Update on Reovirus Infections in Waterfowls
Simple Summary
Abstract
1. Introduction
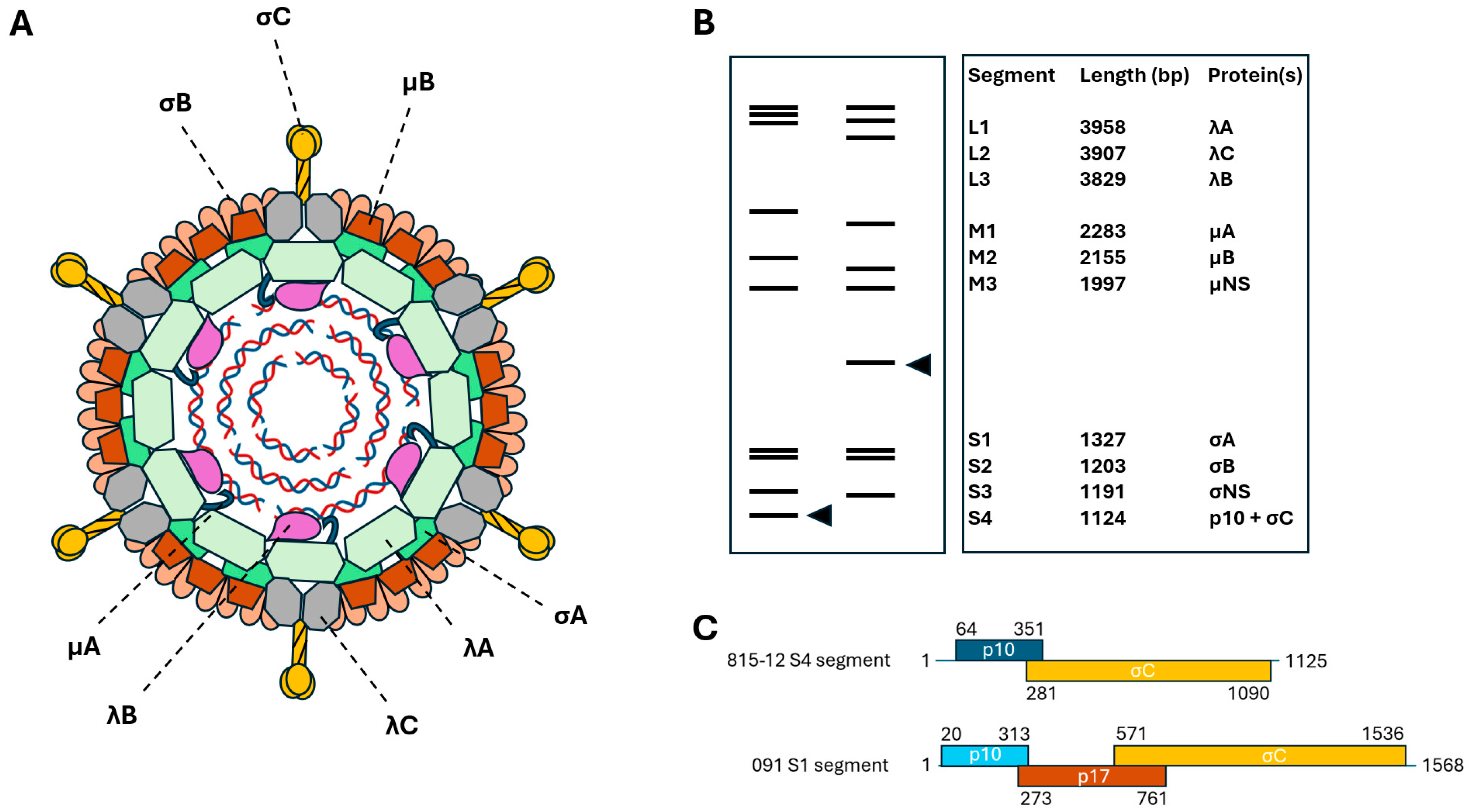
2. Classification, Genomic Features
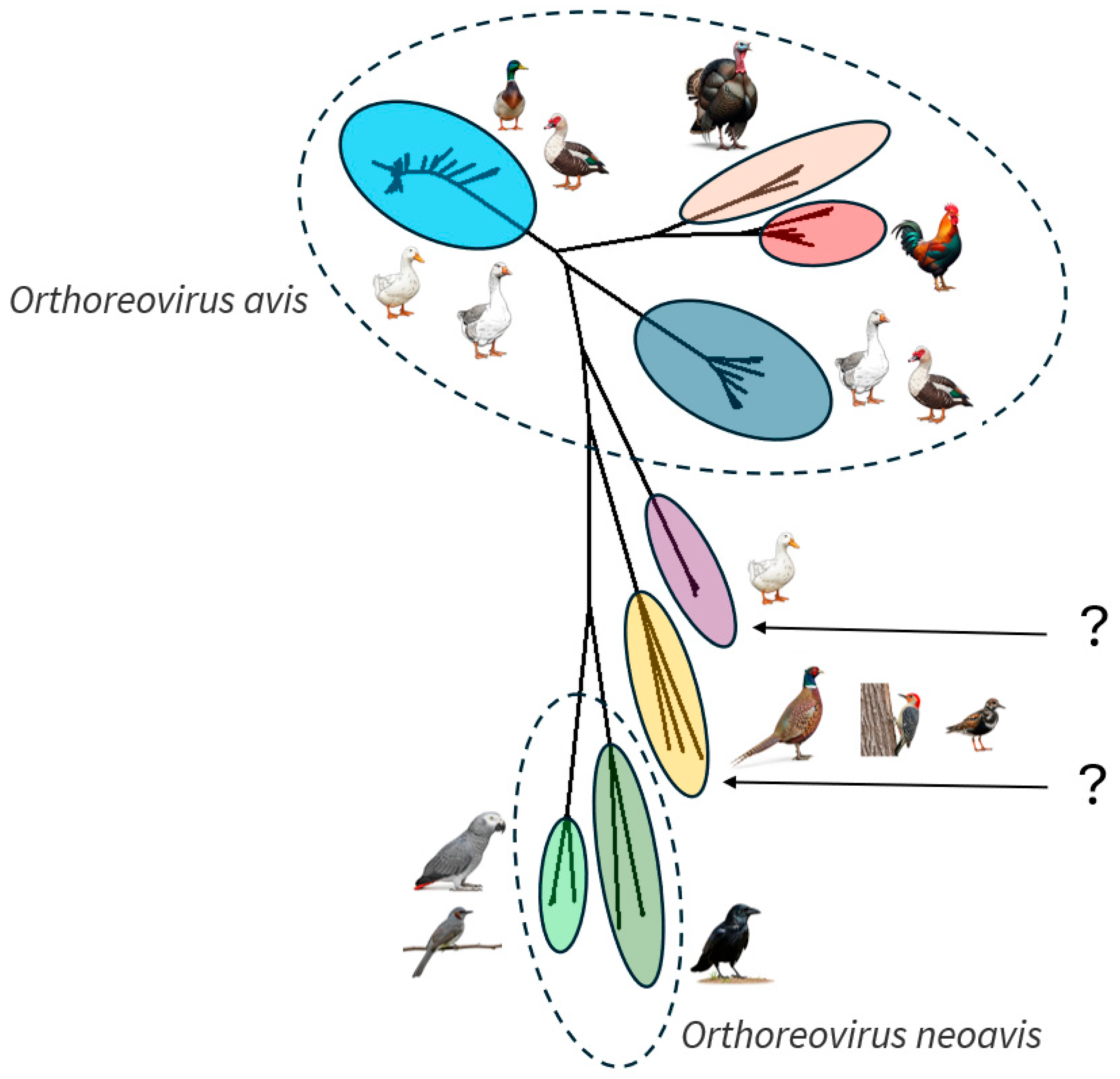
3. Epidemiology, Evolutionary Mechanisms
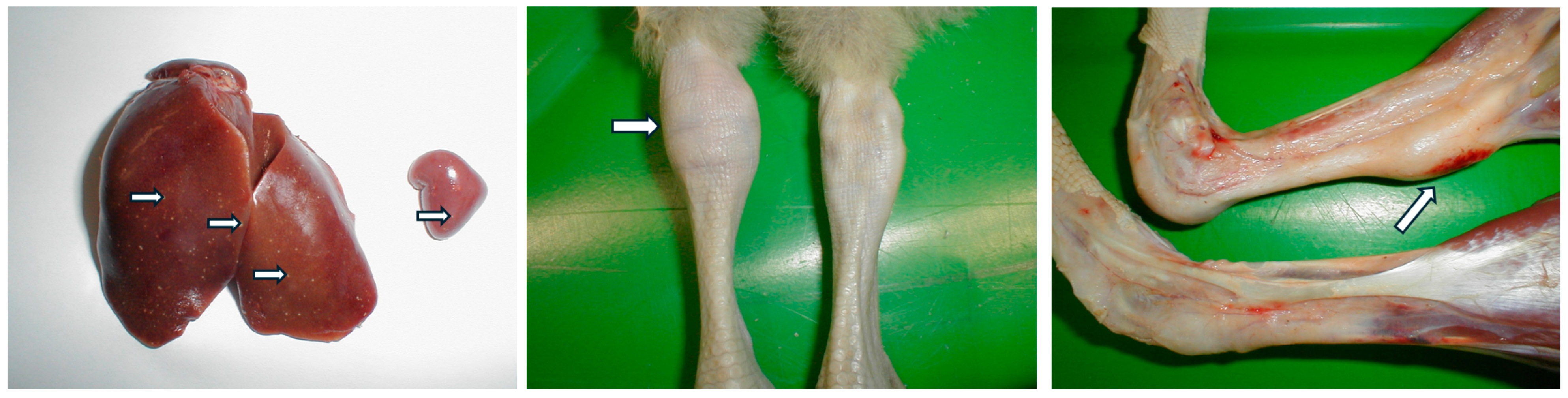
4. Clinical Signs, Pathology
5. Laboratory Diagnosis
6. Disease Control and Prevention, Vaccine Development
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Duck and Goose Meat Market to Keep Growing, Driven by Strong Demand in Asia. Available online: https://www.globaltrademag.com/global-duck-and-goose-meat-market-to-keep-growing-driven-by-strong-demand-in-asia (accessed on 6 July 2025).
