Genome-Wide Association Study of Abdominal and Intramuscular Fat Deposition Traits in Huainan Yellow-Feathered Chickens
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Population and Experimental Design
2.3. Statistical Analysis
2.3.1. Phenotypic Data Statistical Analysis
2.3.2. DNA Extraction and Low Depth Sequencing
2.3.3. Genotyping Data Processing and Population Structure Analysis
2.3.4. GWAS of Four Fat Traits
2.3.5. Candidate Genes Annotation and Functional Enrichment Analysis
3. Results and Discussion
3.1. Statistical Data of Fat Traits and Fat-Related Traits
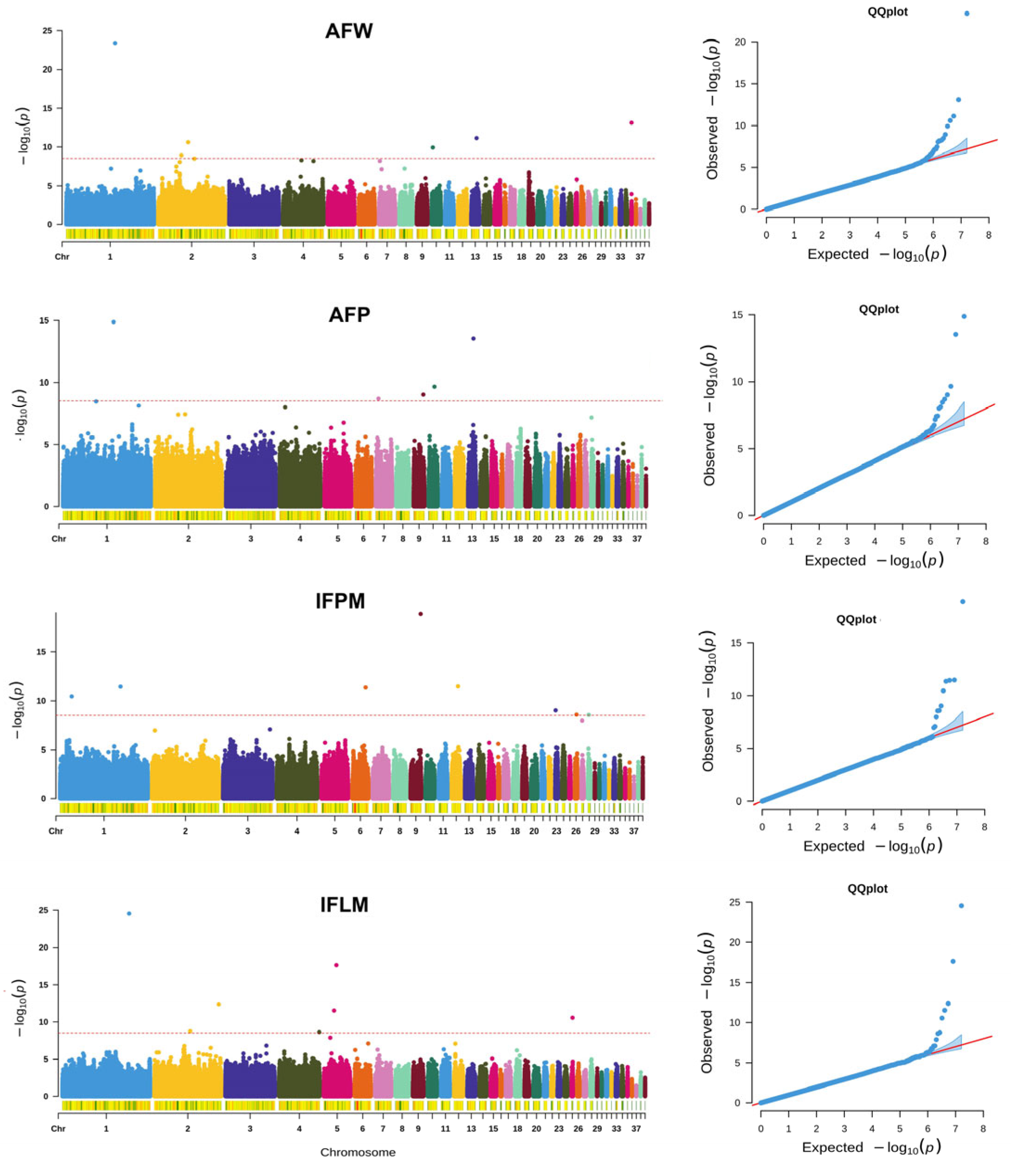
3.2. GWAS for AFW and AFP
3.3. GWAS for IFPM and IFLM
3.4. KEGG Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tian, W.; Nie, R.; Zhang, W.; Ling, Y.; Tian, H.; Zhang, B.; Zhang, H.; Wu, C. Research progress on genetic characteristics and genes regulation of abdominal fat in broiler chickens (Gallus gallus). J. Agric. Biotechnol. 2023, 31, 2387–2400. [Google Scholar] [CrossRef]
- Leng, L.; Zhang, H.; Wang, W.; Wang, S.; Li, H. Research progress of lean broiler selection methods. China Poult. 2019, 41, 102298. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D.; Wang, Y.; Li, G.; Liu, L.; Xu, H. Genome-wide association study identifies genetic markers associated with abdominal fat percentage in chickens. Poult. Sci. 2022, 101, 101789. [Google Scholar] [CrossRef]
- Wang, H.; Li, H.; Wang, Q.; Zhang, X.; Wang, Y.; Liu, Z. Integrated analysis of transcriptome and GWAS reveals candidate genes for abdominal fat deposition in chickens. Poult. Sci. 2022, 101, 101651. [Google Scholar] [CrossRef]
- Zerehdaran, S.; Vereijken, A.L.; van Arendonk, J.A.; van der Waaijt, E.H. Estimation of genetic parameters for fat deposition and carcass traits in broilers. Poult. Sci. 2004, 83, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Cahaner, A.; Nitsan, Z. Evaluation of simultaneous selection for live body weight and against abdominal fat in broilers. Poult. Sci. 1985, 64, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Qi, L.; Xu, Z.; Zhang, D.; Nie, Q.; Zhang, X.; Luo, W. Weighted single-step GWAS identified candidate genes associated with carcass traits in a Chinese yellow-feathered chicken population. Poult. Sci. 2024, 103, 103341. [Google Scholar] [CrossRef]
