Genome-Wide Association Study Reveals Novel Loci and Candidate Genes for Birth Weight in Pigs
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Quality Control
2.2. Statistical Analysis of Environmental and Animal Factors
2.3. Genotyping and Quality Control
2.4. Genome-Wide Association Analysis
2.5. Candidate Gene Functional Annotation
2.6. Validation of SNP Effects and Impacts on Birth Weight
3. Results
3.1. Descriptive Statistics of Birth Weight and Environmental Influences in Pigs
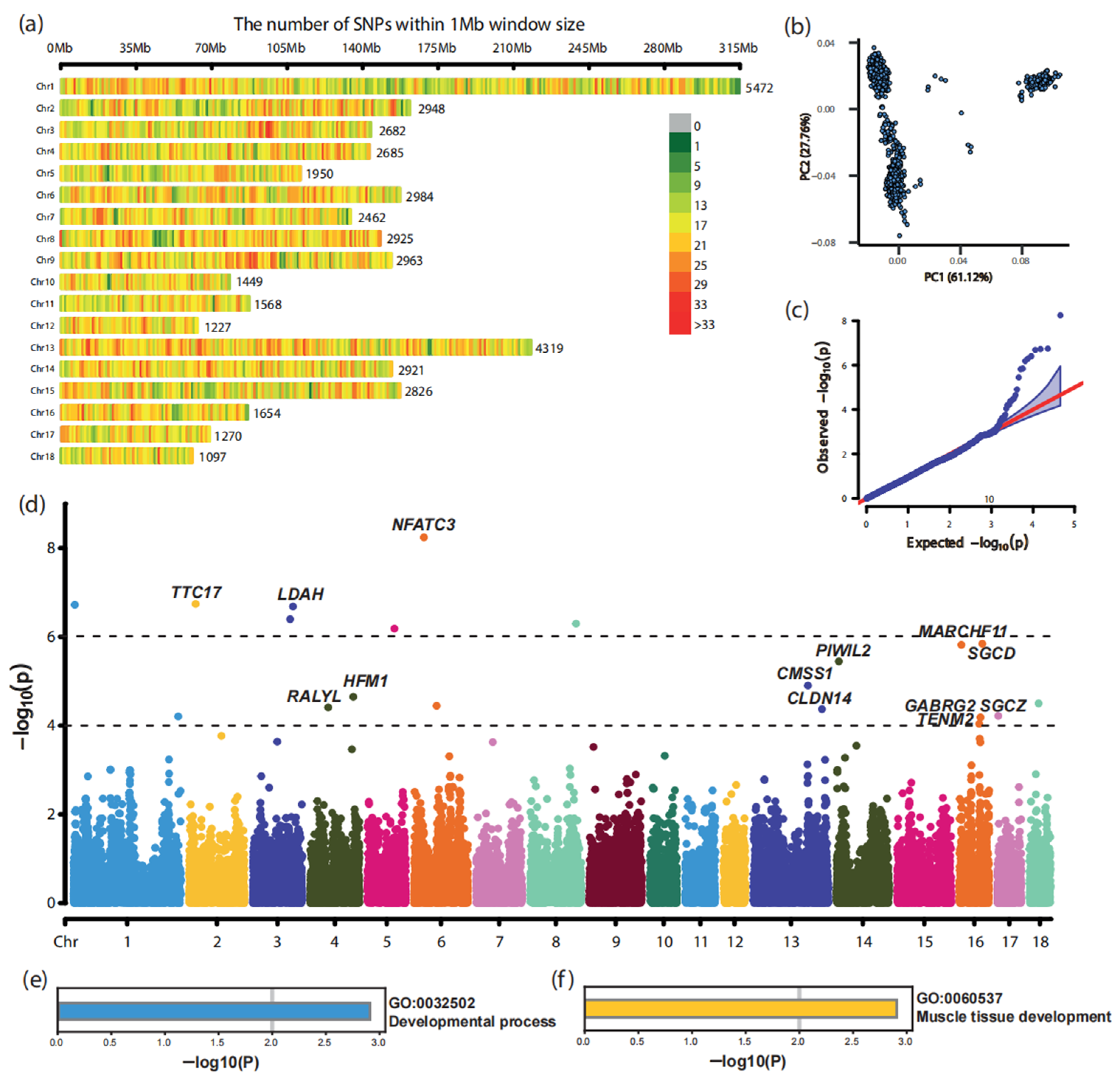
3.2. Genome-Wide Association Study in Landrace Pigs
3.3. Validation of GWAS-Identified SNPs for Birth Weight Across Populations
3.4. Key SNPs and MARCHF11 Gene Associated with Pig Birth Weight
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GWAS | Genome-wide association study |
| MAS | Marker-assisted selection |
| LL | Landrace pigs |
| YY | Yorkshire pigs |
| LSM | Least-squares means |
| MAF | Minor allele frequency |
| PCA | Principal component analysis |
| FarmCPU | Fixed and Random Model Circulating Probability Unification |
| GO | Gene Ontology |
| KW test | Kruskal–Wallis test |
References
- Deng, D.; Wang, H.; Han, K.; Tang, Z.; Li, X.; Liu, X.; Liu, X.; Li, X.; Yu, M. A genome-wide association study reveals candidate genes and regulatory regions associated with birth weight in pigs. Anim. Genet. 2024, 55, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Gondret, F.; Lefaucheur, L.; Louveau, I.; Lebret, B.; Pichodo, X.; Le Cozler, Y. Influence of piglet birth weight on postnatal growth performance, tissue lipogenic capacity and muscle histological traits at market weight. Livest. Prod. Sci. 2005, 93, 137–146. [Google Scholar] [CrossRef]
- Lee, J.B.; Kang, Y.J.; Kim, S.G.; Woo, J.H.; Shin, M.C.; Park, N.G.; Yang, B.C.; Han, S.H.; Han, K.M.; Lim, H.T.; et al. GWAS and Post-GWAS high-resolution mapping analyses identify strong novel candidate genes influencing the fatty acid composition of the longissimus dorsi muscle in pigs. Genes 2021, 12, 1323. [Google Scholar] [CrossRef]
- Zong, W.; Wang, J.; Zhao, R.; Niu, N.; Su, Y.; Hu, Z.; Liu, X.; Hou, X.; Wang, L.; Wang, L.; et al. Associations of genome-wide structural variations with phenotypic differences in cross-bred Eurasian pigs. J. Anim. Sci. Biotechnol. 2023, 14, 136. [Google Scholar] [CrossRef]