- Stoute, S.T. Viral Infections of Waterfowl—Introduction. In Diseases of Poultry, 14th ed.; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., de Wit, S., Grimes, T., Johnson, D., Kromm, M., et al., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2020; pp. 446–449. [Google Scholar]
- Tsai, H.J. Duck hepatitis. In Diseases of Poultry, 14th ed.; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., de Wit, S., Grimes, T., Johnson, D., Kromm, M., et al., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2020; pp. 450–459. [Google Scholar]
- Metwally, S.; Cheng, A. Duck virus enteritis (duck plague). In Diseases of Poultry, 14th ed.; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., de Wit, S., Grimes, T., Johnson, D., Kromm, M., et al., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2020; pp. 460–470. [Google Scholar]
- Palya, V. Parvovirus infections of waterfowl. In Diseases of Poultry, 14th ed.; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., de Wit, S., Grimes, T., Johnson, D., Kromm, M., et al., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2020; pp. 474–484. [Google Scholar]
- Heffels-Redmann, U.; Muller, H.; Kaleta, E.F. Structural and biological characteristics of reoviruses isolated from Muscovy ducks (Cairina moschata). Avian Pathol. 1992, 21, 481–491. [Google Scholar] [CrossRef]
- Malkinson, M.; Perk, K.; Weisman, Y. Reovirus infection of young Muscovy ducks (Cairina moschata). Avian Pathol. 1981, 10, 433–440. [Google Scholar] [CrossRef]
- Palya, V.; Glávits, R.; Dobos-Kovács, M.; Ivanics, E.; Nagy, E.; Bányai, K.; Reuter, G.; Szucs, G.; Dán, A.; Benko, M. Reovirus identified as cause of disease in young geese. Avian Pathol. 2003, 32, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhu, Y.; Li, C.; Liu, G. Outbreak-associated novel duck Reovirus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1209–1211. [Google Scholar] [CrossRef]
- Kaschula, V.R. A new virus disease of the Muscovy duck (Cairina moschata (Linn.)) present in Natal. J. S. Afr. Vet. Med. Assoc. 1950, 21, 18–26. [Google Scholar]
- Yun, T.; Ye, W.; Ni, Z.; Chen, L.; Yu, B.; Hua, J.; Zhang, Y.; Zhang, C. Complete genomic sequence of goose-origin reovirus from China. J. Virol. 2012, 86, 10257. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, S.; Lin, F.; Jiang, B.; Wang, S.; Cheng, X.; Zhu, X.; Zhang, S.; Li, Z.; Cheng, Y. The primary study of pathogen of duck hemorrhagic-necrotic hepatitis. Chin. Agric. Sci. Bull. 2009, 25, 28–31. [Google Scholar] [CrossRef]
- Huang, Y.; Fu, G.; Shi, S.; Wan, C.; Cheng, L.; Jiang, B.; Chen, H.; Lin, F.; Lin, J.; Zhang, D. Isolation and identification of a new pathotype of Muscovy duck reovirus. Chin. J. Vet. Med. 2009, 45, 29–31. [Google Scholar]
- Liu, Q.; Zhang, G.; Huang, Y.; Ren, G.; Chen, L.; Gao, J.; Zhang, D.; Han, B.; Su, W.; Zhao, J.; et al. Isolation and characterization of a reovirus causing spleen necrosis in Pekin ducklings. Vet. Microbiol. 2011, 148, 200–206. [Google Scholar] [CrossRef]
- Farkas, S.L.; Varga-Kugler, R.; Marton, S.; Lengyel, G.; Palya, V.; Bányai, K. Genomic sequence and phylogenetic analyses of two novel orthoreovirus strains isolated from Pekin ducks in 2014 in Germany. Virus Res. 2018, 257, 57–62. [Google Scholar] [CrossRef]
- Varga-Kugler, R.; Marton, S.; Thuma, Á.; Szentpáli-Gavallér, K.; Bálint, Á.; Bányai, K. Candidate ‘Avian orthoreovirus B’: An emerging waterfowl pathogen in Europe and Asia? Transbound. Emerg. Dis. 2022, 69, e3386–e3392. [Google Scholar] [CrossRef]
- Cao, Y.; Sun, M.; Wang, J.; Hu, X.; He, W.; Su, J. Phenotypic and genetic characterisation of an emerging reovirus from Pekin ducks in China. Sci. Rep. 2019, 9, 7784. [Google Scholar] [CrossRef]
- Yamada, S.; Matsuo, K.; Fukuda, T. Susceptibility of duck to Uchida strain of avian reovirus. Nihon Juigaku Zasshi 1979, 41, 551–553. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Tian, J.; Chen, L.; Tang, Y.; Diao, Y. Isolation and genomic characterization of a novel chicken-orign orthoreovirus causing goslings hepatitis. Vet. Microbiol. 2018, 227, 69–77. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Attoui, H.; Bányai, K.; Brussaard, C.P.D.; Danthi, P.; Del Vas, M.; Dermody, T.S.; Duncan, R.; Fāng, Q.; Johne, R.; et al. ICTV Virus Taxonomy Profile: Spinareoviridae 2022. J. Gen. Virol. 2022, 103, 001781. [Google Scholar] [CrossRef]
- Dandár, E.; Farkas, S.L.; Marton, S.; Oldal, M.; Jakab, F.; Mató, T.; Palya, V.; Bányai, K. The complete genome sequence of a European goose reovirus strain. Arch. Virol. 2014, 159, 2165–2169. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Wang, D.; Shi, J.; Jiang, T.; Yuan, Y.; Zhang, D. Complete genomic sequence of a reovirus isolate from Pekin ducklings in China. J. Virol. 2012, 86, 13137. [Google Scholar] [CrossRef]
