Functional and Compositional Changes in Ileal Microbiota in Piglets During the Nursing Period Revealed by 16s rRNA Gene and Metagenomics
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Sample Collection
2.2. Library Construction, Quality Control, and Sequencing
2.3. The 16s rRNA Gene Sequence Analysis
2.4. Metagenomic Library Preparation
2.5. Metagenomic Sequence Analysis
3. Results
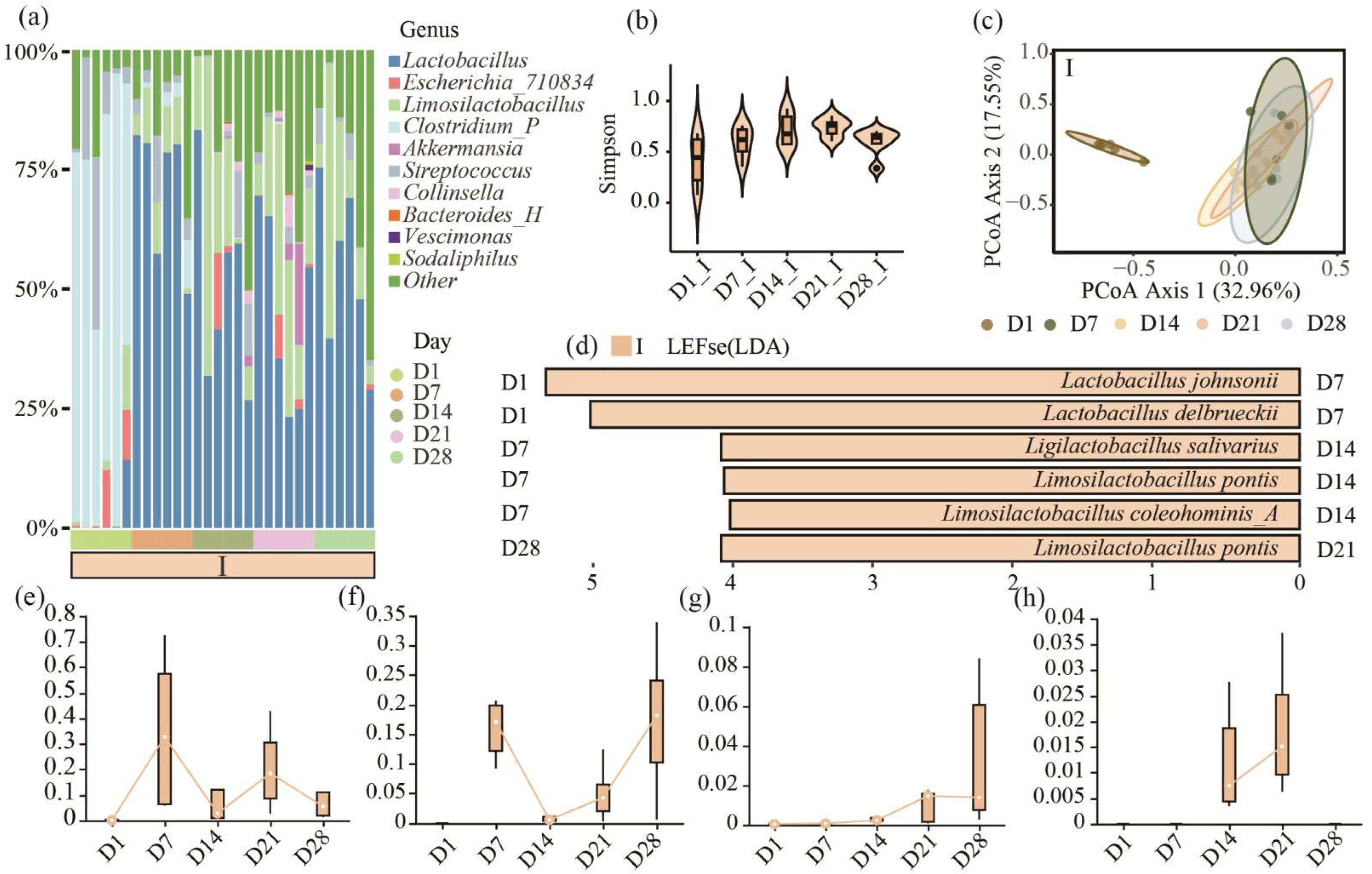
3.1. The 16s rRNA Gene Analysis Reveals the Important Role of Lactobacillus in Ileal Microbiota Dynamics of Piglets During the Nursing Period
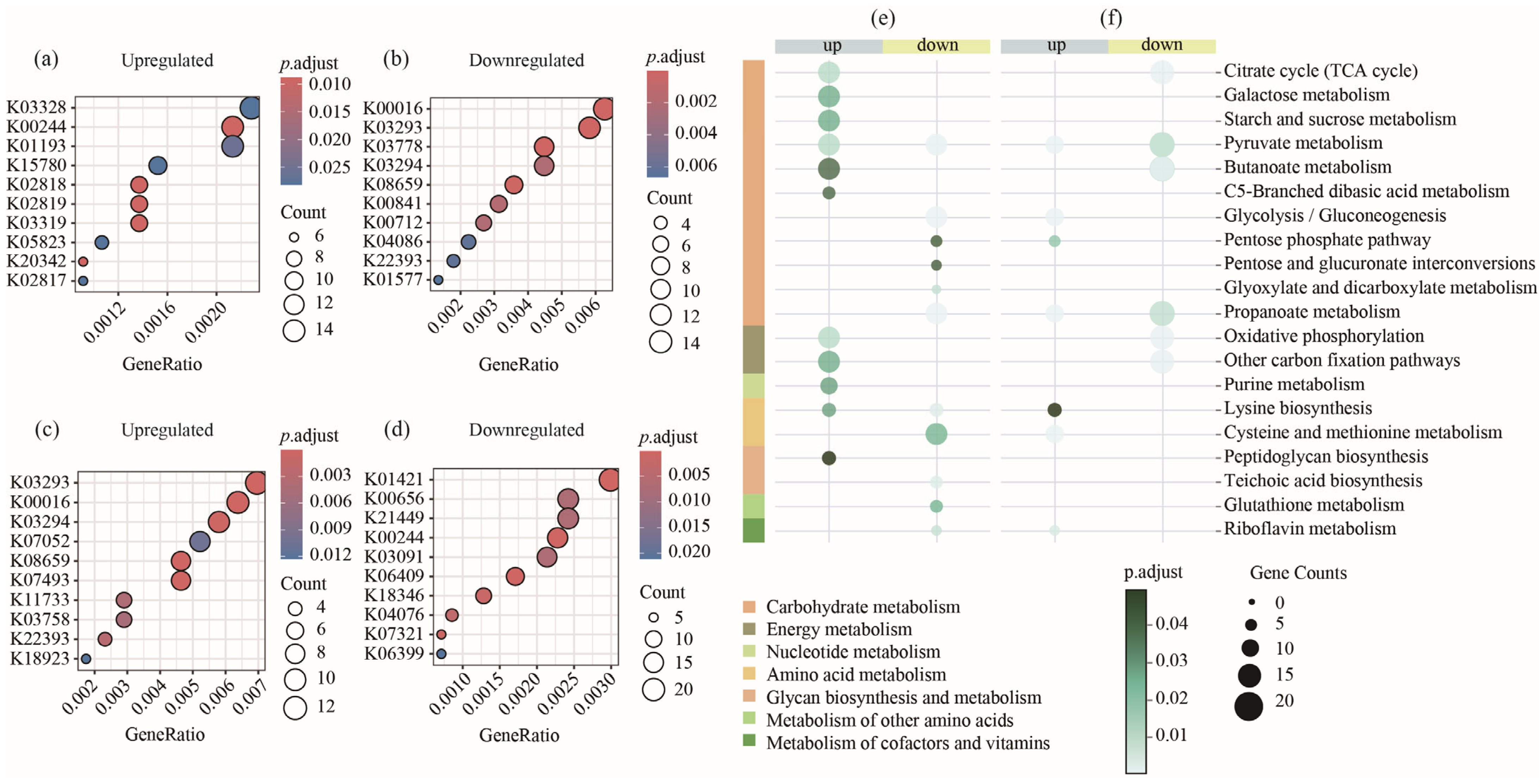
3.2. Metagenomic Analysis of Microbial Functional Changes in Piglets Aged 14–28 Days
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guevarra, R.B.; Lee, J.H.; Lee, S.H.; Seok, M.-J.; Kim, D.W.; Kang, B.N.; Johnson, T.J.; Isaacson, R.E.; Kim, H.B. Piglet Gut Microbial Shifts Early in Life: Causes and Effects. J. Anim. Sci. Biotechnol. 2019, 10, 1. [Google Scholar] [CrossRef]
- Kim, H.B.; Isaacson, R.E. The Pig Gut Microbial Diversity: Understanding the Pig Gut Microbial Ecology through the next Generation High Throughput Sequencing. Vet. Microbiol. 2015, 177, 242–251. [Google Scholar] [CrossRef]
- Saladrigas-García, M.; Durán, M.; D’Angelo, M.; Coma, J.; Pérez, J.F.; Martín-Orúe, S.M. An Insight into the Commercial Piglet’s Microbial Gut Colonization: From Birth towards Weaning. Anim. Microbiome 2022, 4, 68. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, R.; Middelkoop, A.; De Souza, J.G.; Van Veen, L.A.; Gerrits, W.J.J.; Kemp, B.; Bolhuis, J.E.; Kleerebezem, M. Impact of Early-Life Feeding on Local Intestinal Microbiota and Digestive System Development in Piglets. Sci. Rep. 2021, 11, 4213. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, Z.; Yu, L.; Wu, S.; Sun, L.; Wu, S.; Xu, Q.; Cai, S.; Qin, N.; Bao, W. Examination of the Temporal and Spatial Dynamics of the Gut Microbiome in Newborn Piglets Reveals Distinct Microbial Communities in Six Intestinal Segments. Sci. Rep. 2019, 9, 3453. [Google Scholar] [CrossRef]
