Effects of Yeast Culture on Lamb Growth Performance, Rumen Microbiota, and Metabolites
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Culture
2.2. Sample Collection and Processing
2.3. Product Performance
2.4. Determination of Rumen Histomorphometry
2.5. Rumen Fermentation Characteristics
2.6. High-Throughput Sequencing and Analysis
2.7. Metabolomic Analysis of Rumen Fluid
2.8. Statistical Analysis
3. Results
3.1. Growth Performance Analysis
3.2. Differences in VFAs in Sheep Rumen
3.3. Differences in Rumen Organization
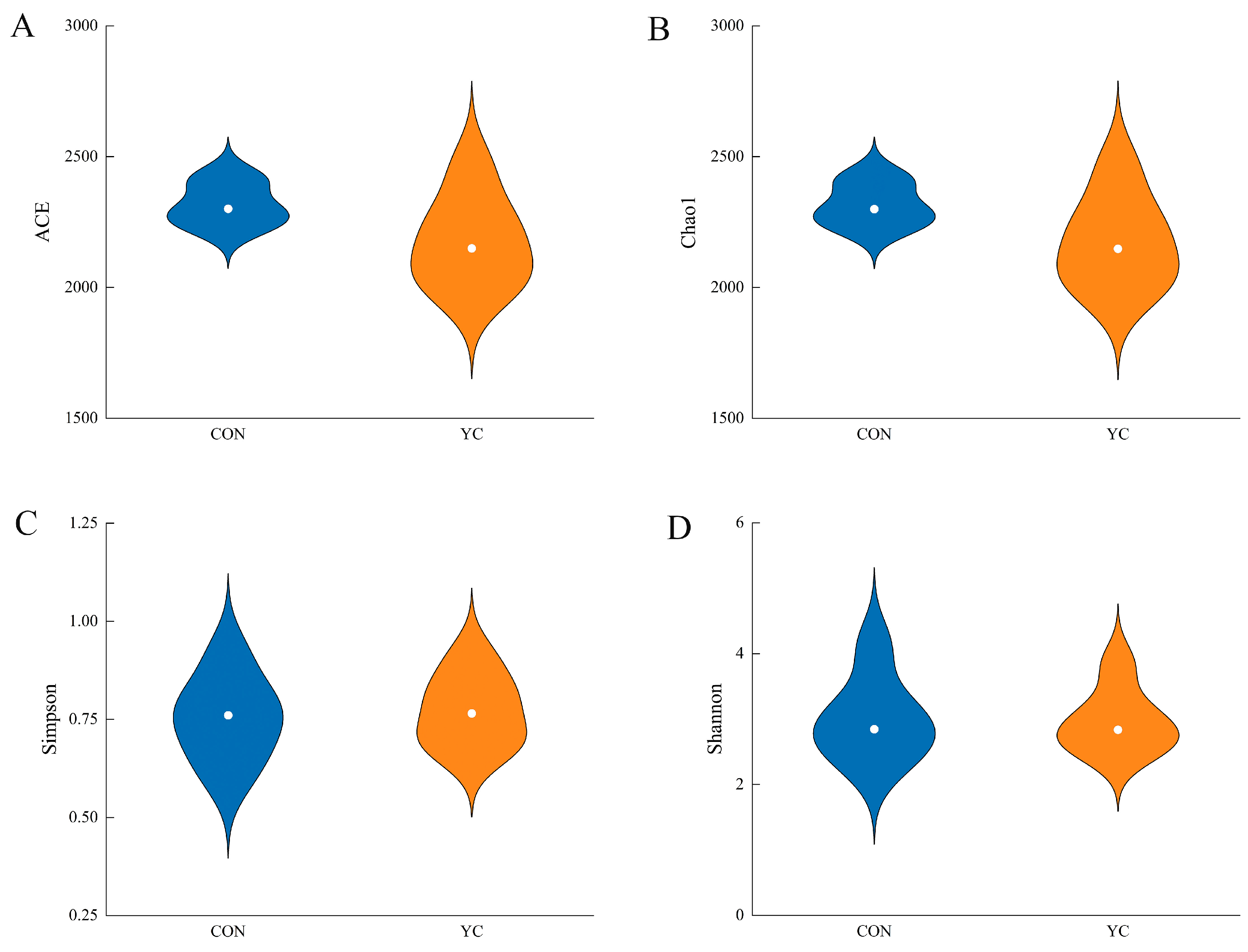
3.4. Analysis of the Alpha Diversity of Rumen Microorganisms
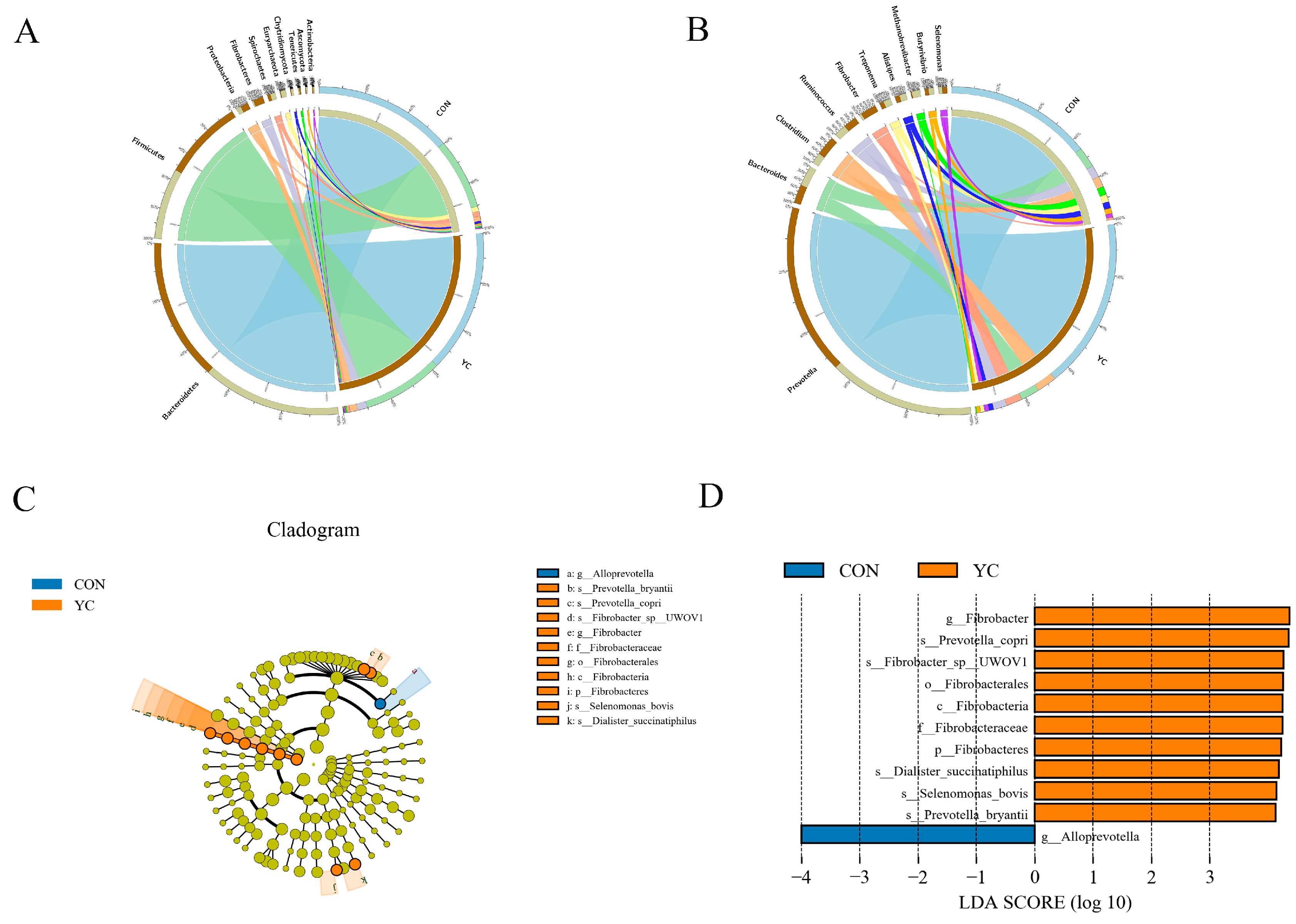
3.5. Differences in the Composition of Rumen Microbial Communities in Sheep
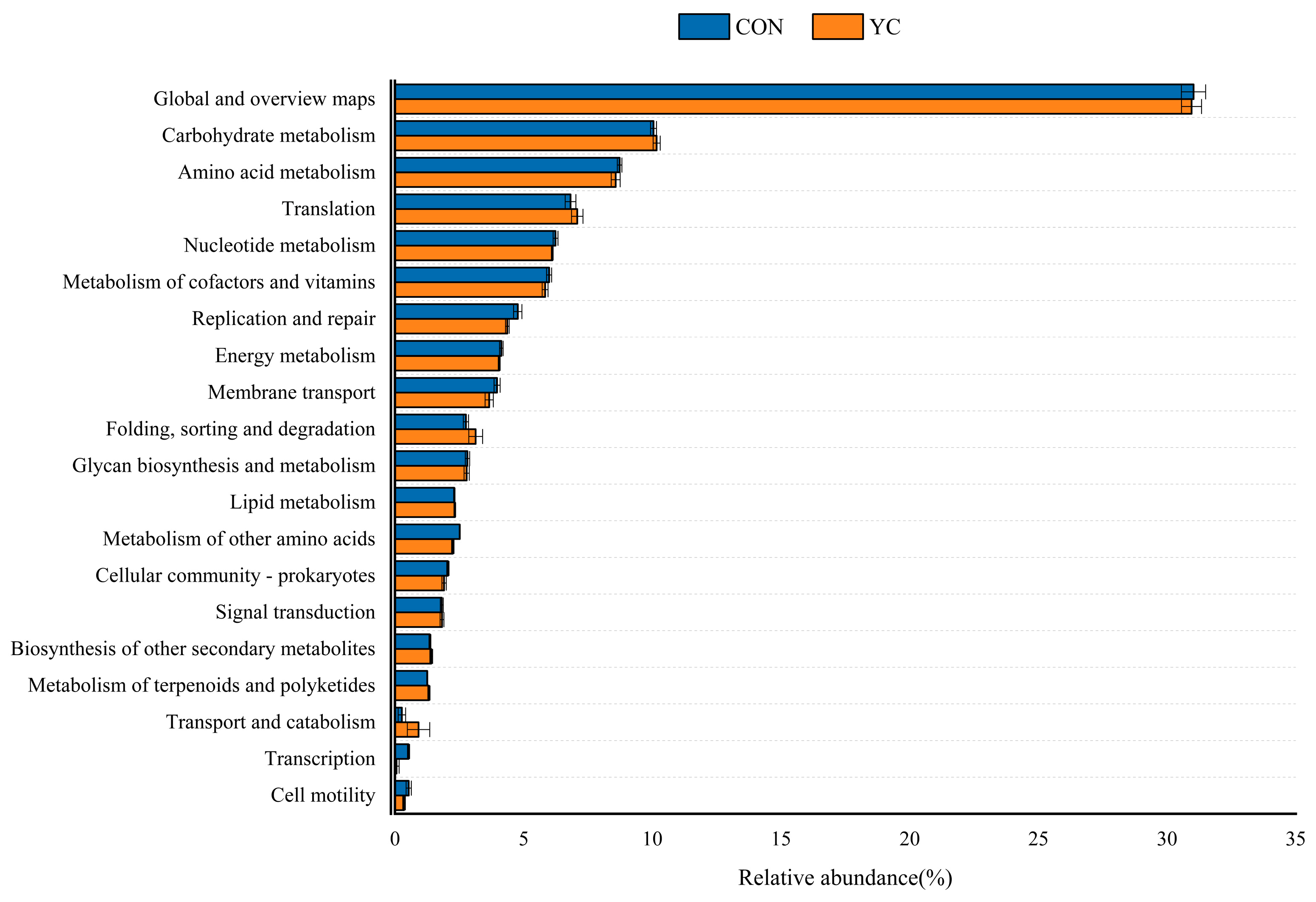
3.6. Differences in CAZy and KEGG Pathways of Sheep Rumen
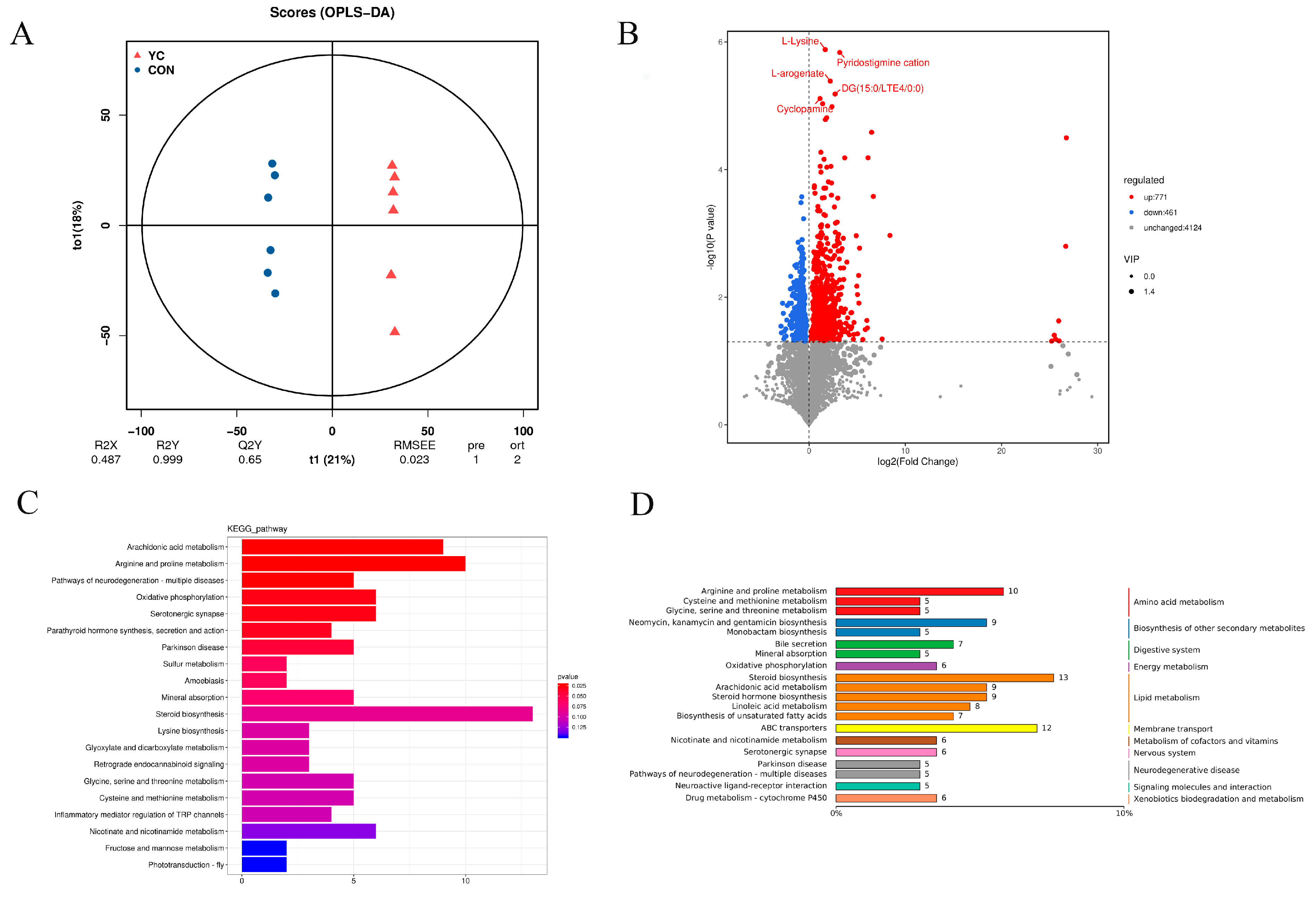
3.7. Differences in Rumen Metabolites
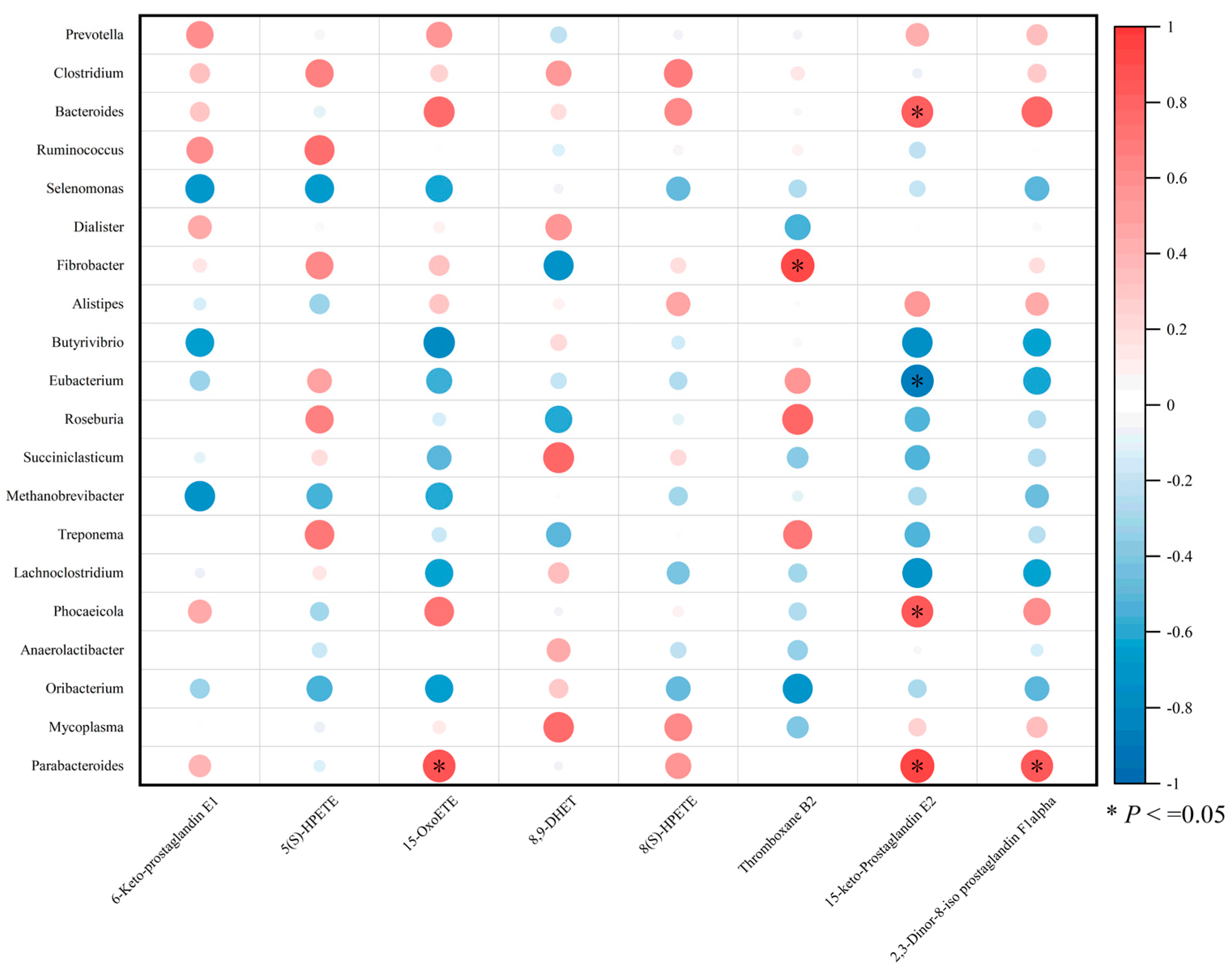
3.8. Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, R.; Zhang, L.; An, X.; Li, J.; Niu, C.; Zhang, J.; Geng, Z.; Xu, T.; Yang, B.; Xu, Z.; et al. Hybridization promotes growth performance by altering rumen microbiota and metabolites in sheep. Front. Vet. Sci. 2024, 11, 1455029. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.P.; Rehemujiang, H.; Zhang, L.D.; Lu, M.L.; Li, C.C.; Hu, L.H.; Wang, Y.L.; Diao, Q.Y.; Xu, G.S. Lycium barbarum (Wolfberry) Branches and Leaves Enhance the Growth Performance and Improve the Rumen Microbiota in Hu Sheep. Animals 2024, 14, 1610. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Shui, Y.; Deng, M.; Guo, Y.; Sun, B.; Liu, G.; Liu, D.; Li, Y. Effects of different dietary energy levels on growth performance, meat quality and nutritional composition, rumen fermentation parameters, and rumen microbiota of fattening Angus steers. Front. Microbiol. 