- Xu, H.; Ma, Y.; Xu, L.L.; Li, Y.; Liu, Y.; Li, Y.; Zhou, X.J.; Zhou, W.; Lee, S.; Zhang, P.; et al. SPA(GRM): Effectively controlling for sample relatedness in large-scale genome-wide association studies of longitudinal traits. Nat. Commun. 2025, 16, 1413. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Gao, N.; Li, X.; El-Ashram, S.; Wang, Z.; Zhu, L.; Jiang, W.; Peng, X.; Zhang, C.; Chen, Y.; et al. Identifying candidate genes associated with sperm morphology abnormalities using weighted single-step GWAS in a Duroc boar population. Theriogenology 2020, 141, 9–15. [Google Scholar] [CrossRef]
- Yuling, Z.; Zhanwei, Z.; Yiyi, L.; Jinyan, H.; Menghao, L.; Xiang, Z.; Linsong, D.; Jian, Y.; Ming, Y.; Enqin, Z.; et al. Genomic prediction based on preselected single-nucleotide polymorphisms from genome-wide association study and imputed whole-genome sequence data annotation for growth traits in Duroc pigs. Evol. Appl. 2024, 17, e13651. [Google Scholar] [CrossRef]
- Wu, Z.; Dou, T.; Bai, L.; Han, J.; Yang, F.; Wang, K.; Han, X.; Qiao, R.; Li, X.L.; Li, X.J. Genomic prediction and genome-wide association studies for additive and dominance effects for body composition traits using 50 K and imputed high-density SNP genotypes in Yunong-black pigs. J. Anim. Breed. Genet. 2024, 141, 124–137. [Google Scholar] [CrossRef]
- Tao, L.; Liu, H.; Adeola, A.C.; Xie, H.B.; Feng, S.T.; Zhang, Y.P. The effects of runs-of-homozygosity on pig domestication and breeding. BMC Genom. 2025, 26, 6. [Google Scholar] [CrossRef]
- Wu, P.; Wang, K.; Zhou, J.; Chen, D.; Jiang, A.; Jiang, Y.; Zhu, L.; Qiu, X.; Li, X.; Tang, G. A combined GWAS approach reveals key loci for socially-affected traits in Yorkshire pigs. Commun. Biol. 2021, 4, 891. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Criado-Mesas, L.; Revilla, M.; Castello, A.; Noguera, J.L.; Fernandez, A.I.; Ballester, M.; Folch, J.M. Identification of strong candidate genes for backfat and intramuscular fatty acid composition in three crosses based on the Iberian pig. Sci. Rep. 2020, 10, 13962. [Google Scholar] [CrossRef]
- Uemoto, Y.; Ichinoseki, K.; Matsumoto, T.; Oka, N.; Takamori, H.; Kadowaki, H.; Kojima-Shibata, C.; Suzuki, E.; Okamura, T.; Aso, H.; et al. Genome-wide association studies for production, respiratory disease, and immune-related traits in Landrace pigs. Sci. Rep. 2021, 11, 15823. [Google Scholar] [CrossRef]