- Lim, J.-A.; Cha, J.; Choi, S.; Kim, J.-H.; Kim, D. Early Colonization of the Intestinal Microbiome of Neonatal Piglets Is Influenced by the Maternal Microbiome. Animals 2023, 13, 3378. [Google Scholar] [CrossRef]
- Jardon, K.M.; Canfora, E.E.; Goossens, G.H.; Blaak, E.E. Dietary Macronutrients and the Gut Microbiome: A Precision Nutrition Approach to Improve Cardiometabolic Health. Gut 2022, 71, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Yan, X.; Wang, H.; Su, Y.; Zhu, W. Effects of Lactic Acid Bacteria-Fermented Formula Milk Supplementation on Ileal Microbiota, Transcriptomic Profile, and Mucosal Immunity in Weaned Piglets. J. Anim. Sci. Biotechnol. 2022, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Kiela, P.R.; Ghishan, F.K. Physiology of Intestinal Absorption and Secretion. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 145–159. [Google Scholar] [CrossRef]
- Nigam, Y.; Knight, J.; Williams, N. Gastrointestinal Tract 4: Anatomy and Role of the Jejunum and Ileum. Nurs. Times 2019, 115, 41–44. [Google Scholar]
- Frese, S.A.; Parker, K.; Calvert, C.C.; Mills, D.A. Diet Shapes the Gut Microbiome of Pigs during Nursing and Weaning. Microbiome 2015, 3, 28. [Google Scholar] [CrossRef]
- Quan, J.; Xu, C.; Ruan, D.; Ye, Y.; Qiu, Y.; Wu, J.; Zhou, S.; Luan, M.; Zhao, X.; Chen, Y. Composition, Function, and Timing: Exploring the Early-Life Gut Microbiota in Piglets for Probiotic Interventions. J. Anim. Sci. Biotechnol. 2023, 14, 143. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Y.; Liu, J.; Wang, H.; Guo, Y.; Du, M.; Cai, C.; Zhao, Y.; Lu, C.; Guo, X. Composition of the Fecal Microbiota of Piglets at Various Growth Stages. Front. Vet. Sci. 2021, 8, 661671. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Han, Y.; Meng, F.; Zhao, J.; Zhou, Z.; Fan, H. Fecal Microbiota Succession of Piglets from Birth to Post-Weaning by 454 Pyrosequencing Analysis. Trans. Tianjin Univ. 2017, 23, 211–220. [Google Scholar] [CrossRef]
- Li, N.; Zuo, B.; Huang, S.; Zeng, B.; Han, D.; Li, T.; Liu, T.; Wu, Z.; Wei, H.; Zhao, J. Spatial Heterogeneity of Bacterial Colonization across Different Gut Segments Following Inter-Species Microbiota Transplantation. Microbiome 2020, 8, 161. [Google Scholar] [CrossRef]