2024, 15, 1378073. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Wang, Y.; Su, S.; Wang, W.; Guo, T.; Hu, Y.; Yin, N.; An, X.; Qi, J.; Xu, X. Supplementation with Complex Phytonutrients Enhances Rumen Barrier Function and Growth Performance of Lambs by Regulating Rumen Microbiome and Metabolome. Animals 2025, 15, 228. [Google Scholar] [CrossRef]
- Liu, T.; Pang, R.; Huang, L.; Mao, T.; Yu, J.; Hua, J.; Zhong, Y.; Ren, C.; Zhang, Z.; Zhu, W. Effects of allicin addition on growth performance, rumen microbiome, and ruminal epithelial proteome of high-grain-fed goats. Anim. Feed Sci. Technol. 2024, 310, 115944. [Google Scholar] [CrossRef]
- Gao, K.; Geng, C. Alterations in the rumen bacterial communities and metabolites of finishing bulls fed high-concentrate diets supplemented with active dry yeast and yeast culture. Front. Microbiol. 2022, 13, 908244. [Google Scholar] [CrossRef]
- Gao, K.; Geng, C. Comparison of rectum fecal bacterial community of finishing bulls fed high-concentrate diets with active dry yeast and yeast culture supplementation. Anim. Biosci. 2023, 36, 63–74. [Google Scholar] [CrossRef]
- Halfen, J.; Carpinelli, N.; Del Pino, F.; Chapman, J.; Sharman, E.; Anderson, J.; Osorio, J. Effects of yeast culture supplementation on lactation performance and rumen fermentation profile and microbial abundance in mid-lactation Holstein dairy cows. J. Dairy Sci. 2021, 104, 11580–11592. [Google Scholar] [CrossRef]
- Suntara, C.; Cherdthong, A.; Wanapat, M.; Uriyapongson, S.; Leelavatcharamas, V.; Sawaengkaew, J.; Chanjula, P.; Foiklang, S. Isolation and Characterization of Yeasts from Rumen Fluids for Potential Use as Additives in Ruminant Feeding. Vet. Sci. 2021, 8, 52. [Google Scholar] [CrossRef]