- Benavente, J.; Martínez-Costas, J. Avian reovirus: Structure and biology. Virus Res. 2007, 123, 105–119. [Google Scholar] [CrossRef]
- Nour, I.; Mohanty, S.K. Avian Reovirus: From Molecular Biology to Pathogenesis and Control. Viruses 2024, 16, 1966. [Google Scholar] [CrossRef]
- Luo, D.; Liu, R.; Weng, L.; Li, K.; Qi, X.; Gao, Y.; Liu, C.; Zhang, Y.; Cui, H.; Pan, Q.; et al. Genomic sequences and pathogenic characteristics of two variant duck reoviruses associated with spleen necrosis. Infect. Genet. Evol. 2021, 92, 104847. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, Y.; Liu, Z.; Li, J.; Han, J.; Liu, Y.; Liu, J.; Chen, H.; Chen, Z. Isolation and characterization of a duck reovirus strain from mature ducks in China. Poult. Sci. 2023, 102, 102345. [Google Scholar] [CrossRef]
- Li, B.; Jiang, X.; Zhang, S.; Wang, Q.; Cui, Y.; Wu, Q.; Jia, W.; Zhang, J.; Diao, Y.; Tang, Y. Isolation and Pathogenicity Analysis of a Novel Orthoreovirus Caused the Outbreak of Duck Viral Arthritis in China. Transbound. Emerg. Dis. 2023, 2023, 8179312. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, X.; Jiang, W.; Fan, X.; Liu, N.; Miao, R.; Zhai, X.; Wei, L.; Jiao, P.; Jiang, S. Research Note: Genetic characterization of novel duck reoviruses from Shandong Province, China in 2022. Poult. Sci. 2023, 102, 102969. [Google Scholar] [CrossRef]
- Yun, T.; Yu, B.; Ni, Z.; Ye, W.; Chen, L.; Hua, J.; Zhang, C. Isolation and genomic characterization of a classical Muscovy duck reovirus isolated in Zhejiang, China. Infect. Genet. Evol. 2013, 20, 444–453. [Google Scholar] [CrossRef]
- Yun, T.; Yu, B.; Ni, Z.; Ye, W.; Chen, L.; Hua, J.; Zhang, C. Genomic characteristics of a novel reovirus from Muscovy duckling in China. Vet. Microbiol. 2014, 168, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gaspard, G.; McMullen, N.; Duncan, R. Polycistronic Genome Segment Evolution and Gain and Loss of FAST Protein Function during Fusogenic Orthoreovirus Speciation. Viruses 2020, 12, 702. [Google Scholar] [CrossRef]
- Kuntz-Simon, G.; Le Gall-Reculé, G.; de Boisséson, C.; Jestin, V. Muscovy duck reovirus sigmaC protein is atypically encoded by the smallest genome segment. J. Gen. Virol. 2002, 83, 1189–1200. [Google Scholar] [CrossRef]
- Bányai, K.; Palya, V.; Benko, M.; Bene, J.; Havasi, V.; Melegh, B.; Szucs, G. The goose reovirus genome segment encoding the minor outer capsid protein, sigma1/sigmaC, is bicistronic and shares structural similarities with its counterpart in Muscovy duck reovirus. Virus Genes 2005, 31, 285–291. [Google Scholar] [CrossRef]
- Farkas, S.L.; Dandár, E.; Marton, S.; Fehér, E.; Oldal, M.; Jakab, F.; Mató, T.; Palya, V.; Bányai, K. Detection of shared genes among Asian and European waterfowl reoviruses in the whole genome constellations. Infect. Genet. Evol. 2014, 28, 55–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yuan, X.; Chen, Y.; Zheng, Q.; Xu, L.; Wu, Y. Endoplasmic Reticulum Stress Mediated MDRV p10.8 Protein-Induced Cell Cycle Arrest and Apoptosis Through the PERK/eIF2α Pathway. Front. Microbiol. 2018, 9, 1327. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Gao, B.; Zhang, S.; Diao, Y.; Tang, Y. Evidence of vertical transmission of novel duck orthoreovirus in ducks. Vet. Microbiol. 2020, 251, 108861. [Google Scholar] [CrossRef]
- Gaudry, D.; Tetkoff, J.; Charles, J.M. A propos d’un nouveau virus isole chez le canard de Barbarie. Bull. Soc. Sei Vet. et Med. Comparée 1972, 74, 137–143. [Google Scholar]
- Wu, B.; Chen, J.; Yao, J.; Chen, Z.; Chen, W.; Li, G.; Zeng, X. Isolation and identification of Muscovy duck reovirus. J. Fujian Agric. Univ. 2001, 30, 227–230. [Google Scholar]
- Wang, D.; Shi, J.; Yuan, Y.; Zheng, L.; Zhang, D. Complete sequence of a reovirus associated with necrotic focus formation in the liver and spleen of Muscovy ducklings. Vet. Microbiol. 2013, 166, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, J.; Dong, J.; Li, L.; Kuang, R.; Sun, M.; Liao, M. Isolation and characterization of a new goose orthoreovirus causing liver and spleen focal necrosis in geese, China. Transbound. Emerg. Dis. 2022, 69, 3028–3034. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, W.; Wang, M.; Yuan, S.; Zhang, X.; Wen, F.; Guo, J.; Mei, K.; Huang, S.; Li, Z. Characterization and pathogenicity evaluation of recombinant novel duck reovirus isolated from Southeast China. Front. Vet. Sci. 2023, 10, 1124999. [Google Scholar] [CrossRef]