- Tang, X.; Xiong, K.; Fang, R.; Li, M. Weaning Stress and Intestinal Health of Piglets: A Review. Front. Immunol. 2022, 13, 1042778. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.M.; Crenshaw, J.D.; Polo, J. The Biological Stress of Early Weaned Piglets. J. Anim. Sci. Biotechnol. 2013, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ma, J.; Jiang, Y.; Deng, L.; Lv, N.; Gao, J.; Cheng, J.; Liang, J.B.; Wang, Y.; Lan, T. Selective Maternal Seeding and Rearing Environment from Birth to Weaning Shape the Developing Piglet Gut Microbiome. Front. Microbiol. 2022, 13, 795101. [Google Scholar] [CrossRef]
- Deng, F.; Han, Y.; Li, M.; Peng, Y.; Chai, J.; Yang, G.; Li, Y.; Zhao, J. HiFi Based Metagenomic Assembly Strategy Provides Accuracy near Isolated Genome Resolution in MAG Assembly. iMetaOmics 2025, e70041. [Google Scholar] [CrossRef]
- Han, Y.; He, J.; Li, M.; Peng, Y.; Jiang, H.; Zhao, J.; Li, Y.; Deng, F. Unlocking the Potential of Metagenomics with the PacBio High-Fidelity Sequencing Technology. Microorganisms 2024, 12, 2482. [Google Scholar] [CrossRef]
- Deng, F.; Wang, C.; Li, D.; Peng, Y.; Deng, L.; Zhao, Y.; Zhang, Z.; Wei, M.; Wu, K.; Zhao, J. The Unique Gut Microbiome of Giant Pandas Involved in Protein Metabolism Contributes to the Host’s Dietary Adaption to Bamboo. Microbiome 2023, 11, 180. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.; He, S.; Dang, C. Assisted Selection of Biomarkers by Linear Discriminant Analysis Effect Size (LEfSe) in Microbiome Data. J. Vis. Exp. (JoVE) 2022, e61715. [Google Scholar]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Davis, M.P.; van Dongen, S.; Abreu-Goodger, C.; Bartonicek, N.; Enright, A.J. Kraken: A Set of Tools for Quality Control and Analysis of High-Throughput Sequence Data. Methods 2013, 63, 41–49. [Google Scholar] [CrossRef]
- Yin, Z.; Wang, K.; Liu, Y.; Li, Y.; He, F.; Yin, J.; Tang, W. Lactobacillus Johnsonii Improves Intestinal Barrier Function and Reduces Post-Weaning Diarrhea in Piglets: Involvement of the Endocannabinoid System. Animals 2024, 14, 493. [Google Scholar] [CrossRef]