- Li, Z.; Hu, Y.; Li, H.; Lin, Y.; Cheng, M.; Zhu, F.; Guo, Y. Effects of yeast culture supplementation on milk yield, rumen fermentation, metabolism, and bacterial composition in dairy goats. Front. Vet. Sci. 2024, 11, 1447238. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, G.; Zhuang, Y.; Chai, J.; Zhang, N. Yeast (Saccharomyces cerevisiae) Culture Promotes the Performance of Fattening Sheep by Enhancing Nutrients Digestibility and Rumen Development. Fermentation 2022, 8, 719. [Google Scholar] [CrossRef]
- Qi, P.; Wang, L. Effect of Adding Yeast Cultures to High-Grain Conditions on Production Performance, Rumen Fermentation Profile, Microbial Abundance, and Immunity in Goats. Animals 2024, 14, 1799. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Xu, J.; Yang, Q.; Sha, Y.; Jiao, T.; Zhao, S. Effects of yeast cultures on meat quality, flavor composition and rumen microbiota in lambs. Curr. Res. Food Sci. 2024, 9, 100845. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Su, M.; Wang, C.; Li, D.; Li, Q.; Liu, Z.; Qi, X.; Wu, Y.; Zhao, Y.; Li, T.; et al. Yeast culture repairs rumen epithelial injury by regulating microbial communities and metabolites in sheep. Front. Microbiol. 2023, 14, 1305772. [Google Scholar] [CrossRef]
- Liu, Y.-Z.; Chen, X.; Zhao, W.; Lang, M.; Zhang, X.-F.; Wang, T.; Farouk, M.H.; Zhen, Y.-G.; Qin, G.-X. Effects of yeast culture supplementation and the ratio of non-structural carbohydrate to fat on rumen fermentation parameters and bacterial-community composition in sheep. Anim. Feed Sci. Technol. 2019, 249, 62–75. [Google Scholar] [CrossRef]
- Majewska, M.P.; Miltko, R.; Bełżecki, G.; Kowalik, B. Population of protozoa and carbohydrate-digesting enzymes in the rumen of sheep fed a diet supplemented with yeast Saccharomyces cerevisiae. Small Rumin. Res. 2021, 205, 106544. [Google Scholar] [CrossRef]
