vedadi, M. Kinetic Characterization of SARS-CoV-2 nsp13 ATPase Activity and Discovery of Small-Molecule Inhibitors. ACS Infect. Dis. 2022, 8, 1533–1542. [Google Scholar] [CrossRef]
- Sommers, J.A.; Loftus, L.N.; Jones, M.P., 3rd; Lee, R.A.; Haren, C.E.; Dumm, A.J.; Brosh, R.M., Jr. Biochemical analysis of SARS-CoV-2 Nsp13 helicase implicated in COVID-19 and factors that regulate its catalytic functions. J. Biol. Chem. 2023, 299, 102980. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Ding, Z.; Fang, P.; Xiao, S.; Fang, L. ATPase and helicase activities of porcine epidemic diarrhea virus nsp13. Vet. Microbiol. 2021, 257, 109074. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Chen, J.; Qiao, C.; Li, C.; Yang, K.; Zhang, Y.; Li, J.; Li, Z. Porcine Epidemic Diarrhea Virus nsp13 Protein Downregulates Neonatal Fc Receptor Expression by Causing Promoter Hypermethylation through the NF-kappaB Signaling Pathway. J. Immunol. 2023, 210, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Mehyar, N. Coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 helicase inhibitors: A systematic review of invitro studies. J. Virus Erad. 2023, 9, 100327. [Google Scholar] [CrossRef]
- Minkoff, J.M.; tenOever, B. Innate immune evasion strategies of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 178–194. [Google Scholar] [CrossRef]
- Wang, P.; Wang, X.; Liu, X.; Sun, M.; Liang, X.; Bai, J.; Jiang, P. Natural Compound ZINC12899676 Reduces Porcine Epidemic Diarrhea Virus Replication by Inhibiting the Viral NTPase Activity. Front. Pharmacol. 2022, 13, 879733. [Google Scholar] [CrossRef] [PubMed]
- Shu, T.; Huang, M.; Wu, D.; Ren, Y.; Zhang, X.; Han, Y.; Mu, J.; Wang, R.; Qiu, Y.; Zhang, D.Y.; et al. SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts. Virol. Sin. 2020, 35, 321–329. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, R.; Chan, J.F.; Zhang, A.J.; Cheng, T.; Chik, K.K.; Ye, Z.W.; Wang, S.; Lee, A.C.; Jin, L.; et al. Metallodrug ranitidine bismuth citrate suppresses SARS-CoV-2 replication and relieves virus-associated pneumonia in Syrian hamsters. Nat. Microbiol. 2020, 5, 1439–1448. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.; McCullagh, M. Role of ATP in the RNA Translocation Mechanism of SARS-CoV-2 NSP13 Helicase. J. Phys. Chem. B 2021, 125, 8787–8796. [Google Scholar] [CrossRef] [PubMed]
- Fairman-Williams, M.E.; Guenther, U.P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef]
- Jia, Z.; Yan, L.; Ren, Z.; Wu, L.; Wang, J.; Guo, J.; Zheng, L.; Ming, Z.; Zhang, L.; Lou, Z.; et al. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. [Google Scholar] [CrossRef]
- Yue, K.; Yao, B.; Shi, Y.; Yang, Y.; Qian, Z.; Ci, Y.; Shi, L. The stalk domain of SARS-CoV-2 NSP13 is essential for its helicase activity. Biochem. Biophys. Res. Commun. 2022, 601, 129–136. [Google Scholar] [CrossRef]
- Parkhe, P.; Verma, S. Evolution, Interspecies Transmission, and Zoonotic Significance of Animal Coronaviruses. Front. Vet. Sci. 2021, 8, 719834. [Google Scholar] [CrossRef]
- Grimes, S.L.; Denison, M.R. The Coronavirus helicase in replication. Virus Res. 2024, 346, 199401. [Google Scholar] [CrossRef]
- Kuzikov, M.; Reinshagen, J.; Wycisk, K.; Corona, A.; Esposito, F.; Malune, P.; Manelfi, C.; Iaconis, D.; Beccari, A.; Tramontano, E.; et al. Drug repurposing screen to identify inhibitors of the RNA polymerase (nsp12) and helicase (nsp13) from SARS-CoV-2 replication and transcription complex. Virus Res. 2024, 343, 199356. [Google Scholar] [CrossRef]
- Perez-Lemus, G.R.; Menendez, C.A.; Alvarado, W.; Bylehn, F.; de Pablo, J.J. Toward wide-spectrum antivirals against coronaviruses: Molecular characterization of SARS-CoV-2 NSP13 helicase inhibitors. Sci. Adv. 2022, 8, eabj4526. [Google Scholar] [CrossRef] [PubMed]