- Yu, K.; Li, Y.; Han, H.; Song, M.; Ma, X.; Liu, C.; Huang, B.; Li, F. Complete genome sequence of an avian reovirus isolated from wild mallard ducks in China. Genome Announc. 2014, 2, e00813-14. [Google Scholar] [CrossRef]
- Zhu, D.; Sun, R.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhao, X.; Yang, Q.; Wu, Y.; Zhang, S.; et al. First isolation and genomic characterization of avian reovirus from black swans (Cygnus atratus) in China. Poult. Sci. 2023, 102, 102947. [Google Scholar] [CrossRef]
- Hollmén, T.; Franson, J.C.; Kilpi, M.; Docherty, D.E.; Hansen, W.R.; Hario, M. Isolation and characterization of a reovirus from common eiders (Somateria mollissima) from Finland. Avian Dis. 2002, 46, 478–484. [Google Scholar] [CrossRef]
- Vibin, J.; Chamings, A.; Klaassen, M.; Alexandersen, S. Metagenomic characterisation of additional and novel avian viruses from Australian wild ducks. Sci. Rep. 2020, 10, 22284. [Google Scholar] [CrossRef]
- Ayalew, L.E.; Ahmed, K.A.; Mekuria, Z.H.; Lockerbie, B.; Popowich, S.; Tikoo, S.K.; Ojkic, D.; Gomis, S. The dynamics of molecular evolution of emerging avian reoviruses through accumulation of point mutations and genetic re-assortment. Virus Evol. 2020, 6, veaa025. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.Q.; Li, C.F.; Bi, Z.L.; Chen, Z.Y.; Meng, C.C.; Wang, G.J.; Ding, C.; Liu, G.Q. Molecular characterization of a novel reovirus isolated from Pekin ducklings in China. Arch. Virol. 2015, 160, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lin, F.; Cheng, X.; Wang, J.; Zhu, X.; Xiao, S.; Zheng, M.; Huang, M.; Chen, S.; Chen, S. The genomic constellation of a novel duck reovirus strain associated with hemorrhagic necrotizing hepatitis and splenitis in Muscovy ducklings in Fujian, China. Mol. Cell. Probes 2020, 53, 101604. [Google Scholar] [CrossRef]
- Sun, X.; Guo, J.; Shen, J.; Guan, M.; Liu, L.; Xie, Y.; Xu, H.; Wang, M.; Ren, A.; Li, W.; et al. Genetics and biological characteristics of duck reoviruses isolated from ducks and geese in China. Vet. Res. 2025, 56, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Shao, J.W.; Li, X.W.; Mei, M.M.; Guo, J.Y.; Li, W.F.; Huang, W.J.; Chi, S.H.; Yuan, S.; Li, Z.L.; et al. Molecular characterization of two novel reoviruses isolated from Muscovy ducklings in Guangdong, China. BMC Vet. Res. 2019, 15, 143. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, D.; Ning, K.; Liang, T.; Wang, M.; Jiang, M.; Zhang, D. A duck reovirus variant with a unique deletion in the sigma C gene exhibiting high pathogenicity in Pekin ducklings. Virus Res. 2016, 215, 37–41. [Google Scholar] [CrossRef]
- Wu, Q.; Ding, M.; Li, C.; Liu, G.; Chen, Z. Construction and characterization of an infectious molecular clone of novel duck reovirus. J. Gen. Virol. 2018, 99, 449–456. [Google Scholar] [CrossRef]
- Wang, H.; Gao, B.; Chen, H.; Diao, Y.; Tang, Y. Isolation and characterization of a variant duck orthoreovirus causing spleen necrosis in Peking ducks, China. Transbound. Emerg. Dis. 2019, 66, 2033–2044. [Google Scholar] [CrossRef]
- Li, N.; Hong, T.; Wang, Y.; Wang, Y.; Yu, K.; Cai, Y.; Liu, S.; Wei, L.; Chai, T. The pathogenicity of novel duck reovirus in Cherry Valley ducks. Vet. Microbiol. 2016, 192, 181–185. [Google Scholar] [CrossRef]
- Varga-Kugler, R.; Palya, V.; Farkas, S.; Bányai, K. Víziszárnyasok reovírus-fertőzései. Magy. Állatorv. Lapja 2020, 142, 387–396. (In Hungarian) [Google Scholar]
- Woźniakowski, G.; Samorek-Salamonowicz, E.; Gaweł, A. Occurrence of reovirus infection in Muscovy ducks (Cairina moschata) in south western Poland. Pol. J. Vet. Sci. 2014, 17, 299–305. [Google Scholar] [CrossRef]
- Kawamura, H.; Tsubahara, H. Common antigenicity of avian reoviruses. Natl. Inst. Anim. Health Q. 1966, 6, 187–193. [Google Scholar]
- Yun, T.; Chen, H.; Yu, B.; Zhang, C.; Chen, L.; Ni, Z.; Hua, J.; Ye, W. Development and application of an indirect ELISA for the detection of antibodies to novel duck reovirus. J. Virol. Methods 2015, 220, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, D.; Liu, M.; Geng, H.; Hu, Q.; Liu, Y.; Liu, N. Characterization of the sigmaB-encoding genes of muscovy duck reovirus: sigmaC-sigmaB-ELISA for antibodies against duck reovirus in ducks. Vet. Microbiol. 2007, 121, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Styś-Fijoł, N.; Kozdruń, W.; Czekaj, H. Detection of Avian Reoviruses in Wild Birds in Poland. J. Vet. Res. 2017, 61, 239–245. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Liu, M.; Shuidong, O.; Hu, Q.L.; Guo, D.C.; Chen, H.Y.; Han, Z. Detection and identification of avian, duck, and goose reoviruses by RT-PCR: Goose and duck reoviruses are part of the same genogroup in the genus Orthoreovirus. Arch. Virol. 2006, 151, 1525–1538. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, S.; Hong, W.; Wang, A.; Zuo, W. A multiplex PCR for detection of six viruses in ducks. J. Virol. Methods 2017, 248, 172–176. [Google Scholar] [CrossRef]