- Wang, X.-L.; Liu, Z.-Y.; Li, Y.-H.; Yang, L.-Y.; Yin, J.; He, J.-H.; Hou, D.-X.; Liu, Y.-L.; Huang, X.-G. Effects of Dietary Supplementation of Lactobacillus Delbrueckii on Gut Microbiome and Intestinal Morphology in Weaned Piglets. Front. Vet. Sci. 2021, 8, 692389. [Google Scholar] [CrossRef]
- Kinara, E.; Mun, J.; Hosseindoust, A.; Tajudeen, H.; Ha, S.; Park, S.R.; Lee, S.; Kim, J. Dietary Supplementation of Lactobacillus Salivarius in Suckling and Weanling Piglets Modulates Intestinal Microbiota, Morphology and Improves Growth Performance. J. Anim. Sci. Technol. 2024, 67, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhao, Q.; Tang, C.; Li, Y.; Zhang, K.; Li, F.; Zhang, J. Butyrate Mitigates Weanling Piglets from Lipopolysaccharide-Induced Colitis by Regulating Microbiota and Energy Metabolism of the Gut–Liver Axis. Front. Microbiol. 2020, 11, 588666. [Google Scholar] [CrossRef] [PubMed]
- Tonel, I.; Pinho, M.; Lordelo, M.M.; Cunha, L.F.; Garres, P.; Freire, J.P.B. Effect of Butyrate on Gut Development and Intestinal Mucosa Morphology of Piglets. Livest. Sci. 2010, 133, 222–224. [Google Scholar] [CrossRef]
- Ma, L.; Lyu, W.; Song, Y.; Chen, K.; Lv, L.; Yang, H.; Wang, W.; Xiao, Y. Anti-inflammatory Effect of Clostridium Butyricum-derived Extracellular Vesicles in Ulcerative Colitis: Impact on Host microRNAs Expressions and Gut Microbiome Profiles. Mol. Nutr. Food Res. 2023, 67, 2200884. [Google Scholar] [CrossRef]
- Zhuang, Y.; Abdelsattar, M.M.; Fu, Y.; Zhang, N.; Chai, J. Butyrate Metabolism in Rumen Epithelium Affected by Host and Diet Regime through Regulating Microbiota in a Goat Model. Anim. Nutr. 2024, 19, 41–55. [Google Scholar] [CrossRef]
- Ma, L.; Tao, S.; Song, T.; Lyu, W.; Li, Y.; Wang, W.; Shen, Q.; Ni, Y.; Zhu, J.; Zhao, J. Clostridium Butyricum and Carbohydrate Active Enzymes Contribute to the Reduced Fat Deposition in Pigs. Imeta 2024, 3, e160. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Guevarra, R.B.; Hong, S.H.; Cho, J.H.; Kim, B.-R.; Shin, J.; Lee, J.H.; Kang, B.N.; Kim, Y.H.; Wattanaphansak, S.; Isaacson, R.E. The Dynamics of the Piglet Gut Microbiome during the Weaning Transition in Association with Health and Nutrition. J. Anim. Sci. Biotechnol. 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Yin, Y.; Duan, Y.; Chen, J. Methionine Metabolism, Functions, and Application in Swine. Anim. Nutr. 2025. [Google Scholar] [CrossRef]