- Shen, L.; Bai, X.; Zhao, L.; Zhou, J.; Chang, C.; Li, X.; Cao, Z.; Li, Y.; Luan, P.; Li, H.; et al. Integrative 3D genomics with multi-omics analysis and functional validation of genetic regulatory mechanisms of abdominal fat deposition in chickens. Nat. Commun. 2024, 15, 19274. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Stanley, B.G.; Willett, V.L., 3rd; Donias, H.W.; Ha, L.H.; Spears, L.C. The lateral hypothalamus: A primary site mediating excitatory amino acid-elicited eating. Brain Res. 1993, 630, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Stanley, B.G.; Willett, V.L., 3rd; Donias, H.W.; Dee, M.G., 2nd; Duva, M.A. Lateral hypothalamic NMDA receptors and glutamate as physiological mediators of eating and weight control. Am. J. Physiol. 1996, 270, R443–R449. [Google Scholar] [CrossRef] [PubMed]
- Adermark, L.; Gutierrez, S.; Lagström, O.; Hammarlund, M.; Licheri, V.; Johansson, M.E. Weight gain and neuroadaptations elicited by high fat diet depend on fatty acid composition. Psychoneuroendocrinology 2021, 126, 105143. [Google Scholar] [CrossRef] [PubMed]
- Montie, E.W.; Fair, P.A.; Bossart, G.D.; Mitchum, G.B.; Houde, M.; Muir, D.C.; Letcher, R.J.; McFee, W.E.; Starczak, V.R.; Stegeman, J.J.; et al. Cytochrome P4501A1 expression, polychlorinated biphenyls and hydroxylated metabolites, and adipocyte size of bottlenose dolphins from the Southeast United States. Aquat. Toxicol. 2008, 86, 397–412. [Google Scholar] [CrossRef]
- Ellero, S.; Chakhtoura, G.; Barreau, C.; Langouët, S.; Benelli, C.; Penicaud, L.; Beaune, P.; de Waziers, I. Xenobiotic-metabolizing cytochromes p450 in human white adipose tissue: Expression and induction. Drug Metab. Dispos. 2010, 38, 679–686. [Google Scholar] [CrossRef]
- Shih, M.C.; Chiu, Y.N.; Hu, M.C.; Guo, I.C.; Chung, B.C. Regulation of steroid production: Analysis of Cyp11a1 promoter. Mol. Cell. Endocrinol. 2011, 336, 80–84. [Google Scholar] [CrossRef]
- Gkouskou, K.G.; Georgiopoulos, G.; Vlastos, I.; Lazou, E.; Chaniotis, D.; Papaioannou, T.G.; Mantzoros, C.S.; Sanoudou, D.; Eliopoulos, A.G. CYP1A2 polymorphisms modify the association of habitual coffee consumption with appetite, macronutrient intake, and body mass index: Results from an observational cohort and a cross-over randomized study. Int. J. Obes. 2022, 46, 162–168. [Google Scholar] [CrossRef]
- Ching, M.E.A.; Hoyeck, M.P.; Basu, L.; Merhi, R.; Poleo-Giordani, E.; van Zyl, E.; Crawley, A.M.; Bruin, J.E. CYP1A1/1A2 enzymes mediate glucose homeostasis and insulin secretion in mice in a sex-specific manner. Am. J. Physiol. Endocrinol. Metab. 2025, 328, E885–E898. [Google Scholar] [CrossRef]