- Yin, Y.W.; Xiong, C.; Shi, K.C.; Xie, S.Y.; Long, F.; Li, J.; Zheng, M.; Wei, X.K.; Feng, S.; Qu, S.; et al. Development and application of a multiplex qPCR assay for the detection of duck circovirus, duck Tembusu virus, Muscovy duck reovirus, and new duck reovirus. Virus Genes 2023, 59, 91–99. [Google Scholar] [CrossRef]
- Zhang, S.; Li, W.; Liu, X.; Li, X.; Gao, B.; Diao, Y.; Tang, Y. A TaqMan-based real-time PCR assay for specific detection of novel duck reovirus in China. BMC Vet. Res. 2020, 16, 306. [Google Scholar] [CrossRef]
- Zheng, M.; Chen, X.; Wang, S.; Wang, J.; Huang, M.; Xiao, S.; Cheng, X.; Chen, S.; Chen, X.; Lin, F.; et al. A TaqMan-MGB real-time RT-PCR assay with an internal amplification control for rapid detection of Muscovy duck reovirus. Mol. Cell. Probes 2020, 52, 101575. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Xie, Z.X.; Xie, L.J.; Deng, X.W.; Xie, Z.Q.; Luo, S.S.; Huang, L.; Huang, J.L.; Zeng, T.T. GeXP analyzer-based multiplex reverse-transcription PCR assay for the simultaneous detection and differentiation of eleven duck viruses. BMC Microbiol. 2015, 15, 247. [Google Scholar] [CrossRef]
- Yan, L.; Song, Y.; Zhai, T.; Qiu, Q.; Wang, J.; Liu, J.; Lv, D.; Huang, X.; Cao, H.; Yang, C.; et al. Establishment of a Visual Gene Chip Method for the Simultaneous Detection of Seven Waterfowl Virus Pathogens. Viruses 2025, 17, 358. [Google Scholar] [CrossRef]
- Wang, C.; Liu, H.; Cheng, J.; Pan, S.; Yang, W.; Wei, X.; Cheng, Y.; Xu, T.; Si, H. One-Step Multiplex Real-Time Fluorescent Quantitative Reverse Transcription PCR for Simultaneous Detection of Four Waterfowl Viruses. Microorganisms 2024, 12, 2423. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, H.; Zheng, X.; Cheng, X.; Wang, S.; You, G.; Zhu, X.; Zheng, M.; Dong, H.; Xiao, S.; et al. Simultaneous detection and differentiation of classical Muscovy duck reovirus and goose-origin Muscovy duck reovirus by RT-qPCR assay with high-resolution melting analysis. Front. Vet. Sci. 2024, 11, 1459898. [Google Scholar] [CrossRef]
- Li, Z.; Cai, Y.; Liang, G.; El-Ashram, S.; Mei, M.; Huang, W.; Li, X.; Li, W.; He, C.; Huang, S. Detection of Novel duck reovirus (NDRV) using visual reverse transcription loop-mediated isothermal amplification (RT-LAMP). Sci. Rep. 2018, 8, 14039. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Huang, Y.; Chen, G.; Shi, M.; Qiao, Y.; Huang, T.; Wei, T.; Mo, M.; He, X.; et al. Rapid and visual detection of the emerging novel duck reovirus by using a specific and sensitive reverse transcription recombinase polymerase amplification method. J. Virol. Methods 2021, 291, 114098. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Lei, D.; Xu, B.; Wang, Z.; Fang, R.; Tang, Y.; Wang, H. High-efficient molecular detection system termed RAA-based CRISPR-Cas13a for novel duck orthoreovirus. Poult. Sci. 2025, 104, 105327. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Zhang, B.; Yu, X.; Zhang, X.; Dou, Y.; Tang, Y.; Diao, Y. Preparation and evaluation of goose reovirus inactivated vaccine. BMC Vet. Res. 2017, 13, 214. [Google Scholar] [CrossRef]
- Huang, D.; Shao, Y.; Wang, R.; Liu, S.; Zhang, S.; Bao, S. Immunogenic cross-reactivity between different serotypes novel duck reoviruses: Evaluation of cross-protection provided by mono or bivalent vaccine. Poult. Sci. 2025, 104, 104900. [Google Scholar] [CrossRef]
- Kuntz-Simon, G.; Blanchard, P.; Cherbonnel, M.; Jestin, A.; Jestin, V. Baculovirus-expressed muscovy duck reovirus sigmaC protein induces serum neutralizing antibodies and protection against challenge. Vaccine 2002, 20, 3113–3122. [Google Scholar] [CrossRef] [PubMed]
- Bi, Z.; Zhu, Y.; Chen, Z.; Li, C.; Wang, Y.; Wang, G.; Liu, G. Induction of a robust immunity response against novel duck reovirus in ducklings using a subunit vaccine of sigma C protein. Sci. Rep. 2016, 6, 39092. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, S.; Chen, X.; Dou, Y.; Yang, X.; Hu, Z.; Wu, S.; Wang, X.; Hu, J.; Liu, X. Single dose of recombinant baculovirus vaccine expressing sigma B and sigma C genes provides good protection against novel duck reovirus challenge in ducks. Poult. Sci. 2025, 104, 104565. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, Z.; Liao, C.; Song, Y.; Zhou, Q.; Shen, H.; Chen, F. Recombinant linear multiple epitopes of σB protein protect Muscovy ducks against novel duck reovirus infection. Front. Vet. Sci. 2024, 11, 1360246. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, C.; Bi, Z.; Chen, Z.; Meng, C.; Wang, G.J.; Ding, C.; Liu, G. Protective immune responses in ducklings induced by a suicidal DNA vaccine of the sigma C gene of novel duck reovirus. Vet. Immunol. Immunopathol. 2015, 165, 88–92. [Google Scholar] [CrossRef]