- Chen, Y.; Li, D.; Dai, Z.; Piao, X.; Wu, Z.; Wang, B.; Zhu, Y.; Zeng, Z. L-Methionine Supplementation Maintains the Integrity and Barrier Function of the Small-Intestinal Mucosa in Post-Weaning Piglets. Amino Acids 2014, 46, 1131–1142. [Google Scholar] [CrossRef]
- Bauchart-Thevret, C.; Cottrell, J.; Stoll, B.; Burrin, D.G. First-Pass Splanchnic Metabolism of Dietary Cysteine in Weanling Pigs. J. Anim. Sci. 2011, 89, 4093–4099. [Google Scholar] [CrossRef] [PubMed]
- Szabó, C.; Kachungwa Lugata, J.; Ortega, A.D.S.V. Gut Health and Influencing Factors in Pigs. Animals 2023, 13, 1350. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Shen, P.; Xu, Z.; Yang, J.; Song, B.; Jiang, H.; Chai, J.; Zhao, J.; Deng, F.; Li, Y. Functional and Compositional Changes in Ileal Microbiota in Piglets During the Nursing Period Revealed by 16s rRNA Gene and Metagenomics. Animals 2025, 15, 3102. https://doi.org/10.3390/ani15213102
Yang B, Shen P, Xu Z, Yang J, Song B, Jiang H, Chai J, Zhao J, Deng F, Li Y. Functional and Compositional Changes in Ileal Microbiota in Piglets During the Nursing Period Revealed by 16s rRNA Gene and Metagenomics. Animals. 2025; 15(21):3102. https://doi.org/10.3390/ani15213102
Chicago/Turabian StyleYang, Boxuan, Pengfei Shen, Zhijian Xu, Jianbo Yang, Bo Song, Hui Jiang, Jianmin Chai, Jiangchao Zhao, Feilong Deng, and Ying Li. 2025. "Functional and Compositional Changes in Ileal Microbiota in Piglets During the Nursing Period Revealed by 16s rRNA Gene and Metagenomics" Animals 15, no. 21: 3102. https://doi.org/10.3390/ani15213102
APA StyleYang, B., Shen, P., Xu, Z., Yang, J., Song, B., Jiang, H., Chai, J., Zhao, J., Deng, F., & Li, Y. (2025). Functional and Compositional Changes in Ileal Microbiota in Piglets During the Nursing Period Revealed by 16s rRNA Gene and Metagenomics. Animals, 15(21), 3102. https://doi.org/10.3390/ani15213102