- Kefala Taye, M.; Dong-Hui, L.; Young-Gyu, C.; Ah-Yeong, S.; Kang-Seok, S. Genome-Wide Association Studies and Runs of Homozygosity Reveals Genetic Markers Associated with Reproductive Performance in Korean Duroc, Landrace, and Yorkshire Breeds. Genes 2024, 15, 1422. [Google Scholar] [CrossRef]
- Bakoev, S.; Getmantseva, L.; Kolosova, M.; Bakoev, F.; Kolosov, A.; Romanets, E.; Shevtsova, V.; Romanets, T.; Kolosov, Y.; Usatov, A. Identifying Significant SNPs of the Total Number of Piglets Born and Their Relationship with Leg Bumps in Pigs. Biology 2024, 13, 1034. [Google Scholar] [CrossRef]
- Hong, Y.; Tan, C.; He, X.; Wu, D.; Zhang, Y.; Song, C.; Wu, Z. Genome-Wide Association Study of Reproductive Traits in Large White Pigs. Animals 2024, 14, 2874. [Google Scholar] [CrossRef]
- Miao, Y.; Zhao, Y.; Wan, S.; Mei, Q.; Wang, H.; Fu, C.; Li, X.; Zhao, S.; Xu, X.; Xiang, T. Integrated analysis of genome-wide association studies and 3D epigenomic characteristics reveal the BMP2 gene regulating loin muscle depth in Yorkshire pigs. PLoS Genet. 2023, 19, e1010820. [Google Scholar] [CrossRef]
- Gonzalez-Prendes, R.; Quintanilla, R.; Canovas, A.; Manunza, A.; Figueiredo Cardoso, T.; Jordana, J.; Noguera, J.L.; Pena, R.N.; Amills, M. Joint QTL mapping and gene expression analysis identify positional candidate genes influencing pork quality traits. Sci. Rep. 2017, 7, 39830. [Google Scholar] [CrossRef]
- Ovilo, C.; Trakooljul, N.; Nunez, Y.; Hadlich, F.; Murani, E.; Ayuso, M.; Garcia-Contreras, C.; Vazquez-Gomez, M.; Rey, A.I.; Garcia, F.; et al. SNP discovery and association study for growth, fatness and meat quality traits in Iberian crossbred pigs. Sci. Rep. 2022, 12, 16361. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Lim, J.H.; Park, H.B. Genome-wide association studies to identify quantitative trait loci and positional candidate genes affecting meat quality-related traits in pigs. J. Anim. Sci. Technol. 2023, 65, 1194–1204. [Google Scholar] [CrossRef]
- He, S.; Wang, Y.; Luo, Y.; Xue, M.; Wu, M.; Tan, H.; Peng, Y.; Wang, K.; Fang, M. Integrated analysis strategy of genome-wide functional gene mining reveals DKK2 gene underlying meat quality in Shaziling synthesized pigs. BMC Genom. 2024, 25, 30. [Google Scholar] [CrossRef]