- Yue, C.; Xie, S.; Zhong, J.; Zhao, H.; Lin, Z.; Zhang, L.; Xu, B.; Luo, Y. SCAMP2/5 as diagnostic and prognostic markers for acute myeloid leukemia. Sci. Rep. 2021, 11, 17012. [Google Scholar] [CrossRef]
- Lin, Y.; Pan, H.; Ding, H.; Ma, W.; Zhang, Z. Role of SCAMP2 and Rab8a in cholesterol transport of primary mouse macrophage-derived foam cells. J. Guangdong Med. Univ. 2022, 40, 132–136. [Google Scholar]
- An, Y.; Han, P.; Zhang, C.; Yue, Y.; Wen, C.; Meng, Y.; Li, H.; Li, X. The role of NUDT3 in lipid accumulation and its functional variants related to backfat thickness in pigs. Int. J. Biol. Macromol. 2025, 307, 141901. [Google Scholar] [CrossRef]
- Zeng, H.; Zhong, Z.; Xu, Z.; Teng, J.; Wei, C.; Chen, Z.; Zhang, W.; Ding, X.; Li, J.; Zhang, Z. Meta-analysis of genome-wide association studies uncovers shared candidate genes across breeds for pig fatness trait. BMC Genom. 2022, 23, 786. [Google Scholar] [CrossRef]
- Kim, Y.M.; Stone, M.; Hwang, T.H.; Kim, Y.G.; Dunlevy, J.R.; Griffin, T.J.; Kim, D.H. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol. Cell 2012, 46, 833–846. [Google Scholar] [CrossRef]
- Kipp, Z.A.; Xu, M.; Bates, E.A.; Lee, W.H.; Kern, P.A.; Hinds, T.D., Jr. Bilirubin Levels Are Negatively Correlated with Adiposity in Obese Men and Women, and Its Catabolized Product, Urobilin, Is Positively Associated with Insulin Resistance. Antioxidants 2023, 12, 170. [Google Scholar] [CrossRef]
- Hinds, T.D., Jr.; Creeden, J.F.; Gordon, D.M.; Stec, D.F.; Donald, M.C.; Stec, D.E. Bilirubin Nanoparticles Reduce Diet-Induced Hepatic Steatosis, Improve Fat Utilization, and Increase Plasma beta-Hydroxybutyrate. Front. Pharmacol. 2020, 11, 594574. [Google Scholar] [CrossRef] [PubMed]
- May, P.; Woldt, E.; Matz, R.L.; Boucher, P. The LDL receptor-related protein (LRP) family: An old family of proteins with new physiological functions. Ann. Med. 2007, 39, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Calvier, L.; Herz, J.; Hansmann, G. Interplay of Low-Density Lipoprotein Receptors, LRPs, and Lipoproteins in Pulmonary Hypertension. JACC Basic Transl. Sci. 2022, 7, 164–180. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.P.; Da, H.; Li, Z.; Kushwaha, P.; Beil, C.; Mei, L.; Xiong, W.C.; Wolfgang, M.J.; Clemens, T.L.; Riddle, R.C. Lrp4 expression by adipocytes and osteoblasts differentially impacts sclerostin’s endocrine effects on body composition and glucose metabolism. J. Biol. Chem. 2019, 294, 6899–6911. [Google Scholar] [CrossRef]