- Boukrouh, S.; Noutfia, A.; Moula, N.; Avril, C.; Hornick, J.-L.; Chentouf, M.; Cabaraux, J.-F. Effects of Sulla Flexuosa Hay as Alternative Feed Resource on Goat’s Milk Production and Quality. Animals 2023, 13, 709. [Google Scholar] [CrossRef]
- Boukrouh, S.; Noutfia, A.; Moula, N.; Avril, C.; Louvieaux, J.; Hornick, J.-L.; Cabaraux, J.-F.; Chentouf, M. Growth performance, carcass characteristics, fatty acid profile, and meat quality of male goat kids supplemented by alternative feed resources: Bitter vetch and sorghum grains. Arch. Anim. Breed. 2024, 67, 481–492. [Google Scholar] [CrossRef]
- Li, X.; Wang, Z.; Qin, W.; Gao, X.; Wu, J.; Zhao, S.; Jiao, T. Effects of oregano essential oil, cobalt and synergistic of both of them on rumen degradation rate and fermentation characteristics for corn silage. Ital. J. Anim. Sci. 2022, 21, 1476–1488. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.-F.; He, Y.; Wu, J.-Y.; Jiang, Y.-X.; Tam, N.F.-Y.; Zhou, H.-W. Comparison of the Levels of Bacterial Diversity in Freshwater, Intertidal Wetland, and Marine Sediments by Using Millions of Illumina Tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef] [PubMed]
- Dubois, P.C.; Trynka, G.; Franke, L.; Hunt, K.A.; Romanos, J.; Curtotti, A.; Zhernakova, A.; Heap, G.A.; Adány, R.; Aromaa, A.; et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010, 42, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Chen, H.; Liu, S.; Li, S.; Li, D.; Li, X.; Xu, Z.; Liu, D. Effects of yeast culture on growth performance, immune function, antioxidant capacity and hormonal profile in Mongolian ram lambs. Front. Vet. Sci. 2024, 11, 1424073. [Google Scholar] [CrossRef]