- Soper, N.; Yardumian, I.; Chen, E.; Yang, C.; Ciervo, S.; Oom, A.L.; Desvignes, L.; Mulligan, M.J.; Zhang, Y.; Lupoli, T.J. A Repurposed Drug Interferes with Nucleic Acid to Inhibit the Dual Activities of Coronavirus Nsp13. ACS Chem. Biol. 2024, 19, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P.M.M.; Olinares, P.D.B.; Maruthi, K.; Eng, E.T.; Vatandaslar, H.; Chait, B.T.; et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell 2020, 182, 1560–1573.e13. [Google Scholar] [CrossRef]
- Newman, J.A.; Douangamath, A.; Yadzani, S.; Yosaatmadja, Y.; Aimon, A.; Brandao-Neto, J.; Dunnett, L.; Gorrie-Stone, T.; Skyner, R.; Fearon, D.; et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021, 12, 4848. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adriaenssens, E.M.; Lefkowitz, E.J.; Oksanen, H.M.; Siddell, S.G.; Zerbini, F.M.; Alfenas-Zerbini, P.; Aylward, F.O.; Dempsey, D.M.; Dutilh, B.E.; et al. Changes to virus taxonomy and the ICTV Statutes ratified by the International Committee on Taxonomy of Viruses (2024). Arch. Virol. 2024, 169, 236. [Google Scholar] [CrossRef]
- Deng, Z.; Lehmann, K.C.; Li, X.; Feng, C.; Wang, G.; Zhang, Q.; Qi, X.; Yu, L.; Zhang, X.; Feng, W.; et al. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res. 2014, 42, 3464–3477. [Google Scholar] [CrossRef]
- Fang, S.; Chen, B.; Tay, F.P.; Ng, B.S.; Liu, D.X. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology 2007, 358, 136–147. [Google Scholar] [CrossRef]
- Saikrishnan, K.; Griffiths, S.P.; Cook, N.; Court, R.; Wigley, D.B. DNA binding to RecD: Role of the 1B domain in SF1B helicase activity. EMBO J. 2008, 27, 2222–2229. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, Y.; Ge, J.; Zheng, L.; Gao, Y.; Wang, T.; Jia, Z.; Wang, H.; Huang, Y.; Li, M.; et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020, 11, 5874. [Google Scholar] [CrossRef]
- Lawal, M.M.; Roy, P.; McCullagh, M. Role of ATP Hydrolysis and Product Release in the Translocation Mechanism of SARS-CoV-2 NSP13. J. Phys. Chem. B 2024, 128, 492–503. [Google Scholar] [CrossRef]
- Grimes, S.L.; Choi, Y.J.; Banerjee, A.; Small, G.; Anderson-Daniels, J.; Gribble, J.; Pruijssers, A.J.; Agostini, M.L.; Abu-Shmais, A.; Lu, X.; et al. A mutation in the coronavirus nsp13-helicase impairs enzymatic activity and confers partial remdesivir resistance. mBio 2023, 14, e0106023. [Google Scholar] [CrossRef] [PubMed]





| Names | Sequences | Nts |
|---|---|---|
| DNA1 | 5′-★-GCGACGTCACGTGCA-3′ | 15 |
| DNA2 | 5′-CAGCTAGACCTGCACGTGACGTCGC-3′ | 25 |
| RNA1 | 5′-★-GCGUCGUAUCGAUCU-3′ | 15 |
| RNA2 | 5′-CAGCUAGACCAGAUCGAUACGACGC-3′ | 25 |
| DNA3 | 5′-★-ACCTGGATTGGTGTCGGTAGAGAACTAGCG -3′ | 30 |
| DNA4 | 5′-TCCCTAGCTTCGCTAGTTCTCTACCGACACCAATCCAGGT-▲-3′ | 40 |
| Trap DNA1 | 5′-GCGACGTCACGTGCA-3′ | 15 |
| Trap DNA2 | 5′-ACCTGGATTGGTGTCGGTAGAGAACTAGCG-3′ | 30 |
| Trap RNA | 5′-GCGUCGUAUCGAUCU-3′ | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.; Tao, L.; Zhou, Q.; Zhang, F.; Zeng, Y. The Stalk and 1B Domains Are Required for Porcine Deltacoronavirus Helicase NSP13 to Separate the Double-Stranded Nucleic Acids, and the Deletion of the ZBD Impairs This Activity. Animals 2025, 15, 865. https://doi.org/10.3390/ani15060865
Wu C, Tao L, Zhou Q, Zhang F, Zeng Y. The Stalk and 1B Domains Are Required for Porcine Deltacoronavirus Helicase NSP13 to Separate the Double-Stranded Nucleic Acids, and the Deletion of the ZBD Impairs This Activity. Animals. 2025; 15(6):865. https://doi.org/10.3390/ani15060865
Chicago/Turabian StyleWu, Chengcheng, Lihan Tao, Quanyong Zhou, Fanfan Zhang, and Yanbing Zeng. 2025. "The Stalk and 1B Domains Are Required for Porcine Deltacoronavirus Helicase NSP13 to Separate the Double-Stranded Nucleic Acids, and the Deletion of the ZBD Impairs This Activity" Animals 15, no. 6: 865. https://doi.org/10.3390/ani15060865
APA StyleWu, C., Tao, L., Zhou, Q., Zhang, F., & Zeng, Y. (2025). The Stalk and 1B Domains Are Required for Porcine Deltacoronavirus Helicase NSP13 to Separate the Double-Stranded Nucleic Acids, and the Deletion of the ZBD Impairs This Activity. Animals, 15(6), 865. https://doi.org/10.3390/ani15060865