- Barbitoff, Y.A.; Serebryakova, E.A.; Nasykhova, Y.A.; Predeus, A.V.; Polev, D.E.; Shuvalova, A.R.; Vasiliev, E.V.; Urazov, S.P.; Sarana, A.M.; Scherbak, S.G.; et al. Identification of Novel candidate markers of type 2 diabetes and obesity in Russia by exome sequencing with a limited sample size. Genes 2018, 9, 415. [Google Scholar] [CrossRef]
- Xie, Y.Y.; Mo, C.L.; Cai, Y.H.; Wang, W.J.; Hong, X.X.; Zhang, K.K.; Liu, Q.F.; Liu, Y.J.; Hong, J.J.; He, T.; et al. Pygo2 Regulates Adiposity and Glucose Homeostasis via β-Catenin-Axin2-GSK3β Signaling Pathway. Diabetes 2018, 67, 2569–2584. [Google Scholar] [CrossRef]
- Cui, H.; Zheng, M.; Zhao, G.; Liu, R.; Wen, J. Identification of differentially expressed genes and pathways for intramuscular fat metabolism between breast and thigh tissues of chickens. BMC Genom. 2018, 19, 55. [Google Scholar] [CrossRef]
- Kochansky, C.J.; Lyman, M.J.; Fauty, S.E.; Vlasakova, K.; D’mello, A.P. Administration of Fenofibrate Markedly Elevates Fabp3 in Rat Liver and Plasma and Confounds Its Use as a Preclinical Biomarker of Cardiac and Muscle Toxicity. Lipids 2018, 53, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Hao, X.; Nie, J.; Zhang, H.; Shang, P.; Zhang, B.; Zhang, H. MUSTN1 and FABP3 interact to regulate adipogenesis and lipid deposition. J. Lipid Res. 2025, 66, 100804. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, S.; Wang, K.; Zi, X.; Zhang, R.; Wang, G.; Kang, J.; Li, Z.; Dou, T.; Ge, C. Physicochemical, nutritional properties and metabolomics analysis fat deposition mechanism of Chahua chicken No. 2 and Yao chicken. Genes 2022, 13, 1358. [Google Scholar] [CrossRef] [PubMed]
- Mead, T.J.; Du, Y.; Nelson, C.M.; Gueye, N.A.; Drazba, J.; Dancevic, C.M.; Vankemmelbeke, M.; Buttle, D.J.; Apte, S.S. ADAMTS9-Regulated Pericellular Matrix Dynamics Governs Focal Adhesion-Dependent Smooth Muscle Differentiation. Cell Rep. 2018, 23, 485–498. [Google Scholar] [CrossRef]
- Graae, A.S.; Grarup, N.; Ribel-Madsen, R.; Lystbæk, S.H.; Boesgaard, T.; Staiger, H.; Fritsche, A.; Wellner, N.; Sulek, K.; Kjolby, M.; et al. ADAMTS9 Regulates Skeletal Muscle Insulin Sensitivity Through Extracellular Matrix Alterations. Diabetes 2019, 68, 502–514. [Google Scholar] [CrossRef]
- Liu, C.T.; Monda, K.L.; Taylor, K.C.; Lange, L.; Demerath, E.W.; Palmas, W.; Wojczynski, M.K.; Ellis, J.C.; Vitolins, M.Z.; Liu, S.; et al. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genet. 2013, 9, e1003681. [Google Scholar] [CrossRef]