- Aschenbach, J.R.; Penner, G.B.; Stumpff, F.; Gäbel, G. Ruminant Nutrition Symposium: Role of fermentation acid absorption in the regulation of ruminal pH. J. Anim. Sci. 2011, 89, 1092–1107. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, L.; Chen, H.; Liu, S.; Li, X.; Li, S.; Ying, C.; Li, X.; Du, R.; Liu, D. Saccharomyces cerevisiae and Kluyveromyces marxianus yeast co-cultures modulate the ruminal microbiome and metabolite availability to enhance rumen barrier function and growth performance in weaned lambs. Anim. Nutr. 2024, 19, 139–152. [Google Scholar] [CrossRef]
- Tun, H.M.; Li, S.; Yoon, I.; Meale, S.J.; Azevedo, P.A.; Khafipour, E.; Plaizier, J.C. Saccharomyces cerevisiae fermentation products (SCFP) stabilize the ruminal microbiota of lactating dairy cows during periods of a depressed rumen pH. BMC Vet. Res. 2020, 16, 237. [Google Scholar] [CrossRef]
- Xue, M.-Y.; Sun, H.-Z.; Wu, X.-H.; Liu, J.-X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef]
- Xue, M.-Y.; Xie, Y.-Y.; Zhong, Y.; Ma, X.-J.; Sun, H.-Z.; Liu, J.-X. Integrated meta-omics reveals new ruminal microbial features associated with feed efficiency in dairy cattle. Microbiome 2022, 10, 32. [Google Scholar] [CrossRef]
- Shao, P.; Sha, Y.; Liu, X.; He, Y.; Wang, F.; Hu, J.; Wang, J.; Li, S.; Chen, X.; Yang, W.; et al. Supplementation with Astragalus Root Powder Promotes Rumen Microbiota Density and Metabolome Interactions in Lambs. Animals 2024, 14, 788. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Kong, L.; Ren, S.; Aschenbach, J.R.; Shen, H. Acid tolerance of lactate-utilizing bacteria of the order Bacteroidales contributes to prevention of ruminal acidosis in goats adapted to a high-concentrate diet. Anim. Nutr. 2023, 14, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhaxi, Y.; Ciwang, R.; Wang, Z.; Liu, M. Responses of rumen microorganisms and metabolites to different roughage of domesticated Tibetan sheep. Front. Microbiol. 2023, 14, 1247609. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, Y.; Qi, X.; Liu, Z.; Wang, C.; Ma, X.; Ma, Y. Prickly Ash Seeds Improve the Ruminal Epithelial Development and Growth Performance of Hu Sheep by Modulating the Rumen Microbiota and Metabolome. Microorganisms 2024, 12, 2242. [Google Scholar] [CrossRef]