- Li, Y.; Li, B.; Yang, M.; Han, H.; Chen, T.; Wei, Q.; Miao, Z.; Yin, L.; Wang, R.; Shen, J. Genome-wide association study and fine mapping reveals candidate genes for birth weight of Yorkshire and Landrace pigs. Front. Genet. 2020, 11, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.X.; Gao, G.X.; Zhou, Y.; Guo, C.X.; Li, B.; El-Ashram, S.; Li, Z.L. Genome-wide association studies uncover genes associated with litter traits in the pig. Animal 2022, 16, 100672. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xiao, J.; Jiang, Y.; Wang, Y.; Cao, M.; Wei, J.; Yu, T.; Ding, X.; Yang, G. Genome-Wide Association Study on Reproductive Traits Using Imputation-Based Whole-Genome Sequence Data in Yorkshire Pigs. Genes 2023, 14, 861. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: New York, NY, USA, 2016; pp. 1–20. [Google Scholar]
- Bai, Y.; Zhang, J.B.; Xue, Y.; Peng, Y.L.; Chen, G.; Fang, M.Y. Differential expression of CYB5A in Chinese and European pig breeds due to genetic variations in the promoter region. Anim. Genet. 2015, 46, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Yuan, M.; Zhan, F.; Song, M.; Shang, P.; Yang, F.; Li, X.; Qiao, R.; Han, X.; et al. Genome-Wide Association Studies and Runs of Homozygosity to Identify Reproduction-Related Genes in Yorkshire Pig Population. Genes 2023, 14, 2133. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Cam, T.L.; Shashaank, V.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Stange, K.; Miersch, C.; Sponder, G.; Rontgen, M. Low birth weight influences the postnatal abundance and characteristics of satellite cell subpopulations in pigs. Sci. Rep. 2020, 10, 6149. [Google Scholar] [CrossRef]
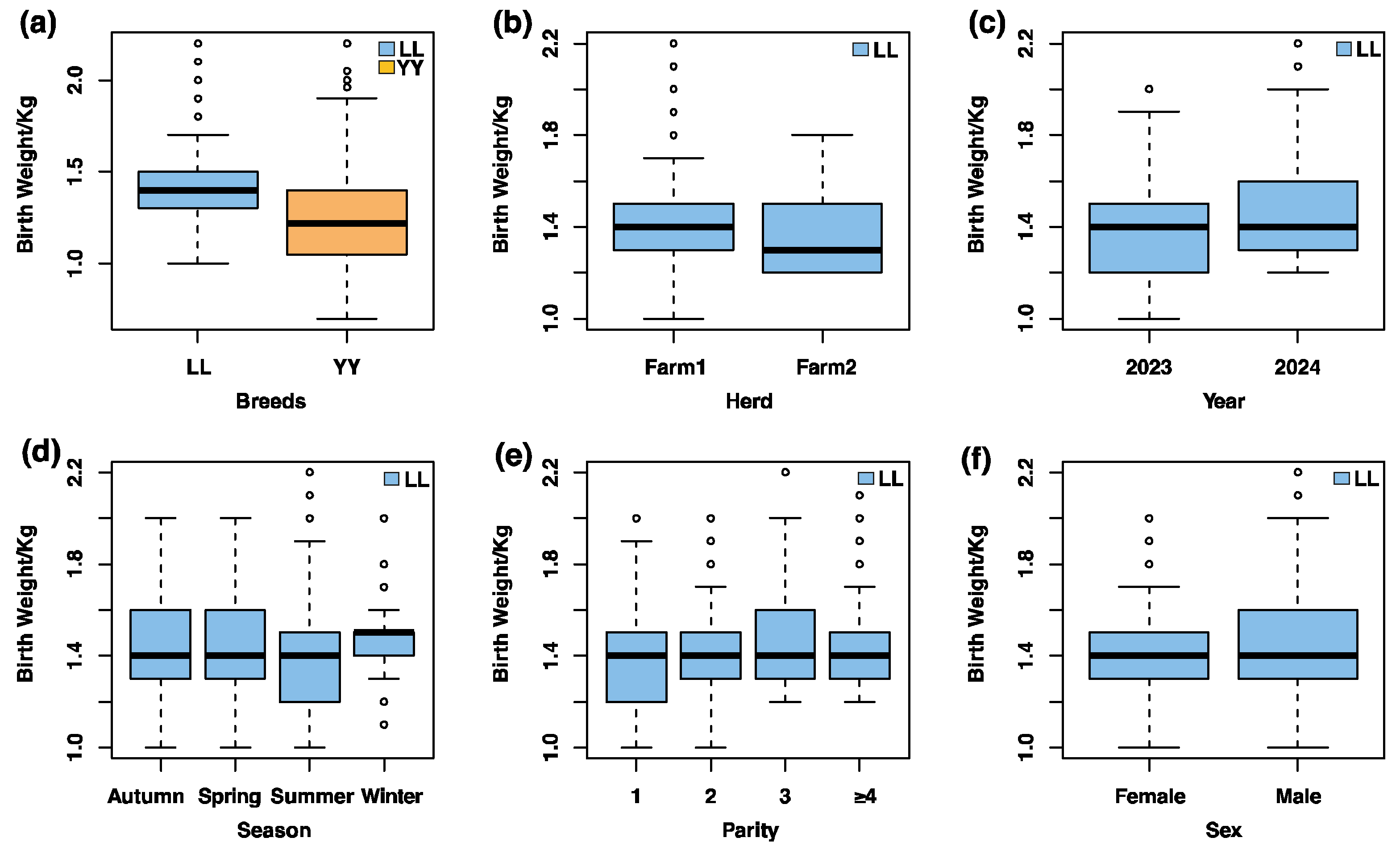


| Species | Count | Mean | SD 1 | Median | Min | Max | CV 2 (%) |
|---|---|---|---|---|---|---|---|
| Landrace | 1125 | 1.42 | 0.20 | 1.40 | 0.80 | 2.20 | 13.73 |
| Yorkshire | 998 | 1.22 | 0.25 | 1.22 | 0.70 | 2.20 | 20.66 |
| Total | 2123 | 1.33 | 0.24 | 1.30 | 0.70 | 2.20 | 18.34 |
| Population | Breeds | Herd | Year | Season | Parity | Sex |
|---|---|---|---|---|---|---|
| Landrace | -- | ** | ** | *** | *** | *** |
| Yorkshire | -- | *** | *** | ns | ns | -- |
| Total | *** | *** | * | *** | ns | *** |