- Friedrich, F.W.; Reischmann, S.; Schwalm, A.; Unger, A.; Ramanujam, D.; Münch, J.; Müller, O.J.; Hengstenberg, C.; Galve, E.; Charron, P.; et al. FHL2 expression and variants in hypertrophic cardiomyopathy. Basic Res. Cardiol. 2014, 109, 451. [Google Scholar] [CrossRef]
- Clemente-Olivo, M.P.; Hernández-Quiles, M.; Sparrius, R.; van der Stoel, M.M.; Janssen, V.; Habibe, J.J.; van den Burg, J.; Jongejan, A.; Alcaraz-Sobrevals, P.; van Es, R.; et al. Early adipogenesis is repressed through the newly identified FHL2-NFAT5 signaling complex. Cell. Signal. 2023, 104, 110587. [Google Scholar] [CrossRef]
- Clemente-Olivo, M.P.; Habibe, J.J.; Vos, M.; Ottenhoff, R.; Jongejan, A.; Herrema, H.; Zelcer, N.; Kooijman, S.; Rensen, P.C.N.; van Raalte, D.H.; et al. Four-and-a-half LIM domain protein 2 (FHL2) deficiency protects mice from diet-induced obesity and high FHL2 expression marks human obesity. Metabolism 2021, 121, 154815. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Y.; Wang, Y.; Zhao, G.; Wen, J.; Cui, H. Transcriptome analysis reveals steroid hormones biosynthesis pathway involved in abdominal fat deposition in broilers. J. Integr. Agr. 2024, 23, 3118–3128. [Google Scholar] [CrossRef]
- Mattioli, A.V. Sex-specific impacts of caffeine on body composition: Commentary on a retrospective cohort study. J. Int. Soc. Sports Nutr. 2025, 22, 2454633. [Google Scholar] [CrossRef] [PubMed]
- Sima, A.; Manolescu, D.C.; Bhat, P. Retinoids and retinoid-metabolic gene expression in mouse adipose tissues. Biochem. Cell Biol. 2011, 89, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Haduch, A.; Bromek, E.; Kuban, W.; Daniel, W.A. The Engagement of Cytochrome P450 Enzymes in Tryptophan Metabolism. Metabolites 2023, 13, 629. [Google Scholar] [CrossRef]
- Berry, D.C.; DeSantis, D.; Soltanian, H.; Croniger, C.M.; Noy, N. Retinoic acid upregulates preadipocyte genes to block adipogenesis and suppress diet-induced obesity. Diabetes 2012, 61, 1112–1121. [Google Scholar] [CrossRef]
- Lin, W.; Chow, S.K.H.; Cui, C.; Liu, C.; Wang, Q.; Chai, S.; Wong, R.M.Y.; Zhang, N.; Cheung, W.H. Wnt/beta-catenin signaling pathway as an important mediator in muscle and bone crosstalk: A systematic review. J. Orthop. Translat. 2024, 47, 63–73. [Google Scholar] [CrossRef]