- Wang, K.; Jiang, M.; Chen, Y.; Huang, Y.; Cheng, Z.; Datsomor, O.; Jama, S.M.; Zhu, L.; Li, Y.; Zhao, G.; et al. Changes in the rumen development, rumen fermentation, and rumen microbiota community in weaned calves during steviol glycosides treatment. Front. Microbiol. 2024, 15, 1395665. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 2016, 24, 151–157. [Google Scholar] [CrossRef]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms 2022, 11, 1. [Google Scholar] [CrossRef]
- Carpinelli, N.; Halfen, J.; Trevisi, E.; Chapman, J.; Sharman, E.; Anderson, J.; Osorio, J. Effects of peripartal yeast culture supplementation on lactation performance, blood biomarkers, rumen fermentation, and rumen bacteria species in dairy cows. J. Dairy Sci. 2021, 104, 10727–10743. [Google Scholar] [CrossRef]
- Sirisan, V.; Pattarajinda, V.; Vichitphan, K.; Leesing, R. Isolation, identification and growth determination of lactic acid-utilizing yeasts from the ruminal fluid of dairy cattle. Lett. Appl. Microbiol. 2013, 57, 102–107. [Google Scholar] [CrossRef]
- Ogunade, I.; Schweickart, H.; McCoun, M.; Cannon, K.; McManus, C. Integrating 16S rRNA Sequencing and LC–MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle. Animals 2019, 9, 28. [Google Scholar] [CrossRef]
- Ma, H.; Dong, A.; Xu, Y.; Wu, Q.; Lambo, M.T.; Zhang, Y.; Dou, X.; Li, Y. Regulatory effects of high concentrate diet synergistically fermented with cellulase and lactic acid bacteria: Ruminal fermentation, methane production, and rumen microbiome. Anim. Feed Sci. Technol. 2025, 319, 116194. [Google Scholar] [CrossRef]