- Yan, H.; Xu, G.; Zhu, Y.; Xie, Z.; Zhang, R.; Jiang, S. Isolation and characterization of a naturally attenuated novel duck reovirus strain as a live vaccine candidate. Vet. Microbiol. 2021, 261, 109214. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Zhang, H.; Zhang, X.; Wang, H.; Liu, Z.; Qiao, H.; Lv, Y.; Bian, C. Identification and molecular characterization of novel duck reoviruses in Henan Province, China. Front. Vet. Sci. 2023, 10, 1137967. [Google Scholar] [CrossRef] [PubMed]
- Nowak, T.; Kwiecinski, A.; Kwiecinski, P.; Tomczyk, G.; Wodz, K. Detection and identification of avian reovirus in young geese (Anser anser domestica) in Poland. Animals 2022, 12, 3346. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liang, J.; Shi, M.; Chen, G.; Huang, Y.; Zhang, Y.; Zhao, Z.; Wang, M.; Li, M.; Mo, M.; et al. The diagnosis and successful replication of a clinical case of duck spleen necrosis disease: An experimental co-infection of an emerging unique reovirus and Salmonella indiana reveals the roles of each of the pathogens. Vet. Microbiol. 2020, 246, 108723. [Google Scholar] [CrossRef]

| Study | Methodology | Target Virus(es) | Target WRV Gene | Sensitivity and Specificity |
|---|---|---|---|---|
| Zhang et al., 2015 [66] | GeXP analyzer-based multiplex reverse-transcription PCR assay | AIV, DHV, DTMUV, EDSV, DEV, NDV, DuCV, MDRV, MDPV | σA gene | 103 copies (when all 12 plasmids were present) Specific, no amplification products from EC, S, SA, PM, IBV, MG |
| Wang et al., 2017 [62] | Multiplex PCR | DHAV-1, DPV, DTMUV, MDPV, MDRV, AIV | σC gene | 10 copies/µL (single template), 100 copies/µL (combined template) Specific, no cross-reaction with other viruses |
| Li et al., 2018 [70] | Visual reverse transcription loop-mediated isothermal amplification (RT-LAMP) | NDRV | σB gene | 200 fg virus-positive RNA extract Specific, no cross-reaction with MDRV, ARV, DPV, DHAV, NDV, AIV, DTMUV |
| Zheng et al., 2020 [65] | TaqMan-MGB real-time RT-PCR assay with an internal amplification control | MDRV | σB gene | 2.83 × 101 copies/µL Specific, no cross-reaction with DHAV-1, DPV, DTMUV, MDPV, APMV-1, ARV, NDRV |
| Zhang et al., 2020 [64] | TaqMan-based real-time PCR assay | NDRV | σA gene | 10 copies/µL (Ct values: 38.3) Specific, no cross-reaction with AIV, DTMUV, GPV, NGPV, DHAV-1, DHAV-3, DuCV, DPV |
| Wang et al., 2021 [71] | Reverse transcription recombinase polymerase amplification (RT-RPA) | NDRV | σB gene | 4.14 × 102 copies/µL Specific, no cross-reaction with MDRV, ARV, DHAV, NDV, DTMUV, DPV, IBDV, IBV, aMPV |
| Yin et al., 2023 [63] | Multiplex digital PCR | DTMUV, DuCV, NDRV | σB gene | 1.3 copies/µL Specific, no cross-reaction with MDRV, MDPV, GPV, AIV, NDV |
| Xu et al., 2024 [69] | RT-qPCR assay with high-resolution melting analysis | C-MDRV, Go-MDRV | σNS gene (C-MDRV), λA gene (Go-MDRV) | C-MDRV: 5.14 × 101 copies/µL, Go-MDRV: 6.18 × 101 copies/µL Specific, no cross-reaction with GPV, MDPV, DAdV, DTMUV, DEV, NDRV, DHAV-1, DPMV |
| Wang et al., 2024 [68] | One-Step Multiplex Real-Time Fluorescent Quantitative Reverse Transcription PCR | DTMUV, DHV, MDRV, MDPV | σB gene | 2.68 × 101 copies/µL Specific, no cross-reaction with FADV, IBDV, IBV, ILTV, HPg, DuCV, GoAstV, MG |
| Jiang et al., 2025 [72] | RAA-based CRISPR-Cas13a molecular detection system | NDRV | σC gene | 1 copy/µL Specific, no cross-reaction with DHAV, TMUV, NGPV |
| Yan et al., 2025 [67] | Visual Gene Chip Method | GPV, DEV, MDPV, DHAV-1, DHAV-3, DTMUV, NDRV | σC gene | 1 copy/µL for single and mixed samples Specific, no cross-reaction with other pathogens |
| Study | Vaccine Candidate | Vaccine Type/Route of Administration | Vaccine Antigen (Dosage) | Immune Response | Clinical Protection/ Lesions/Virus Load/Virus Shedding |
|---|---|---|---|---|---|
| Wang et al., 2025 [77] | rBac-σB | Subunit (Baculovirus-expressed) IM | σB protein (160 ng, single dose) | Induced σB antibodies | No significant weight loss after challenge for all four vaccines. Reduced gross lesions, milder or no histological lesions for recombinant vaccines; somewhat more severe lesions for inactivated vaccine. Reduced viral replication and shedding for recombinant vaccines partly for inactivated vaccine |
| rBac-σC | Subunit (Baculovirus-expressed) IM | σC protein (160 ng, single dose) | Induced stronger σC antibody response and higher VN titer than inactivated vaccine | ||