- Zhang, X.; He, L.; Wang, L.; Wang, Y.; Yan, E.; Wan, B.; Zeng, Q.; Zhang, P.; Zhao, X.; Yin, J. CLIC5 promotes myoblast differentiation and skeletal muscle regeneration via the BGN-mediated canonical Wnt/β-catenin signaling pathway. Sci. Adv. 2024, 10, eadq6795. [Google Scholar] [CrossRef]
- Sinanoglou, V.J.; Mantis, F.; Miniadis-Meimaroglou, S.; Symeon, G.K.; Bizelis, I.A. Effects of caponisation on lipid and fatty acid composition of intramuscular and abdominal fat of medium-growth broilers. Br. Poult. Sci. 2011, 52, 310–317. [Google Scholar] [CrossRef]
- Zhang, Z.; Liao, Q.; Sun, Y.; Pan, T.; Liu, S.; Miao, W.; Li, Y.; Zhou, L.; Xu, G. Lipidomic and Transcriptomic Analysis of the Longissimus Muscle of Luchuan and Duroc Pigs. Front. Nutr. 2021, 8, 667622. [Google Scholar] [CrossRef]
- Wigger, D.; Schumacher, F.; Schneider-Schaulies, S.; Kleuser, B. Sphingosine 1-phosphate metabolism and insulin signaling. Cell. Signal. 2021, 82, 109959. [Google Scholar] [CrossRef]
- Tang, Y.; Yin, L.; Liu, L.; Chen, Q.; Lin, Z.; Zhang, D.; Wang, Y.; Liu, Y. Comparative Analysis of Different Proteins and Metabolites in the Liver and Ovary of Local Breeds of Chicken and Commercial Chickens in the Later Laying Period. Int. J. Mol. Sci. 2023, 24, 14394. [Google Scholar] [CrossRef]
| Traits a | Nb | Max | Min | Mean | SD | CV (%) |
|---|---|---|---|---|---|---|
| CW | 211 | 2606 | 860 | 1615.27 | 291.95 | 18.07 |
| LVW | 211 | 84.02 | 21.88 | 35.59 | 9.33 | 26.22 |
| AFW | 211 | 122.9 | 14.18 | 53.79 | 27.88 | 51.82 |
| HEW | 211 | 16.79 | 4.65 | 8.99 | 2.38 | 26.51 |
| LVR | 211 | 6.26 | 1.37 | 2.26 | 0.68 | 30.07 |
| AFP | 211 | 0.84 | 6.4 | 3.18 | 1.46 | 45.89 |
| HER | 211 | 0.96 | 0.32 | 0.56 | 0.11 | 19.15 |
| IFPM | 211 | 10.158 | 1.09 | 4.19 | 1.15 | 27.54 |
| IFLM | 211 | 16.38 | 3.39 | 7.48 | 2.15 | 28.75 |
| CW | LVW | AFW | HEW | LVR | AFP | HER | IFPM | IFLM | |
|---|---|---|---|---|---|---|---|---|---|
| CW | 1 | 0.281 ** | 0.341 ** | 0.701 ** | −0.402 ** | 0.264 ** | −0.031 | −0.097 | 0.085 |
| LVW | 1 | −0.126 | 0.347 ** | 0.743 ** | −0.167 * | 0.207 ** | 0.169 * | 0.007 | |
| AFW | 1 | −0.026 | −0.331 ** | 0.989 ** | −0.337 ** | 0.093 | 0.276 ** | ||
| HEW | 1 | −0.133 | −0.083 | 0.875 ** | −0.064 | 0.039 | |||
| LVR | 1 | −0.329 ** | 0.240 ** | 0.242 ** | −0.047 | ||||
| AFP | 1 | −0.345 ** | 0.102 | 0.275 ** | |||||
| HER | 1 | 0.008 | −0.011 | ||||||
| IFPM | 1 | 0.157 * | |||||||
| IFLM | 1 |
| Traits | Chr | nSNP | Position (Mb) | nGene | Effect a | Gene |