- Bature, I.; Xiaohu, W.; Ding, X. The roles of phytogenic feed additives, trees, shrubs, and forages on mitigating ruminant methane emission. Front. Vet. Sci. 2024, 11, 1475322. [Google Scholar] [CrossRef] [PubMed]
- Ungerfeld, E.M. Metabolic Hydrogen Flows in Rumen Fermentation: Principles and Possibilities of Interventions. Front. Microbiol. 2020, 11, 589. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Marin, S.B.; Betancur-Murillo, C.L.; Isaza, G.A.; Mesa, H.; Jovel, J. Lower methane emissions were associated with higher abundance of ruminal Prevotella in a cohort of Colombian buffalos. BMC Microbiol. 2020, 20, 364. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef]
- Bai, J.; Tang, L.; Bi, Y.; Li, M. Multi-omics insights into the energy compensation of rumen microbiota of grazing yaks in cold season. Front. Microbiol. 2024, 15, 1467841. [Google Scholar] [CrossRef]
- Liu, X.; Sha, Y.; Lv, W.; Cao, G.; Guo, X.; Pu, X.; Wang, J.; Li, S.; Hu, J.; Luo, Y. Multi-Omics Reveals That the Rumen Transcriptome, Microbiome, and Its Metabolome Co-regulate Cold Season Adaptability of Tibetan Sheep. Front. Microbiol. 2022, 13, 859601. [Google Scholar] [CrossRef]

| Items | Diets (%) | Nutrient Levels | |
|---|---|---|---|
| Corn | 32.0 | Digestive energy DE/(MJ/kg) | 11.02 |
| Wheat bran | 6.0 | Moisture content/% | 11.76 |
| Grass meal | 10.0 | CP/% | 16.43 |
| Wheatgrass | 10.0 | Ash/% | 8.16 |
| Soybean meal (43%) | 5.0 | Salinity/% | 0.96 |
| Cottonseed meal (46%) | 5.0 | Ca/% | 1.23 |
| Bran | 8.0 | P/% | 0.49 |
| Corn germ meal | 15.0 | NDF/% | 37.84 |
| DDGS (distiller dried grains with solubles) | 5.0 | ADF% | 26.28 |
| Premix a | 4.0 | NFE (g/kg) b | 617.92 |
| Items | CON | 3% YC | 6% YC | 9% YC | 12% YC | p Value |
|---|---|---|---|---|---|---|
| Initial weight (kg) | 27.33 ± 0.58 | 27.77 ± 0.81 | 28.23 ± 0.82 | 28.47 ± 0.72 | 27.5 ± 0.57 | 0.764 |