| SNP | Chr. 1 | Position/bp | Allele | p-Value | Candidate Gene |
|---|---|---|---|---|---|
| CNC10010096 | 1 | 4,264,479 | A/G | 1.90 × 10−7 | -- |
| CNCB10001505 | 1 | 307,515,291 | T/C | 6.21 × 10−5 | -- |
| CNC10020434 | 2 | 18,828,505 | A/G | 1.81 × 10−7 | TTC17 |
| CNC10032309 | 3 | 117,474,270 | T/G | 2.07 × 10−7 | LDAH |
| CNC10032128 | 3 | 109,118,738 | T/C | 4.00 × 10−7 | -- |
| CNC10042365 | 4 | 125,588,650 | G/A | 2.25 × 10−5 | HFM1 |
| CNCB10003289 | 4 | 52,145,174 | G/A | 3.89 × 10−5 | RALYL |
| CNC10051462 | 5 | 78,270,522 | A/G | 6.53 × 10−7 | -- |
| CNC10060592 | 6 | 28,761,105 | T/A | 5.73 × 10−9 | NFATC3 |
| CNC10061395 | 6 | 66,110,498 | T/C | 3.58 × 10−5 | -- |
| CNC10082700 | 8 | 133,591,911 | C/A | 5.04 × 10−7 | -- |
| CNCB10009227 | 13 | 159,079,583 | T/C | 1.25 × 10−5 | CMSS1 |
| CNCB10009475 | 13 | 200,184,861 | T/C | 4.26 × 10−5 | CLDN14 |
| CNCB10009612 | 14 | 6,581,366 | G/A | 3.57 × 10−6 | PIWIL2 |
| CNCB10011512 | 16 | 66,786,864 | T/G | 1.43 × 10−6 | SGCD |
| CNC10160101 | 16 | 5,379,864 | C/T | 1.52 × 10−6 | MARCHF11 |
| CNCB10011490 | 16 | 61,520,473 | T/C | 6.53 × 10−5 | GABRG2 |
| CNC10161130 | 16 | 57,760,728 | A/G | 9.15 × 10−5 | TENM2 |
| CNC10170045 | 17 | 2,300,590 | T/C | 6.04 × 10−5 | SGCZ |
| CNC10180515 | 18 | 26,725,722 | G/A | 3.16 × 10−5 | -- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Zeng, Y.; Tian, Y.; Cheng, L.; Li, W.; Cheng, S.; Wang, J.; Li, L. Genome-Wide Association Study Reveals Novel Loci and Candidate Genes for Birth Weight in Pigs. Animals 2025, 15, 825. https://doi.org/10.3390/ani15060825
Liu J, Zeng Y, Tian Y, Cheng L, Li W, Cheng S, Wang J, Li L. Genome-Wide Association Study Reveals Novel Loci and Candidate Genes for Birth Weight in Pigs. Animals. 2025; 15(6):825. https://doi.org/10.3390/ani15060825
Chicago/Turabian StyleLiu, Jiajia, Yue Zeng, Yu Tian, Linghua Cheng, Wenchao Li, Shunfeng Cheng, Junjie Wang, and Lan Li. 2025. "Genome-Wide Association Study Reveals Novel Loci and Candidate Genes for Birth Weight in Pigs" Animals 15, no. 6: 825. https://doi.org/10.3390/ani15060825
APA StyleLiu, J., Zeng, Y., Tian, Y., Cheng, L., Li, W., Cheng, S., Wang, J., & Li, L. (2025). Genome-Wide Association Study Reveals Novel Loci and Candidate Genes for Birth Weight in Pigs. Animals, 15(6), 825. https://doi.org/10.3390/ani15060825