| rBac-σB+σC | Subunit (Baculovirus-expressed) IM | σB+σC proteins (160 ng each, single dose) | Better protective efficacies than single subunit formulas and inactivated | ||
| Inactivated NDRV | Whole virus inactivated IM | NDRV WYC strain (5 × 10−4.5 EID50) | Showed lower σC and VN antibody levels compared to subunit vaccines | ||
| Chen et al., 2024 [78] | Recombinant MLBE of σB protein | Subunit (E. coli expressed) | Multiple linear B cell epitopes (MLBEs) of σB protein (10, 20, and 40 µg) | 100% serum conversion rate when using higher protein amount (20 and 40 µg) | 100% protection, no clinical signs or gross/histopathological lesions, reduced viral load and viral shedding for 20 and 40 µg doses |
| Huang et al., 2025 [74] | Inactivated Monovalent DE13 | Whole virus inactivated SC | NDRV DE13 strain (108 TCID50/mL) | For monovalent vaccines, virus neutralizing titer against homologous virus significantly higher than heterologous For bivalent vaccine, rapid immunity (within 2 weeks), high neutralizing antibodies in breeders, persisting for 80 days in serum and yolk | For monovalent vaccines, 100% clinical protection against homologous virus challenge, partial (13.3% to 26.7%) protection against heterologous virus For bivalent vaccine, 100% protection against both challenge viruses. Passive immunity in offspring provides 100% protection until 21 days of age. |
| Inactivated Monovalent WL01 | Whole virus inactivated SC | NDRV WL01 strain (108 TCID50/mL) | |||
| Inactivated Bivalent (DE13 + WL01) | Whole virus inactivated SC | NDRV DE13 + WL01 strains (108 TCID50/mL of each strain) | |||
| Yan et al., 2021 [80] | Naturally Attenuated N20 strain | Live attenuated vaccine IM | N20 strain (100 ELD50/mL) | Generated NDRV-specific antibodies at D6. Sera neutralized different NDRVs. | 100% protection against virulent N19 challenge. No clinical signs, gross/histopathological lesions, or body weight loss. Reduced viral replication and shedding |
| Bi et al., 2016 [76] | Recombinant σC protein (TH11) | Subunit (Baculovirus-expressed) SC | Purified σC protein (5 µg σC protein, two doses, 2-week intervals) | Robust humoral and cellular immune responses. High neutralizing antibodies. Elevated IFN-γ and IL-4 levels | 100% protection against lethal challenge. Mild signs of illness. Reduced viremia, decreased virus replication |
| Zhu et al., 2015 [79] | Suicidal DNA vaccine of σC gene | DNA vaccine (SFV replicon based) IM | σC gene of NDRV (100 µg as primary, 200 µg as booster) | Induced NDRV-specific antibodies, neutralizing antibodies, IFN-γ and IL-4. | 100% protection against challenge. No clinical signs or mortality. No lesions. Undetectable levels of virus by RT-PCR. Reduced viremia, decreased virus replication |
| Niu et al., 2017 [73] | Inactivated GRV Vaccine | Whole virus inactivated IM | GRV JS-01 strain (0.2 mL or 0.5 mL allantoic fluid) | Antibodies detectable at 6 days post-vaccination, peaked at 3 weeks, ELISA values of serum antibody correlates with protection | At 12 dpi with 0.5 mL vaccine, 100% protection observed. Reduced or no lesions. Lower viral shedding in vaccinated groups. |
| Kuntz-Simon et al., 2002 [75] | Baculovirus-expressed σC | Subunit (Baculovirus-expressed) SC | σC protein (DRV-89330 strain) (1 × 107 infected cell equivalents (1st dose), 1.5 × 107 (2nd dose) | Elicited DRV-specific neutralizing antibodies. Antibodies detected in 100% of sera | For both vaccines, partial or full prevention of clinical signs and reduced severity of tenosynovitis For the bivalent vaccine, less lesions than σC alone. |
| Baculovirus-expressed σC+σB | Subunit (Baculovirus-expressed) SC | σC + σB proteins (DRV-89330 strain) (1 × 107 infected cell equivalents (1st dose), 1.5 × 107 (2nd dose) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farkas, S.L.; Lanszki, Z.; Malik, Y.S.; Martella, V.; Palya, V.; Bányai, K. Update on Reovirus Infections in Waterfowls. Animals 2025, 15, 3053. https://doi.org/10.3390/ani15203053
Farkas SL, Lanszki Z, Malik YS, Martella V, Palya V, Bányai K. Update on Reovirus Infections in Waterfowls. Animals. 2025; 15(20):3053. https://doi.org/10.3390/ani15203053
Chicago/Turabian StyleFarkas, Szilvia L., Zsófia Lanszki, Yashpal S. Malik, Vito Martella, Vilmos Palya, and Krisztián Bányai. 2025. "Update on Reovirus Infections in Waterfowls" Animals 15, no. 20: 3053. https://doi.org/10.3390/ani15203053
APA StyleFarkas, S. L., Lanszki, Z., Malik, Y. S., Martella, V., Palya, V., & Bányai, K. (2025). Update on Reovirus Infections in Waterfowls. Animals, 15(20), 3053. https://doi.org/10.3390/ani15203053