| Final weight (kg) | 42.9 ± 0.25 b | 44.87 ± 1.11 | 45 ± 0.89 | 46.57 ± 0.80 a | 45.02 ± 0.39 | 0.040 |
| ADG (g/d) | 277.98 ± 24.13 | 305.36 ± 27.50 | 299.40 ± 25.59 | 323.21 ± 28.19 | 312.80 ± 29.18 | 0.232 |
| ADFI (kg/d) | 1.61 ± 0.09 | 1.62 ± 0.07 | 1.59 ± 0.1 | 1.67 ± 0.13 | 1.66 ± 0.09 | 0.977 |
| F/G | 5.79 ± 0.31 | 5.31 ± 0.24 | 5.32 ± 0.35 | 5.17 ± 0.41 | 5.30 ± 0.28 | 0.706 |
| Items | CON | 9% YC | p Value |
|---|---|---|---|
| pH | 6.65 ± 0.16 | 6.44 ± 0.15 | 0.346 |
| NH3-N (mg/dL) | 8.70 ± 2.50 | 5.93 ± 0.55 | 0.051 |
| TVFA (mmol/L) | 67.41 ± 2.30 b | 81.91 ± 3.47 a | 0.006 |
| Acetate (mmol/L) | 41.86 ± 0.73 | 47.79 ± 3.13 | 0.119 |
| Propionate (mmol/L) | 14.30 ± 1.59 b | 19.11 ± 0.99 a | 0.002 |
| Butyrate (mmol/L) | 8.69 ± 1.13 b | 11.91 ± 0.76 a | 0.039 |
| Isobutyric acid (mmol/L) | 0.73 ± 0.08 | 0.78 ± 0.05 | 0.644 |
| Valerate (mmol/L) | 0.711 ± 0.02 | 0.78 ± 0.04 | 0.151 |
| Isovalerate (mmol/L) | 1.12 ± 0.20 | 1.54 ± 0.14 | 0.124 |
| A/P | 2.96 ± 0.13 | 2.51 ± 0.12 | 0.028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Li, X.; Fan, Q.; Zhao, S.; Jiao, T. Effects of Yeast Culture on Lamb Growth Performance, Rumen Microbiota, and Metabolites. Animals 2025, 15, 738. https://doi.org/10.3390/ani15050738
Xu J, Li X, Fan Q, Zhao S, Jiao T. Effects of Yeast Culture on Lamb Growth Performance, Rumen Microbiota, and Metabolites. Animals. 2025; 15(5):738. https://doi.org/10.3390/ani15050738
Chicago/Turabian StyleXu, Jinlong, Xiongxiong Li, Qingshan Fan, Shengguo Zhao, and Ting Jiao. 2025. "Effects of Yeast Culture on Lamb Growth Performance, Rumen Microbiota, and Metabolites" Animals 15, no. 5: 738. https://doi.org/10.3390/ani15050738
APA StyleXu, J., Li, X., Fan, Q., Zhao, S., & Jiao, T. (2025). Effects of Yeast Culture on Lamb Growth Performance, Rumen Microbiota, and Metabolites. Animals, 15(5), 738. https://doi.org/10.3390/ani15050738