|---|---|---|---|---|---|---|
| AFW | 1 | 1 | 109,198,052 | 4 | 14.75 | PRDM15, C2CD2, ZBTB21, UMODL |
| 2 | 1 | 51,692,642 | 3 | 11.83 | BLVRA, VOPP1, LANCL2 | |
| 2 | 1 | 66,761,782 | 1 | −7.05 | GMDS | |
| 10 | 1 | 66,761,782 | 16 | −12.54 | SCAMP2, ULK3, CPLX3, CSK, CYP1A2, CYP1A1, EDC3, CLK3, ARID3B, ACTG1L, UBL7, SEMA7A, CYP11A1, STRA6, ISLR, ISLR2 | |
| 13 | 1 | 2,535,341 | 3 | 26.69 | FAM114A2, GRIA1, MFAP3 | |
| 35 | 1 | 11,945,109 | 1 | 30.32 | PA28_beta | |
| AFP | 1 | 1 | 112,969,661 | 3 | 0.87 | OTC, TSPAN7, RPGR |
| 7 | 1 | 5,897,982 | 4 | 0.76 | TRAF3IP1, USP40, UGT1A1, SH3BP4 | |
| 9 | 1 | 21,433,493 | - | −0.63 | - | |
| 10 | 1 | 13,199,669 | 6 | −1.75 | ACAN, AEN, DET1, RPS11, NUDIX, NTRK3 | |
| 13 | 1 | 11,945,109 | 3 | 1.93 | FAM114A2, GRIA1, MFAP3 | |
| IFLM | 1 | 1 | 149,287,395 | - | 2.76 | - |
| 2 | 1 | 79,956,861 | 16 | −1.31 | OTC, RPGR, NUDIX, NTRK3, UGT1A1, TSPAN7, MFAP3, ACAN, AEN, DET1, FAM114A2, SH3BP4, RPS11, USP40, TRAF3IP1, NUDIX | |
| 2 | 1 | 144,069,823 | 1 | 1.33 | KCNK9, TRAPPC9 | |
| 4 | 1 | 89,896,431 | - | 0.78 | - | |
| 5 | 1 | 22,865,617 | 3 | 2.35 | CKAP5, LRP4, C11orf49 | |
| 5 | 1 | 28,039,569 | 1 | 0.98 | RAD51B | |
| 25 | 1 | 22,865,617 | 20 | 0.66 | UBE2Q1, CHRNB2, ADAR, KCNN3, PBXIP1, PYGO2, SHC1, CKS1B, FLAD1, LOC112530287, ZBTB7B, HCN3, KHDC4, DCST2, LOC107050229, LOC107049672, FDPS, SCAMP3, CLK2, ASH1L | |
| IFPM | 1 | 1 | 25,946,497 | - | 1.18 | - |
| 1 | 1 | 135,236,335 | 4 | 1.03 | MRPS9, TGFBRAP1, C2orf49, FHL2 | |
| 6 | 1 | 29,803,881 | 4 | 1.33 | VAX1, KCNK18, HSPA12A, SHTN1 | |
| 9 | 1 | 22,557,616 | 6 | 1.38 | VEPH1, GMPS, LEKR1, TIPARP, SSR3, KCNAB1 | |
| 12 | 1 | 13,972,290 | 3 | 0.63 | PRICKLE2, ADAMTS9, CCNL1 | |
| 23 | 1 | 1,059,926 | 3 | 0.28 | ZCCHC17, FABP3, SERINC2 | |
| 26 | 1 | 3,635,010 | 9 | 0.24 | LRIG2, MAGI3, TAFA3, WNT2B, ST7L, CAPZA1, MOV10, RHOC, PPM1J | |
| 28 | 1 | 1,544,929 | 15 | 1.10 | UNC13A, MYO5B, PLPP2, LOC100857637, NFIC, FZR1, LOC100858505, PIP5K1C, TBXA2R, HMG20B, DOHH, MFSD12, LOC101748203, C19orf71, CACTIN |
| Traits | ID | Description | p-Value | Key Genes |
|---|---|---|---|---|
| AFW | ko00232 | Caffeine metabolism | 4.29 × 104 | CYP1A2, CYP1A1 |
| ko00140 | Steroid hormone biosynthesis | 1.19 × 103 | CYP1A2, CYP1A1, CYP11A1 | |
| ko04913 | Ovarian steroidogenesis | 1.87 × 103 | CYP1A2, CYP1A1, CYP11A1 | |
| ko00591 | Linoleic acid metabolism | 8.05 × 103 | CYP1A2, CYP1A1 | |
| ko00830 | Retinol metabolism | 1.32 × 102 | CYP1A2, CYP1A1 | |
| ko00980 | Metabolism of xenobiotics by cytochrome P450 | 1.33 × 102 | CYP1A2, CYP1A1 | |
| ko00982 | Drug metabolism-cytochrome P450 | 1.33 × 102 | CYP1A2, CYP1A1 | |
| ko00380 | Tryptophan metabolism | 1.57 × 102 | CYP1A2, CYP1A1 | |
| ko05204 | Chemical carcinogenesis-DNA adducts | 2.03 × 102 | CYP1A2, CYP1A1 | |
| AFP | ko00220 | Arginine biosynthesis | 1.66 × 102 | OTC |
| ko00053 | Ascorbate and aldarate metabolism | 1.76 × 102 | UGT1A1 | |
| ko00040 | Pentose and glucuronate interconversions | 2.17 × 102 | UGT1A1 | |
| ko00860 | Porphyrin metabolism | 2.47 × 102 | UGT1A1 | |
| ko05033 | Nicotine addiction | 4.19 × 102 | GRIA1 | |
| ko00830 | Retinol metabolism | 4.49 × 102 | UGT1A1 | |
| IJPM | ko04011 | MAPK signaling pathway-yeast | 6.72 × 103 | RHOC, PIP5K1C |
| ko04310 | Wnt signaling pathway | 9.21 × 103 | PRICKLE2, WNT2B, RHOC | |
| ko04072 | Phospholipase D signaling pathway | 1.36 × 102 | PLPP2, RHOC, PIP5K1C | |
| ko04144 | Endocytosis | 4.12 × 102 | CAPZA1, RHOC, PIP5K1C | |
| ko04666 | Fc gamma R-mediated phagocytosis | 4.24 × 102 | PLPP2, PIP5K1C | |
| ko05231 | Choline metabolism in cancer | 431 × 102 | PLPP2, PIP5K1C | |
| IJLM | ko00511 | Other glycan degradation | 2.631 × 105 | LOC107050229, SCAMP3 |
| ko00600 | Sphingolipid metabolism | 4.01 × 104 | LOC107050229, SCAMP3 | |
| ko00900 | Terpenoid backbone biosynthesis | 1.35 × 103 | FDPS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Z.; Li, Y.; Liu, J.; Chen, R.; Zhu, H.; Lei, M. Genome-Wide Association Study of Abdominal and Intramuscular Fat Deposition Traits in Huainan Yellow-Feathered Chickens. Animals 2025, 15, 3342. https://doi.org/10.3390/ani15223342
Dai Z, Li Y, Liu J, Chen R, Zhu H, Lei M. Genome-Wide Association Study of Abdominal and Intramuscular Fat Deposition Traits in Huainan Yellow-Feathered Chickens. Animals. 2025; 15(22):3342. https://doi.org/10.3390/ani15223342
Chicago/Turabian StyleDai, Zichun, Yaxin Li, Jie Liu, Rong Chen, Huanxi Zhu, and Mingming Lei. 2025. "Genome-Wide Association Study of Abdominal and Intramuscular Fat Deposition Traits in Huainan Yellow-Feathered Chickens" Animals 15, no. 22: 3342. https://doi.org/10.3390/ani15223342
APA StyleDai, Z., Li, Y., Liu, J., Chen, R., Zhu, H., & Lei, M. (2025). Genome-Wide Association Study of Abdominal and Intramuscular Fat Deposition Traits in Huainan Yellow-Feathered Chickens. Animals, 15(22), 3342. https://doi.org/10.3390/ani15223342






